Published online Jun 15, 2023. doi: 10.4251/wjgo.v15.i6.1019
Peer-review started: January 6, 2023
First decision: January 23, 2023
Revised: February 6, 2023
Accepted: April 27, 2023
Article in press: April 27, 2023
Published online: June 15, 2023
Processing time: 159 Days and 19.4 Hours
The distal-less homeobox (DLX) gene family plays an important role in the development of several tumors. However, the expression pattern, prognostic and diagnostic value, possible regulatory mechanisms, and the relationship between DLX family genes and immune infiltration in colon cancer have not been systematically reported.
We aimed to comprehensively analyze the biological role of the DLX gene family in the pathogenesis of colon cancer.
Colon cancer tissue and normal colon tissue samples were collected from the Cancer Genome Atlas and Gene Expression Omnibus databases. Wilcoxon rank sum test and t-test were used to assess DLX gene family expression between colon cancer tissue and unpaired normal colon tissue. cBioPortal was used to analyze DLX gene family variants. R software was used to analyze DLX gene expression in colon cancer and the relationship between DLX gene family expression and clinical features and correlation heat map. The survival package and Cox regression module were used to assess the prognostic value of the DLX gene family. The pROC package was used to analyze the diagnostic value of the DLX gene family. R software was used to analyze the possible regulatory mechanisms of DLX gene family members and related genes. The GSVA package was used to analyze the relationship between the DLX gene family and immune infiltration. The ggplot2, the survminer package, and the clusterProfiler package were used for visualization.
DLX1/2/3/4/5 were significantly aberrantly expressed in colon cancer patients. The expression of DLX genes were associated with M stage, pathologic stage, primary therapy outcome, residual tumor, lymphatic invasion, T stage, N stage, age, perineural invasion, and history of colon polyps. DLX5 was independently correlated with the prognosis of colon cancer in multivariate analysis. DLX1/2/3/4/5/6 were involved in the development and progression of colon cancer by participating in immune infiltration and associated pathways, including the Hippo signaling pathway, the Wnt signaling pathway, several signaling pathways regulating the pluripotency of stem cells, and Staphylococcus aureus infection.
The results of this study suggest a possible role for the DLX gene family as potential diagnostic or prognostic biomarkers and therapeutic targets in colon cancer.
Core Tip: The distal-less homeobox (DLX) gene family plays an important role in the pathogenesis of several tumors. However, the expression pattern, prognostic and diagnostic value, possible regulatory mechanisms, and the relationship between DLX family genes and immune infiltration in colon cancer have not been systematically reported. In this study, we aimed to investigate the expression level, clinical significance, and relationship between DLX genes and immune infiltration in colon cancer to establish an adequate scientific basis for clinical decision making and risk management. The DLX gene family holds promise as a potential diagnostic or prognostic biomarker and therapeutic target for colon cancer.
- Citation: Chen YC, Li DB, Wang DL, Peng H. Comprehensive analysis of distal-less homeobox family gene expression in colon cancer. World J Gastrointest Oncol 2023; 15(6): 1019-1035
- URL: https://www.wjgnet.com/1948-5204/full/v15/i6/1019.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i6.1019
Colon cancer comprises a widely study group of tumors whose incidence is on the rise. Approximately 10% of all cancer deaths are caused by colon cancer and related complications[1]. Colon adenocarcinoma (COAD) is the most common, accounting for 98% of colon cancer cases[2]. Colon cancer has a high recurrence rate after treatment, with 42% of patients recurring within 5 y and a median time from recurrence to death of 12 mo[3]. Unfortunately, about 20% of colon cancer patients are diagnosed with stage IV each year[4]. Therefore, exploring novel molecular markers is of great clinical significance to improve the diagnosis and treatment of colon cancer.
The distal-less homeobox (DLX) gene is a homolog of Drosophila distal-less and consists of 6 members, including DLX1, DLX2, DLX3, DLX4, DLX5, and DLX6[5]. DLX1 can be used to identify prostate cancer for early diagnosis[6]. Overexpression of DLX2 has been associated with poor prognosis in hepatocellular carcinoma (HCC)[7]. High expression of DLX2 has been shown to be a poor prognostic marker for patients with glioblastoma multiforme[8]. DLX3 has been demonstrated as a key regulator of the STAT3 signaling network that maintains skin homeostasis[9]. DLX4 can also be used as a prognostic marker for HCC[10]. DLX5 has been shown to be a potential diagnostic biomarker and therapeutic target for oral squamous cell carcinoma (OSCC)[11]. DLX6 has been shown to promote cell proliferation and survival in OSCC[12]. To our knowledge, no studies have systematically assessed the role of the DLX gene family in colon cancer using bioinformatics methods. In this study, we aimed to investigate the expression level, clinical significance, and relationship between DLX family genes and immune infiltration in colon cancer to establish an adequate scientific basis for clinical decision making and risk management.
The cBio Cancer Genomics Portal (cBioPortal) (http://cbioportal.org) was used to study mutations in DLX genes in colon cancer[13]. Queries for visualization and analysis were performed using the following entries: (1) Cancer type: COAD; (2) 2 selected studies: COAD (CaseCCC, PNAS 2015), colon cancer (CPTAC-2 Prospective, Cell 2019); (3) Molecular profile: Mutations and copy number alterations (CNAs); (4) Selection of patients/case sets: All samples (139); and (5) Input genes: DLX1(ENSG
R software (version 3.6.3) was used for statistical analysis and visualization[14,15]. The R packages used included ggplot2 (version 3.3.3) for visualization. UCSC XENA (https://xenabrowser.net/datapages/) RNAseq data were uniformly processed by the Toil process into TPM (transcripts per million reads) format for the Cancer Genome Atlas (TCGA) and GTEx[16]. Data for colon cancer were extracted from the TCGA and corresponding normal tissue data were extracted from GTEx. RNAseq data were in TPM format and log2 transformed for expression comparisons between samples. The data filtering condition was set to retain paired samples.
Correlation between every 2 genes of the DLX family was assessed using a Pearson’s correlation coefficient. The R package used was mainly ggplot2 (version 3.3.3). The filter condition was set to remove data from the normal/control groups (of note, not every item had a normal/control group).
The R package used was the basic R package[17]. Grouping was based on the median.
The survminer package (version 0.4.9) was used for visualizing survival data, and the survival package (version 3.2-10) allowed statistical analysis of survival data. Subgroups included 0-50 and 50-100. The prognosis types were OS, progression-free interval (PFS), and disease specific survival (DSS). Supplementary data were prognostic data from the reference literature[18]. The filter condition was set to remove data from the normal/control groups (of note, not every item had a normal/control group) and keep the data for clinical information.
The R package used was the survivor package (version 3.2-10). Statistical analysis was performed using the Cox regression module. Prognosis types were OS, PFS, and DSS, and included variables were DLX1, DLX2, DLX3, DLX4, DLX5, and DLX6. Supplementary data were prognostic data from the reference literature[18]. The filter condition was set to remove data from the normal/control groups (of note, not every item had a normal/control group) and keep the data for clinical information.
Two R packages were used: the pROC package (for analysis) and ggplot2 package (version 3.3.3). Clinical variables were “tumor” and “normal”. UCSC XENA (https://xenabrowser.net/datapages/) RNAseq data were uniformly processed by the Toil process into TPM format for TCGA and GTEx[16]. Data for colon cancer were extracted from TCGA and corresponding normal tissue data were extracted from GTEx. The RNAseq data were in TPM format and log2 transformed for expression comparison between samples. Data were not filtered. The horizontal coordinate was the false positive rate and the vertical coordinate was the true positive rate.
The R package used was the stat package (version 3.6.3) (base package). The TCGA colon cancer project provided the RNAseq data in level 3 HTSeq-FPKM format. The TPM format was converted to FPKM, and log2 transformation was applied to the transformed data. The control/normal groups were removed from the results (of note, not all projects had control/normal groups).
The R packages used were mainly ggplot2 package (version 3.3.3) and clusterProfiler package (version 3.14.3).
The R package used was the GSVA package (version 1.34.0)[19]. For immune infiltration, the GSVA package had a built-in algorithm, ssGSEA. Immune cells included were activated dendritic cells (aDCs), B-cells, CD8 T-cells, cytotoxic cells, dendritic cells (DCs), eosinophils, immature DCs (iDCs), macrophages, mast cells, neutrophils, natural killer (NK) CD56bright cells, NK CD56dim cells, NK cells, plasmacytoid DCs (pDCs), T-cells, T helper (Th) cells, T central memory cells, T effector memory (Tem) cells, T follicular helper (TFH) cells, T gamma delta (Tgd) cells, Th1 cells, Th17 cells, Th2 cells, and regulatory T (Treg) cells[20]. The data filtering condition was set to remove the control/normal group (of note, not all projects had control/normal groups). Markers for 24 immune cells were obtained from the reference literature[21].
To further verify the accuracy of the TCGA database, we downloaded colon cancer samples from the Gene Expression Omnibus database for analysis. The 30 colon cancer tissue samples and 30 normal colon tissue samples contained in GSE74062 were used for DLX gene expression analysis.
All statistical analyses were performed using R software (v.3.6.3). The Wilcoxon rank sum test, chi-square test, and Fisher exact test were used to analyze the relationship between clinical characteristics and DLX genes. P values less than 0.05 were considered statistically significant.
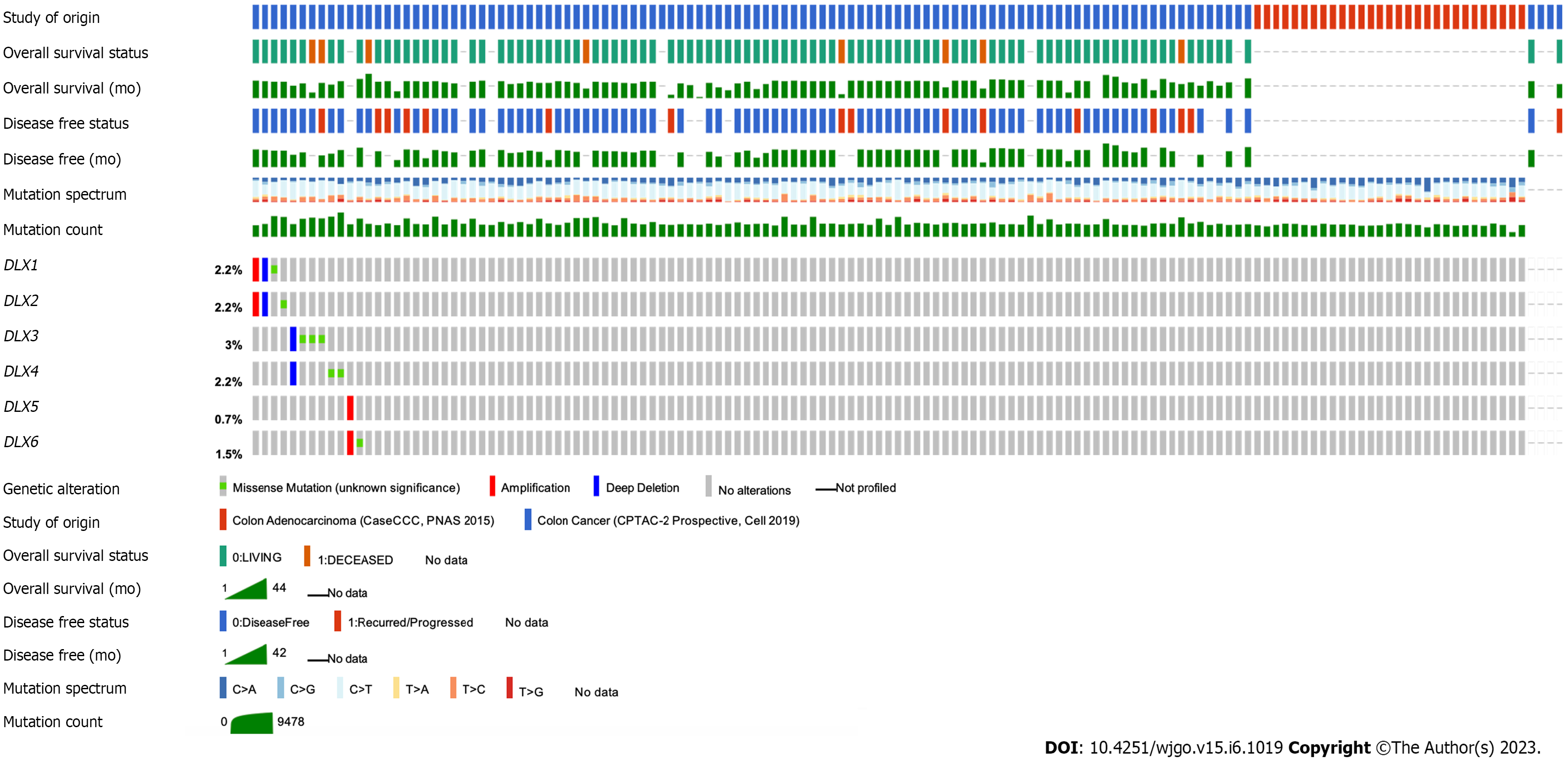
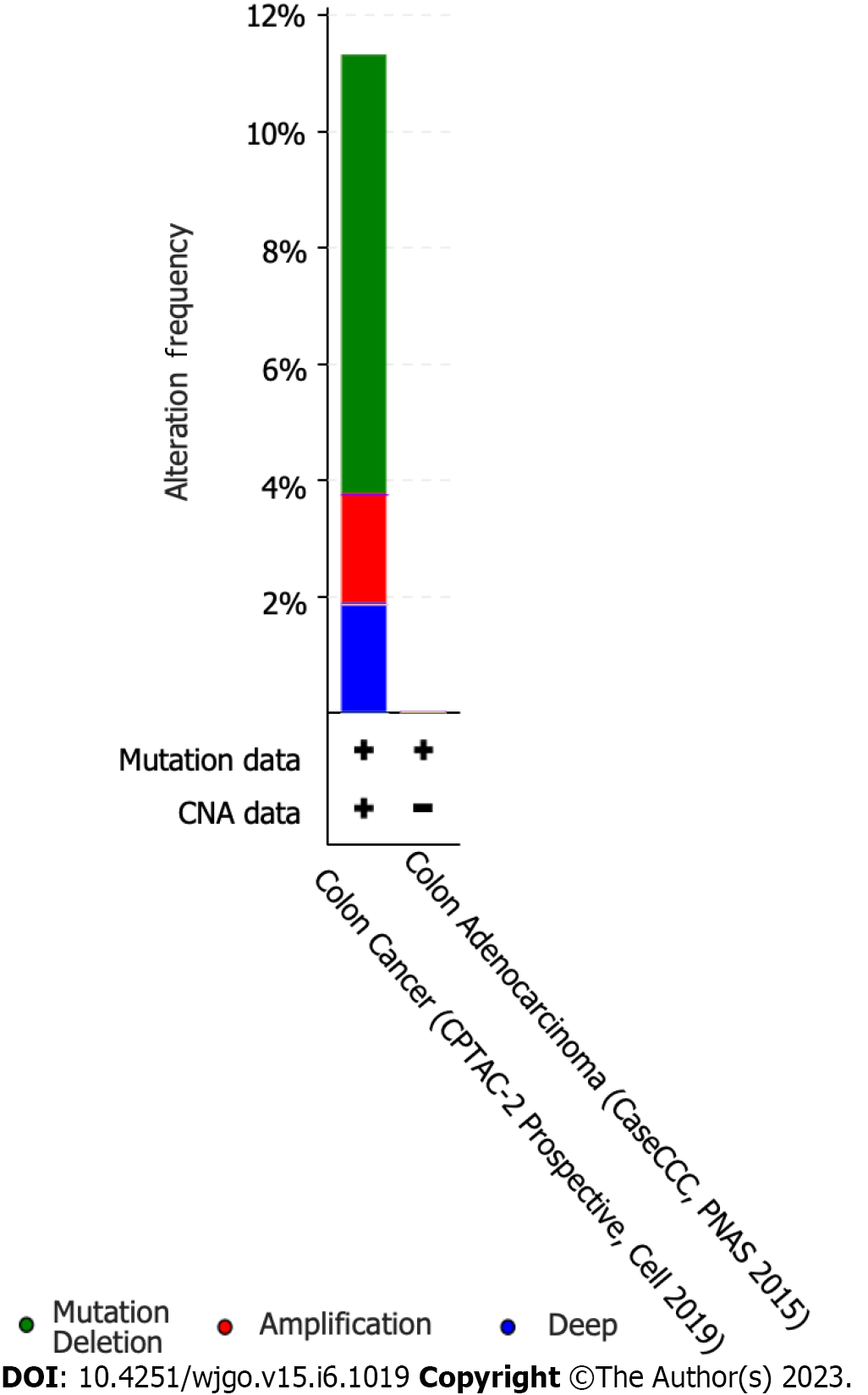
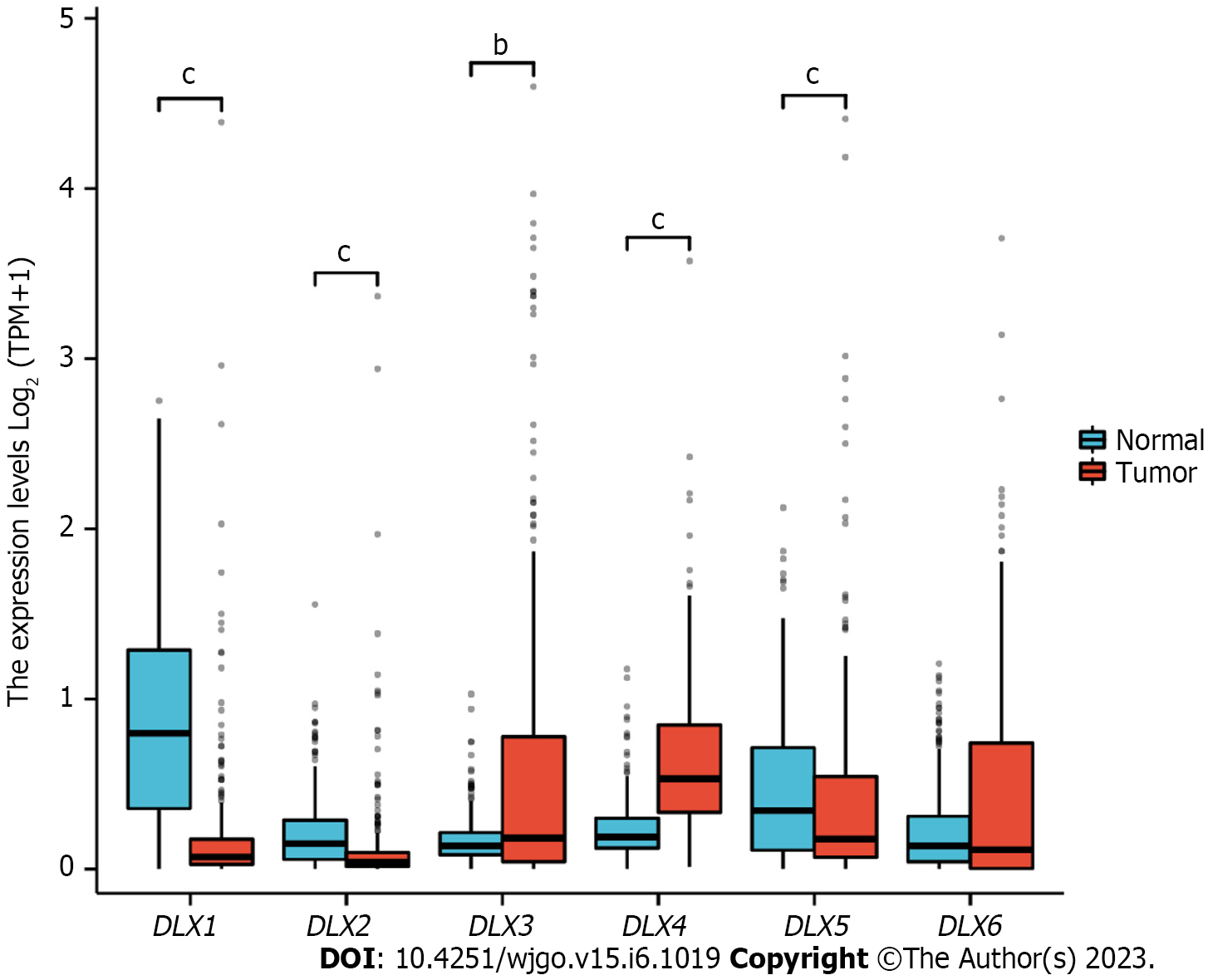
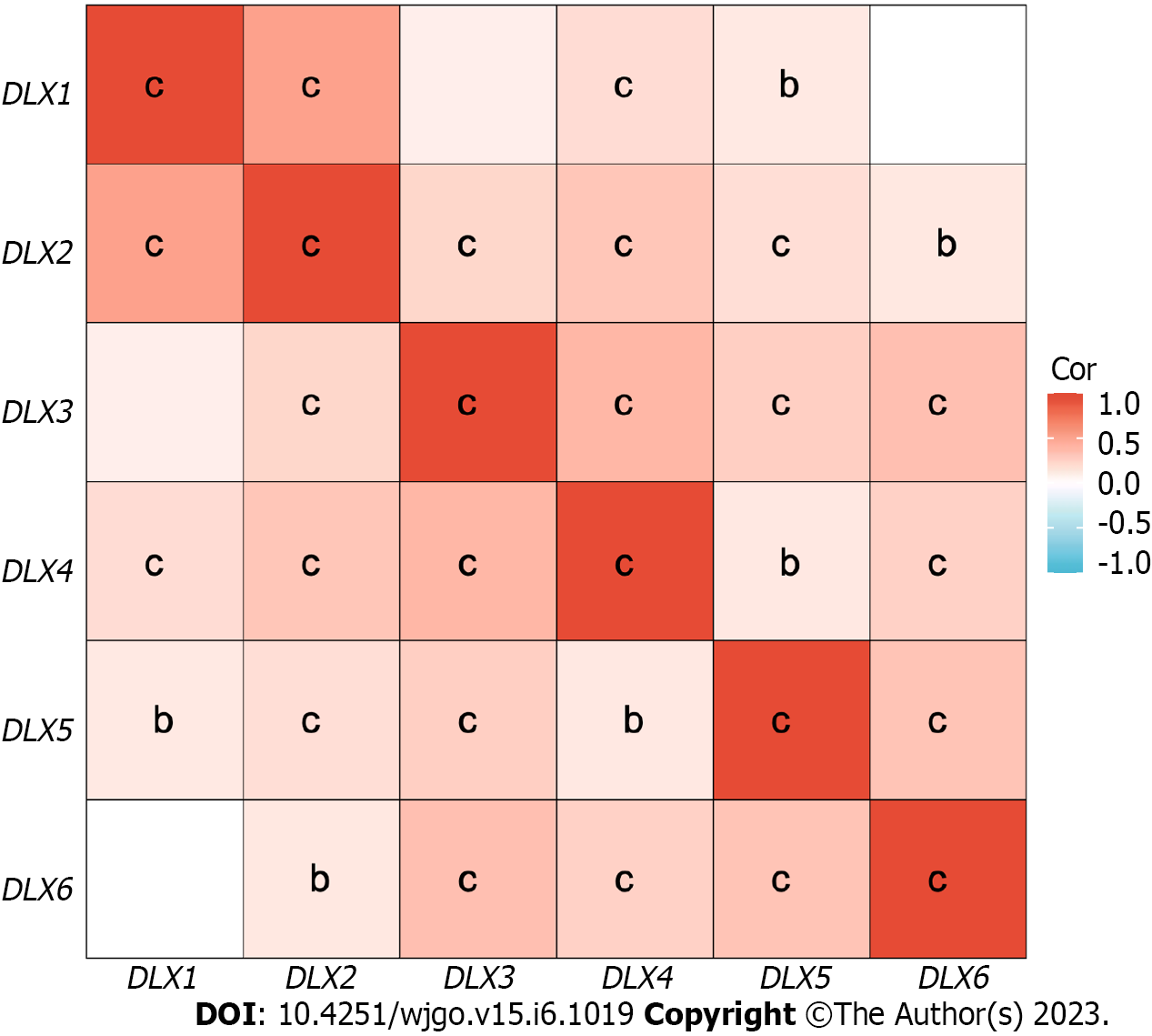
The cBioPortal online tool was used to analyze the expression of DLX family genes in colon cancer patients. Alterations in the expression of DLX genes in colon cancer ranged from 0.7% to 3% (Figure 1). The mutation data, CNA data, and deep deletion from the 2 studies are depicted in Figure 2. The analysis of DLX gene expression was performed based on 41 colon cancer tissue samples and 41 paired samples of normal colon tissue (Figure 3). The results showed that the expression level of DLX1 in colon cancer was significantly lower than that in normal colon tissue (0.199 ± 0.026 vs 0.867 ± 0.031; P < 0.001). The expression level of DLX2 in colon cancer was significantly lower than that in normal colon tissue (0.129 ± 0.020 vs 0.211 ± 0.011; P = 0.0074). The expression level of DLX3 in colon cancer was significantly higher than that in normal colon tissue (0.593 ± 0.052 vs 0.171 ± 0.008; P < 0.001). The expression level of DLX4 in colon cancer was significantly higher than in normal colon tissue (0.635 ± 0.027 vs 0.229 ± 0.009; P < 0.001). The expression level of DLX5 in colon cancer was significantly lower than that in normal colon tissue (0.416 ± 0.036 vs 0.463 ± 0.022; P < 0.001). There was no significant difference in DLX6 expression in colon cancer compared to normal colon tissue (0.229 ± 0.014 vs 0.449 ± 0.037; P = 0.554). We examined the correlation between DLX genes using Pearson correlation analysis. There was no significant correlation between DLX1 and DLX3, DLX1 and DLX6; there was a significant positive correlation between other DLX genes (Figure 4).
Clinical characteristics data and gene expression data for 478 colon cancer samples were downloaded from the TCGA database (Supplementary Table 1). DLX2 expression was associated with M stage (P = 0.005), pathologic stage (P = 0.014), primary therapy outcome (P = 0.036), residual tumor (P = 0.002), and lymphatic invasion (P = 0.013). DLX3 expression was associated with N stage (P < 0.001), M stage (P < 0.001), pathologic stage (P < 0.001), height (P = 0.045), and residual tumor (P < 0.001). DLX5 expression was associated with T stage (P < 0.001), N stage (P < 0.001), M stage (P = 0.005), pathologic stage (P < 0.001), primary therapy outcome (P = 0.005), age (P < 0.001), perineural invasion (P = 0.023), lymphatic invasion (P < 0.001), and history of colon polyps (P = 0.009). However, the expression of DLX1, DLX4, and DLX6 did not significantly correlate with any clinical characteristic of colon cancer patients.
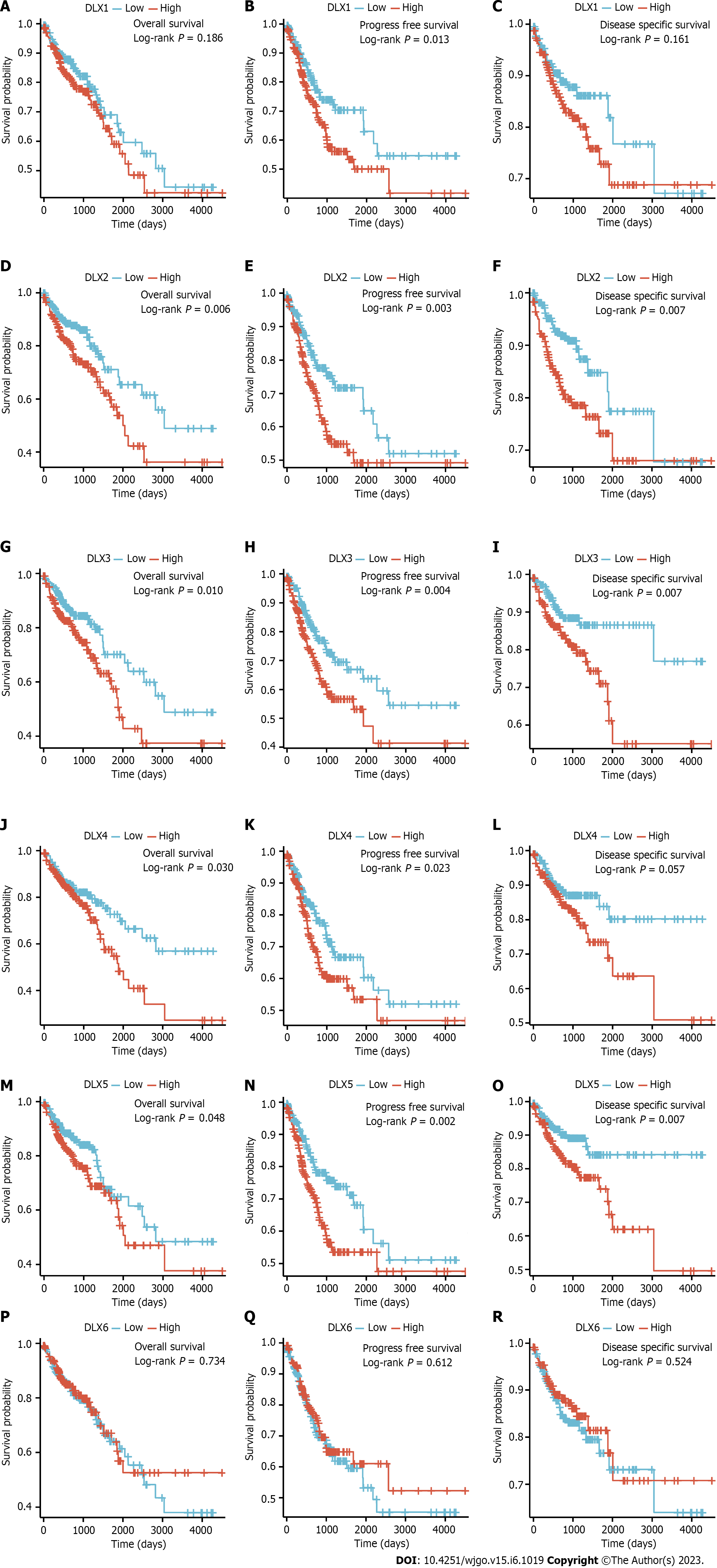
A low expression of DLX1 was associated with PFS (P = 0.013); a low expression of DLX2 was associated with OS (P = 0.006), PFS (P = 0.003), and DSS (P = 0.007); a high expression of DLX3 was associated with OS (P = 0.010), PFS (P = 0.004), and DSS (P = 0.007); a high expression of DLX4 was associated with OS (P = 0.030) and PFS (P = 0.023); a low expression of DLX5 was associated with poor OS (P = 0.048), PFS (P = 0.002), and DSS (P = 0.007). However, a high expression of DLX6 was not significantly associated with prognosis in colon cancer (Figure 5).
Univariate Cox regression analysis for OS showed that DLX2 (P = 0.007), DLX3 (P = 0.011), DLX4 (P = 0.031), and DLX5 (P = 0.049) were associated with OS, and DLX1 (P = 0.014), DLX2 (P = 0.003), DLX3 (P = 0.004), DLX4 (P = 0.024), and DLX5 (P = 0.002) were associated with PFS. DLX2 (P = 0.008), DLX3 (P = 0.008), and DLX5 (P = 0.009) were associated with DSS. DLX5 was independently correlated with PFS (P = 0.012) and DSS (P = 0.035) in multivariate analysis (Table 1).
| Survival | Characteristics | Total, n | Univariate analysis | Multivariate analysis | ||
| HR (95%CI) | P value | HR (95%CI) | P value | |||
| Overall | DLX1 (low vs high) | 477 | 1.299 (0.880-1.917) | 0.186 | ||
| DLX2 (low vs high) | 477 | 1.736 (1.167-2.584) | 0.007 | 1.502 (0.988-2.285) | 0.057 | |
| DLX3 (low vs high) | 477 | 1.668 (1.123-2.476) | 0.011 | 1.374 (0.900-2.099) | 0.142 | |
| DLX4 (low vs high) | 477 | 1.538 (1.039-2.276) | 0.031 | 1.197 (0.783-1.830) | 0.405 | |
| DLX5 (low vs high) | 477 | 1.485 (1.001-2.202) | 0.049 | 1.334 (0.893-1.993) | 0.159 | |
| DLX6 (low vs high) | 477 | 0.935 (0.634-1.379) | 0.734 | |||
| Progression-free | DLX1 (low vs high) | 477 | 1.557 (1.094-2.214) | 0.014 | 1.316 (0.901-1.921) | 0.155 |
| DLX2 (low vs high) | 477 | 1.715 (1.201-2.449) | 0.003 | 1.317 (0.883-1.964) | 0.178 | |
| DLX3 (low vs high) | 477 | 1.670 (1.174-2.376) | 0.004 | 1.365 (0.937-1.990) | 0.105 | |
| DLX4 (low vs high) | 477 | 1.497 (1.054-2.125) | 0.024 | 1.181 (0.813-1.715) | 0.382 | |
| DLX5 (low vs high) | 477 | 1.742 (1.217-2.492) | 0.002 | 1.588 (1.105-2.283) | 0.012 | |
| DLX6 (low vs high) | 477 | 0.914 (0.646-1.294) | 0.613 | |||
| Disease specific | DLX1 (low vs high) | 461 | 1.426 (0.865-2.349) | 0.164 | ||
| DLX2 (low vs high) | 461 | 2.014 (1.202-3.376) | 0.008 | 1.666 (0.971-2.857) | 0.064 | |
| DLX3 (low vs high) | 461 | 2.007 (1.202-3.349) | 0.008 | 1.570 (0.909-2.713) | 0.106 | |
| DLX4 (low vs high) | 461 | 1.617 (0.981-2.664) | 0.059 | 1.179 (0.692-2.011) | 0.545 | |
| DLX5 (low vs high) | 461 | 2.011 (1.193-3.390) | 0.009 | 1.765 (1.039-2.998) | 0.035 | |
| DLX6 (low vs high) | 461 | 0.852 (0.520-1.395) | 0.524 | |||
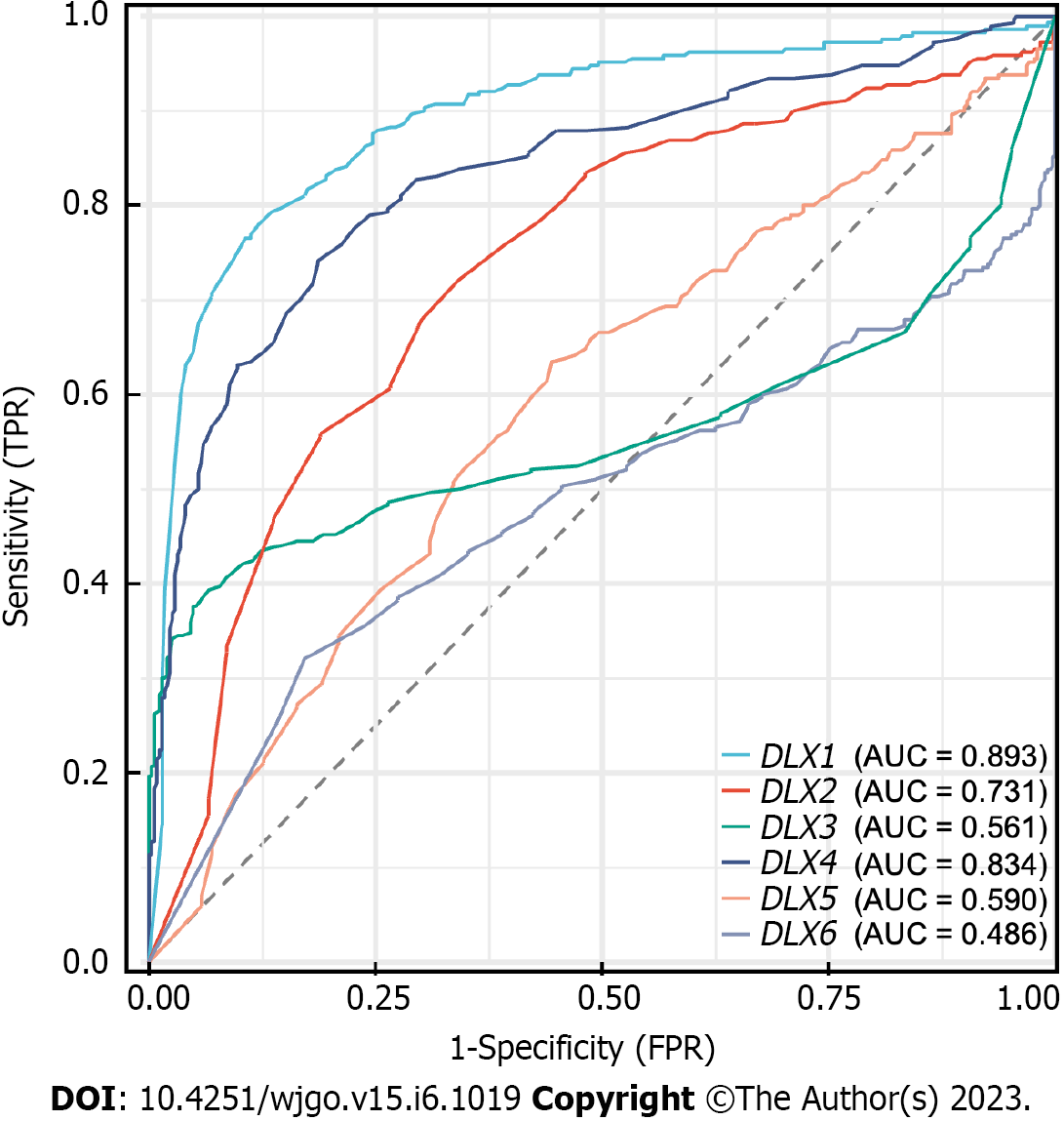
DLX1 had some accuracy in diagnosing normal and tumor outcomes [area under curve (AUC) = 0.893; 95%CI: 0.867-0.920]. DLX2 also had some accuracy in diagnosing normal and tumor outcomes (AUC = 0.731; 95%CI: 0.691-0.771), while DLX3 had a lower accuracy in diagnosing these outcomes (AUC = 0.561; 95%CI: 0.512-0.611). DLX4 also had some accuracy in diagnosing normal and tumor outcomes (AUC = 0.834; 95%CI: 0.802-0.867), while DLX5 had low accuracy in diagnosing these outcomes (AUC = 0.590; 95%CI: 0.546-0.635). Lastly, DLX6 had poor accuracy in diagnosing normal and tumor outcomes (AUC = 0.486; 95%CI: 0.439-0.534) (Figure 6).
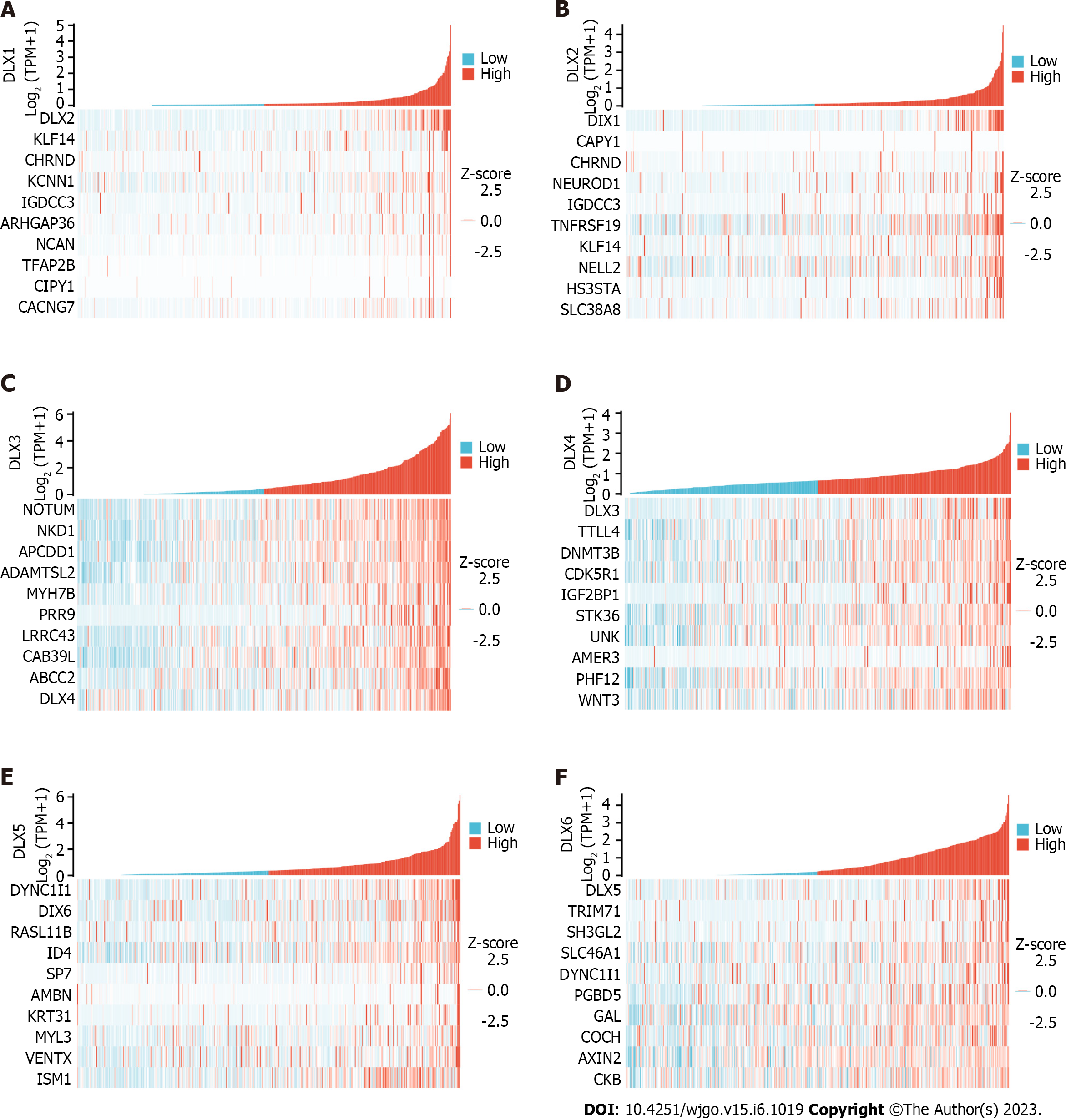
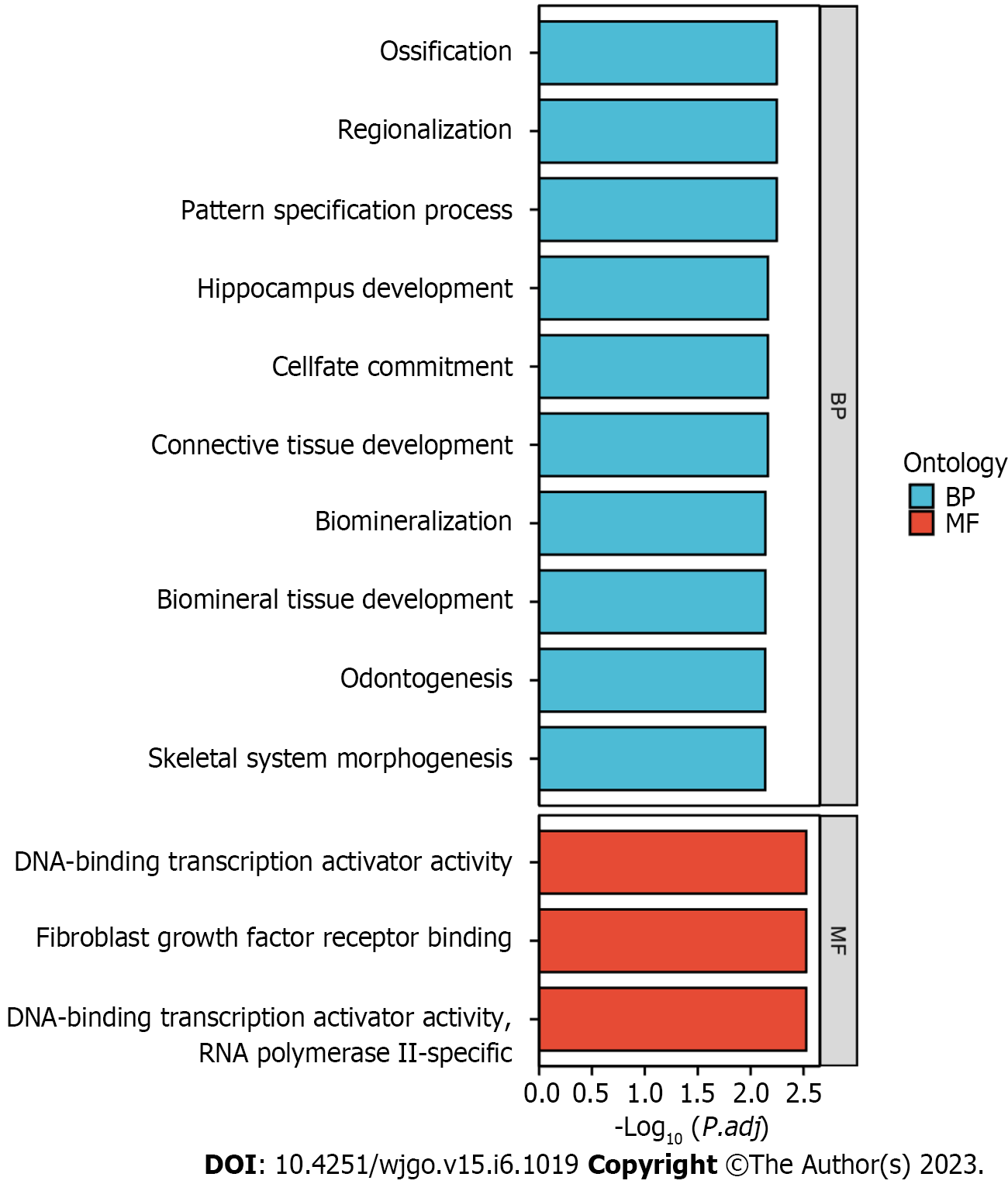
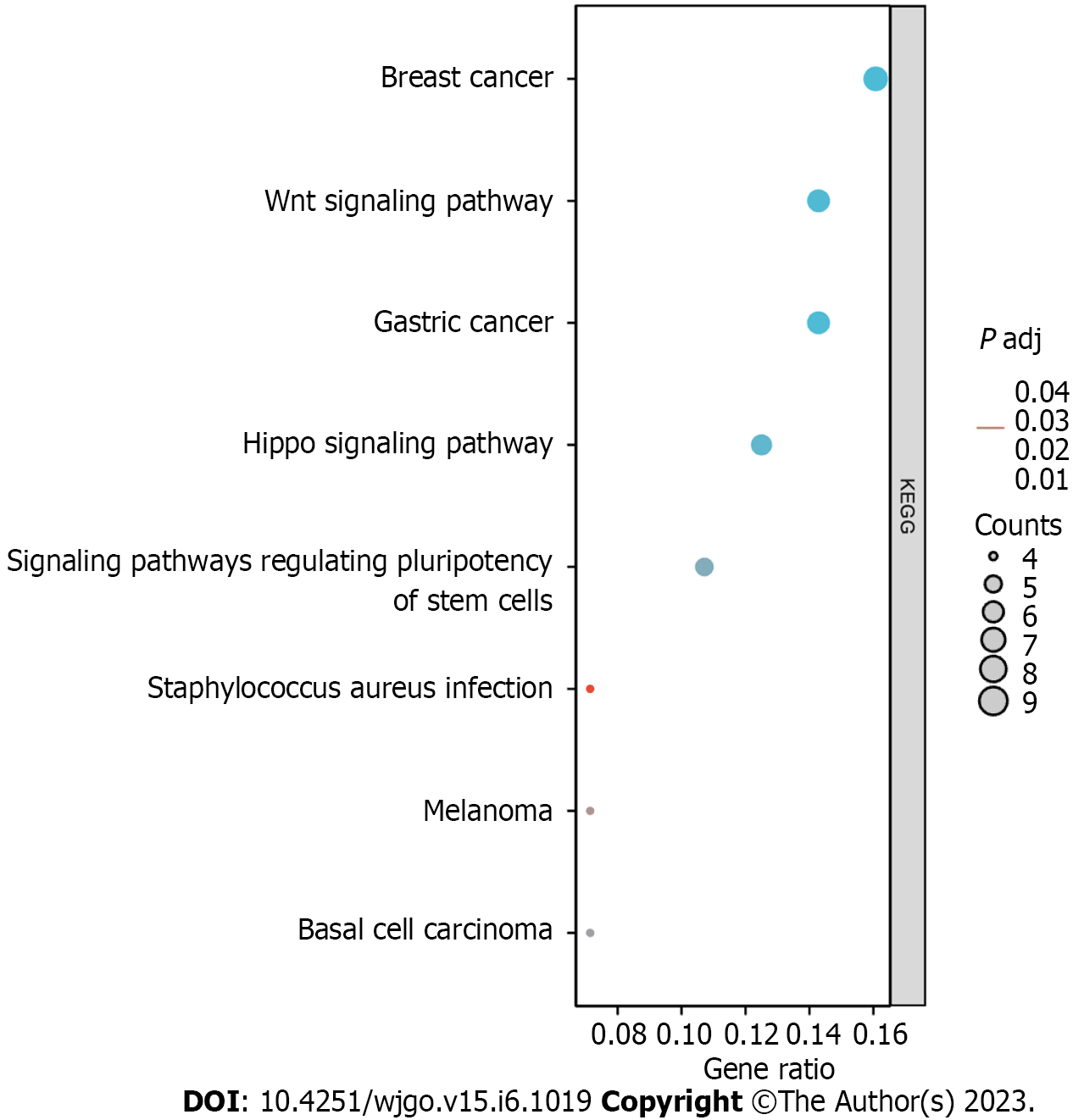
The top 10 significantly associated genes for each DLX gene are shown in the single gene co-expression heat map (Figure 7). Genes significantly associated with DLX1 included DLX2, KLF14, CHRND, KCNN1, IGDCC3, ARHGAP36, NCAN, TFAP2B, CNPY1, and CACNG7. Genes significantly associated with DLX2 included DLX1, CNPY1, CHRND, NEUROD1, IGDCC3, TNFRSF19, KLF14, NELL2, HS3ST4, and SLC38A8. Genes significantly associated with DLX3 included NOTUM, NKD1, APCDD1, ADAMTSL2, MYH7B, PRR9, LRRC43, CAB39L, ABCC2, and DLX4. Genes significantly associated with DLX4 included DLX3, TTLL4, DNMT3B, CDK5R1, IGF2BP1, STK36, UNK, AMER3, PHF12, and WNT3. Genes significantly associated with DLX5 included DYNC1I1, DLX6, RASL11B, ID4, SP7, AMBN, KRT31, MYL3, VENTX, and ISM1. Genes significantly associated with DLX6 included DLX5, TRIM71, SH3GL2, SLC46A1, DYNC1I1, PGBD5, GAL, COCH, AXIN2, and CKB. The top 30 genes significantly associated with each DLX gene (147 in total) were analyzed for Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment (Supplementary Table 2). The top biological processes included pattern specification, regionalization, ossification, connective tissue development, cell fate commitment, hippocampus development, biomineral tissue development, biomineralization, skeletal system morphogenesis, and odontogenesis. The significantly related molecular functions included DNA-binding transcription activator activity, RNA polymerase II-specificity, fibroblast growth factor receptor binding, DNA-binding transcription activator activity (Figure 8 and Supplementary Table 3). The significantly related pathways included the Hippo signaling pathway, the Wnt signaling pathway, and signaling pathways regulating the pluripotency of stem cells and Staphylococcus aureus infection (Figure 9 and Supplementary Table 3).
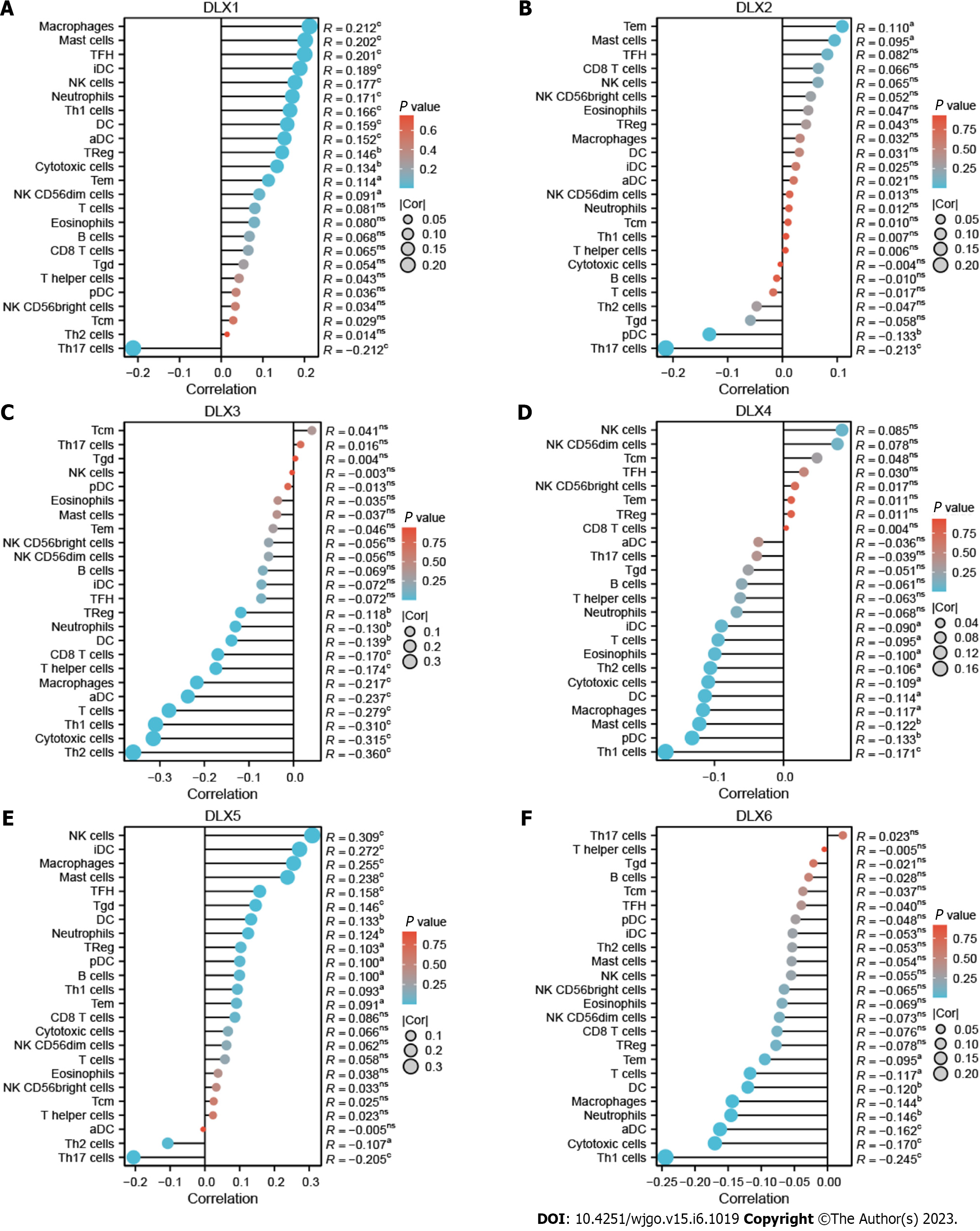
There was a correlation between DLX gene expression and immune cells in colon cancer (Figure 10). DLX1 gene expression positively correlated with some tumor-infiltrating immune cells (TIICs), including aDCs, cytotoxic cells, DCs, eosinophils, iDCs, macrophages, mast cells, neutrophils, NK CD56dim cells, NK cells, Tem cells, TFH cells, Tgd cells, Th1 cells, and Treg cells; DLX1 expression negatively correlated with Th17 cells. DLX2 gene expression positively correlated with mast cells and TFH cells and negatively correlated with pDCs and Th17 cells. DLX3 gene expression negatively correlated with some TIICs, including aDCs, CD8 T-cells, cytotoxic cells, DCs, macrophages, neutrophils, T-cells, Th cells, Th1 cells, Th2 cells, and Treg cells. DLX4 gene expression positively correlated with NK cells and negatively correlated with some TIICs, including cytotoxic cells, DCs, macrophages, pDCs, Th1 cells, and Th2 cells. DLX5 gene expression positively correlated with some TIICs, including B-cells, CD8 T-cells, DCs, iDCs, macrophages, mast cells, neutrophils, NK cells, pDCs, Tem cells, TFH cells, Tgd cells, and Treg cells; DLX5 expression negatively correlated with Th17 cells and Th2 cells. DLX6 gene expression negatively correlated with some TIICs, including aDCs, cytotoxic cells, DCs, macrophages, neutrophils, NK CD56dim cells, T-cells, Tem cells, and Th1 cells.
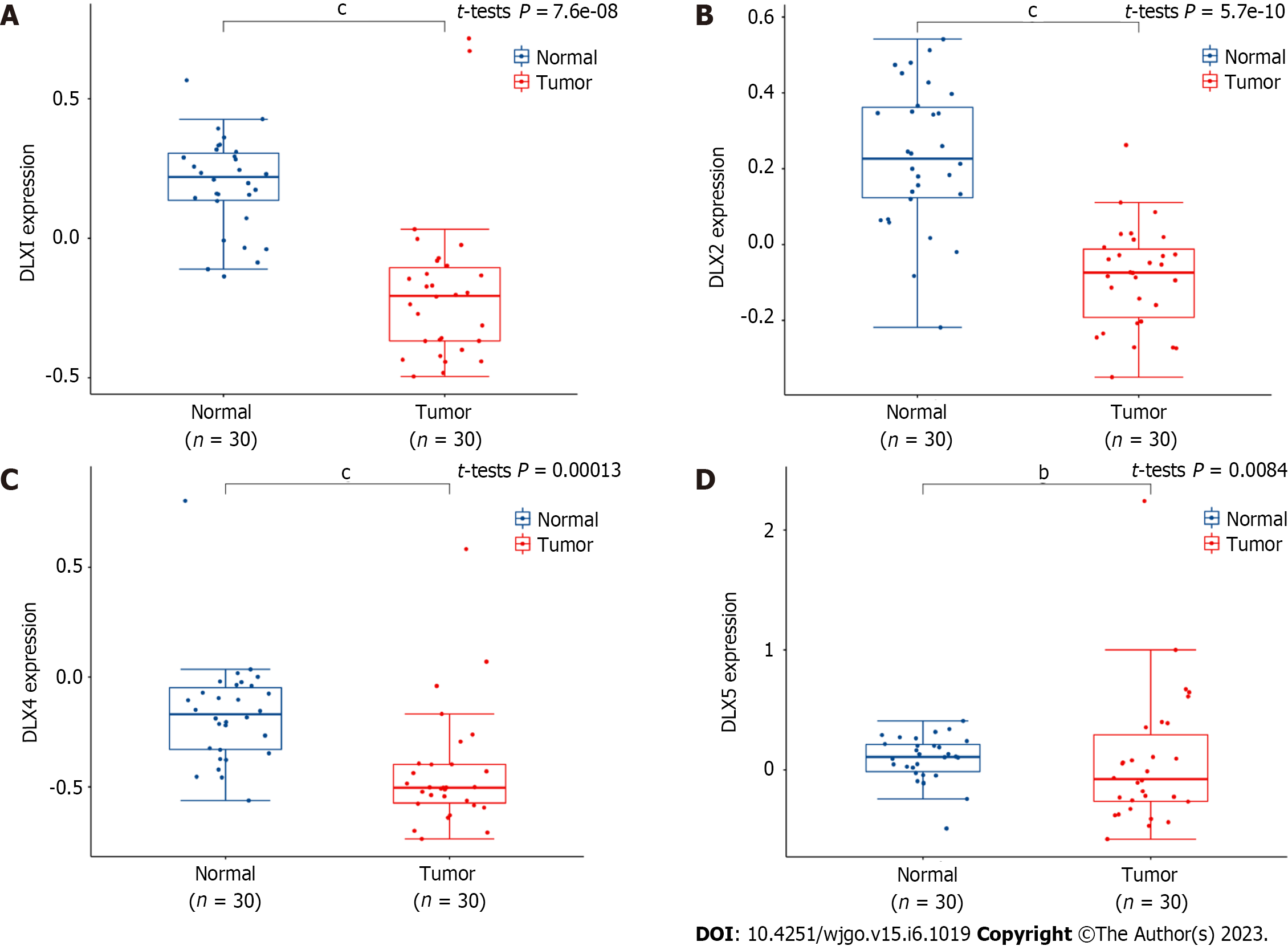
Compared to normal colon, DLX1 (P = 7.6e-08), DLX2 (P = 5.7e-08), DLX4 (P = 0.00013), and DLX5 (P = 0.0084) were aberrantly expressed in colon cancer tissue. However, DLX3 and DLX6 were not aberrantly expressed in colon cancer (Figure 11).
DLX1 has been shown to be significantly upregulated in prostate cancer tissues and cells[22]. DLX2 is known to be significantly upregulated in HCC tissues and cell lines[7,23], and its expression in gastric cancer has been shown to significantly correlated with tumor size, depth of infiltration, lymph node metastasis, and tumor-lymph node metastasis stage[24]. DLX4 has been demonstrated to be upregulated in nasopharyngeal carcinoma (NPC) cell lines[25], and its expression was shown to be elevated in HCC and correlated significantly with tumor size, histopathological classification, and serum alpha-fetoprotein[10]. DLX5 has been shown to be upregulated in OSCC tissues and cell lines, and has been associated with advanced TNM staging, lymph node metastasis, poor cell differentiation, and tumor location[11]. DLX6 has been shown to be upregulated in oral cancer and has been associated with advanced tumor stage and poor prognosis[12]. In this study, DLX1/2/3/4/5 were aberrantly expressed in colon cancer tissue samples. The expression of DLX family genes was associated with M stage, pathologic stage, primary therapy outcome, residual tumor, lymphatic invasion, T stage, N stage, age, perineural invasion, and history of colon polyps. In the multivariate analysis, DLX5 was independently related to PFS and OS. In diagnosing the outcome of normal and tumor tissues, DXL1/2/4 had some accuracy.
MiR-129-5p has been shown to impede the biological function of cancer cells by inhibiting DLX1 expression[26]. DLX1, a key target of FOXM1, has been shown to promote ovarian cancer aggressiveness by enhancing transforming growth factor (TGF)-β/SMAD4 signaling[27]. Circ_HIPK3 has been demonstrated to promote HCC progression by mediating the miR-582-3p/DLX2 pathway[23]. In tumor cells, DLX2/3/4 can be involved in the control of fenretinide (4HPR)-mediated apoptosis[28]. DLX3 has been shown to be downregulated by miR-133[29]. The homology domain protein DLX4 has been shown to promote NPC progression through the upregulation of YB-1[25]. DLX5 regulation of CCND1 affected the progression of OSCC[11]. DLX5 has been shown to promote osteosarcoma progression through activation of the NOTCH signaling pathway[30]. DLX6 has been demonstrated to regulate OSCC cell proliferation through the EGFR-CCND1 axis[12]. In this study, the DLX gene family is suggested to be involved in the development and progression of colon cancer by participating in several pathways, including breast cancer, gastric cancer, the Hippo signaling pathway, the Wnt signaling pathway, and signaling pathways regulating the pluripotency of stem cells, basal cell carcinoma, melanoma, and Staphylococcus aureus infection. Dlx-2 is involved in TGF-β- and Wnt-induced inhibition of mitochondria by epithelial-mesenchymal transition, glycolytic conversion, and Snail activation[31]. However, the specific mechanisms by which the DLX gene family mediates the pathways involved in the development of colon cancer need to be further investigated.
Immune-related mechanisms play an important role in the development of colon cancer, and immunotherapeutic strategies are considered a promising direction for the treatment of this disease[32]. Another important aspect of the current study was that the expression of the DLX gene family correlated with different levels of immune infiltration. Here, the expression levels of DLX family genes were negatively correlated with some TIICs, and positively correlated with other TIICs. The DLX gene family plays an important role in the recruitment and regulation of immune infiltrating cells in colon cancer.
The present study has several limitations. Firstly, colon cancer shows strong heterogeneity, and the mRNA expression levels in the TCGA database are the average mRNA expression levels for all cell types within various colon tumors. Single-cell sequencing is needed to further elucidate the role of DLX genes in colon cancer and its subtypes. Secondly, our study findings are not confirmed by biological or molecular experiments.
DLX1/2/3/4/5 were significantly aberrantly expressed in colon cancer tissue samples. DLX 2/3/5 were associated with M stage, pathologic stage, primary therapy outcome, residual tumor, lymphatic invasion, T stage, N stage, age, perineural invasion, and history of colon polyps. DLX5 was independently correlated with the prognosis of colon cancer in multivariate analysis. DLX1/2/4 had some accuracy in diagnosing normal and tumor conditions. The DLX gene family may be involved in the development and progression of colon cancer by participating in immune infiltration and pathways, including the Hippo signaling pathway, the Wnt signaling pathway, and signaling pathways regulating the pluripotency of stem cells and Staphylococcus aureus infection. The results of this study suggest a role for DLX family genes as a potential diagnostic or prognostic biomarkers and therapeutic targets in colon cancer.
The distal-less homeobox (DLX) gene family plays an important role in several tumors. However, the role of DLX gene family in colon cancer is not yet clear.
The aim of this study was to investigate the role of the DLX gene family in colon cancer and to establish a sound scientific basis for clinical decision making and risk management.
In this study, we aimed to comprehensively analyze the biological role of the DLX gene family in colon cancer.
Colon cancer and normal colon tissue samples were collected from the Cancer Genome Atlas (TCGA) and Gene Expression Omnibus databases. We used Wilcoxon rank sum test and t-test to assess DLX gene family expression between colon cancer tissue samples and unpaired normal colon tissue samples, cBioPortal to analyze DLX gene family variants, R software (version 3.6.3) to analyze DLX gene expression in colon cancer and the relationship between DLX gene family expression and clinical features and correlation heat map, the survival package [version 3.2-10] and Cox regression module to assess the prognostic value of the DLX gene family, the pROC package [version 1.17.0.1] to analyze the diagnostic value of the DLX gene family, R software (version 3.6.3) to analyze the possible regulatory mechanisms of DLX gene family members and related genes, the GSVA package [version 1.34.0] to analyze the relationship between the DLX gene family and immune infiltration, and the ggplot2 [version 3.3.3], the survminer package [version 0.4.9], and the clusterProfiler package [version 3.14.3] for visualization.
Expression levels of DLX1/2/3/4/5 were significantly abnormal in tissue from patients with colon cancer. DLX gene family expression in colon cancer was significantly associated with clinical characteristics, including M stage, pathological stage, primary treatment outcome, residual tumor, lymphatic invasion, T stage, N stage, age, peripheral invasion, and history of colonic polyps. Results of the multivariate Cox analysis showed DLX5 to be an independent prognostic factor in patients with colon cancer. DLX1/2/3/4/5/6 may be involved in the development and progression of colon cancer through mediation of multiple pathways, including the Hippo signaling pathway, the Wnt signaling pathway, and signaling pathways regulating the pluripotency of stem cells. DLX1/2/3/4/5/6 are associated with immune infiltration.
DLX family genes may function as potential diagnostic or prognostic biomarkers and therapeutic targets for colon cancer.
It may be possible to use DLX family genes as a diagnostic or prognostic biomarkers or therapeutic targets for colon cancer.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Abdalla AN, Saudi Arabia; Zhang W, China S-Editor: Wang JJ L-Editor: Filipodia P-Editor: Zhang XD
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55820] [Article Influence: 7974.3] [Reference Citation Analysis (132)] |
| 2. | Alibolandi M, Rezvani R, Farzad SA, Taghdisi SM, Abnous K, Ramezani M. Tetrac-conjugated polymersomes for integrin-targeted delivery of camptothecin to colon adenocarcinoma in vitro and in vivo. Int J Pharm. 2017;532:581-594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 3. | Sargent DJ, Wieand HS, Haller DG, Gray R, Benedetti JK, Buyse M, Labianca R, Seitz JF, O'Callaghan CJ, Francini G, Grothey A, O'Connell M, Catalano PJ, Blanke CD, Kerr D, Green E, Wolmark N, Andre T, Goldberg RM, De Gramont A. Disease-free survival versus overall survival as a primary end point for adjuvant colon cancer studies: individual patient data from 20,898 patients on 18 randomized trials. J Clin Oncol. 2005;23:8664-8670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 491] [Cited by in RCA: 522] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 4. | Zhou Y, Zang Y, Yang Y, Xiang J, Chen Z. Candidate genes involved in metastasis of colon cancer identified by integrated analysis. Cancer Med. 2019;8:2338-2347. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 5. | Cohen SM, Brönner G, Küttner F, Jürgens G, Jäckle H. Distal-less encodes a homoeodomain protein required for limb development in Drosophila. Nature. 1989;338:432-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 306] [Cited by in RCA: 301] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 6. | Leyten GH, Hessels D, Smit FP, Jannink SA, de Jong H, Melchers WJ, Cornel EB, de Reijke TM, Vergunst H, Kil P, Knipscheer BC, Hulsbergen-van de Kaa CA, Mulders PF, van Oort IM, Schalken JA. Identification of a Candidate Gene Panel for the Early Diagnosis of Prostate Cancer. Clin Cancer Res. 2015;21:3061-3070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 162] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 7. | Liu J, Cui X, Qu L, Hua L, Wu M, Shen Z, Lu C, Ni R. Overexpression of DLX2 is associated with poor prognosis and sorafenib resistance in hepatocellular carcinoma. Exp Mol Pathol. 2016;101:58-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 8. | Yan ZH, Bao ZS, Yan W, Liu YW, Zhang CB, Wang HJ, Feng Y, Wang YZ, Zhang W, You G, Zhang QG, Jiang T. Upregulation of DLX2 confers a poor prognosis in glioblastoma patients by inducing a proliferative phenotype. Curr Mol Med. 2013;13:438-445. [PubMed] |
| 9. | Bhattacharya S, Kim JC, Ogawa Y, Nakato G, Nagle V, Brooks SR, Udey MC, Morasso MI. DLX3-Dependent STAT3 Signaling in Keratinocytes Regulates Skin Immune Homeostasis. J Invest Dermatol. 2018;138:1052-1061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 10. | Gao Y, Li Z, Guo X, Liu Y, Zhang K. DLX4 as a prognostic marker for hepatocellular carcinoma. Neoplasma. 2014;61:318-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 11. | Zhang J, Wu J, Chen Y, Zhang W. Dlx5 promotes cancer progression through regulation of CCND1 in oral squamous cell carcinoma (OSCC). Biochem Cell Biol. 2021;99:424-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 12. | Liang J, Liu J, Deng Z, Liu Z, Liang L. DLX6 promotes cell proliferation and survival in oral squamous cell carcinoma. Oral Dis. 2022;28:87-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 13. | Chen J, Tang H, Li T, Jiang K, Zhong H, Wu Y, He J, Li D, Li M, Cai X. Comprehensive Analysis of the Expression, Prognosis, and Biological Significance of OVOLs in Breast Cancer. Int J Gen Med. 2021;14:3951-3960. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 14. | Lin Z, Huang W, Yi Y, Li D, Xie Z, Li Z, Ye M. LncRNA ADAMTS9-AS2 is a Prognostic Biomarker and Correlated with Immune Infiltrates in Lung Adenocarcinoma. Int J Gen Med. 2021;14:8541-8555. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 15. | Liang W, Lu Y, Pan X, Zeng Y, Zheng W, Li Y, Nie Y, Li D, Wang D. Decreased Expression of a Novel lncRNA FAM181A-AS1 is Associated with Poor Prognosis and Immune Infiltration in Lung Adenocarcinoma. Pharmgenomics Pers Med. 2022;15:985-998. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 16. | Vivian J, Rao AA, Nothaft FA, Ketchum C, Armstrong J, Novak A, Pfeil J, Narkizian J, Deran AD, Musselman-Brown A, Schmidt H, Amstutz P, Craft B, Goldman M, Rosenbloom K, Cline M, O'Connor B, Hanna M, Birger C, Kent WJ, Patterson DA, Joseph AD, Zhu J, Zaranek S, Getz G, Haussler D, Paten B. Toil enables reproducible, open source, big biomedical data analyses. Nat Biotechnol. 2017;35:314-316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 819] [Cited by in RCA: 864] [Article Influence: 108.0] [Reference Citation Analysis (0)] |
| 17. | Yang D, Liu M, Jiang J, Luo Y, Wang Y, Chen H, Li D, Wang D, Yang Z. Comprehensive Analysis of DMRT3 as a Potential Biomarker Associated with the Immune Infiltration in a Pan-Cancer Analysis and Validation in Lung Adenocarcinoma. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 18. | Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, Omberg L, Wolf DM, Shriver CD, Thorsson V; Cancer Genome Atlas Research Network, Hu H. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell. 2018;173:400-416.e11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2534] [Cited by in RCA: 2360] [Article Influence: 337.1] [Reference Citation Analysis (0)] |
| 19. | Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7222] [Cited by in RCA: 9274] [Article Influence: 772.8] [Reference Citation Analysis (0)] |
| 20. | Lu X, Jing L, Liu S, Wang H, Chen B. miR-149-3p Is a Potential Prognosis Biomarker and Correlated with Immune Infiltrates in Uterine Corpus Endometrial Carcinoma. Int J Endocrinol. 2022;2022:5006123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 21. | Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, Bruneval P, Fridman WH, Becker C, Pagès F, Speicher MR, Trajanoski Z, Galon J. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782-795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1792] [Cited by in RCA: 2955] [Article Influence: 246.3] [Reference Citation Analysis (0)] |
| 22. | Sun B, Fan Y, Yang A, Liang L, Cao J. MicroRNA-539 functions as a tumour suppressor in prostate cancer via the TGF-β/Smad4 signalling pathway by down-regulating DLX1. J Cell Mol Med. 2019;23:5934-5948. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 23. | Zhang H, Dai Q, Zheng L, Yuan X, Pan S, Deng J. Knockdown of circ_HIPK3 inhibits tumorigenesis of hepatocellular carcinoma via the miR-582-3p/DLX2 axis. Biochem Biophys Res Commun. 2020;533:501-509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 24. | Tang P, Huang H, Chang J, Zhao GF, Lu ML, Wang Y. Increased expression of DLX2 correlates with advanced stage of gastric adenocarcinoma. World J Gastroenterol. 2013;19:2697-2703. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 27] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 25. | Ling Z, Long X, Li J, Feng M. Homeodomain protein DLX4 facilitates nasopharyngeal carcinoma progression via up-regulation of YB-1. Genes Cells. 2020;25:466-474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 26. | Li Q, Xu K, Tian J, Lu Z, Pu J. MiR-129-5p/DLX1 signalling axis mediates functions of prostate cancer during malignant progression. Andrologia. 2021;53:e14230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 27. | Chan DW, Hui WW, Wang JJ, Yung MM, Hui LM, Qin Y, Liang RR, Leung TH, Xu D, Chan KK, Yao KM, Tsang BK, Ngan HY. DLX1 acts as a crucial target of FOXM1 to promote ovarian cancer aggressiveness by enhancing TGF-β/SMAD4 signaling. Oncogene. 2017;36:1404-1416. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 46] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 28. | Ferrari N, Paleari L, Palmisano GL, Tammaro P, Levi G, Albini A, Brigati C. Induction of apoptosis by fenretinide in tumor cell lines correlates with DLX2, DLX3 and DLX4 gene expression. Oncol Rep. 2003;10:973-977. [PubMed] |
| 29. | Qadir AS, Lee J, Lee YS, Woo KM, Ryoo HM, Baek JH. Distal-less homeobox 3, a negative regulator of myogenesis, is downregulated by microRNA-133. J Cell Biochem. 2019;120:2226-2235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 30. | Zhang X, Bian H, Wei W, Wang Q, Chen J, Hei R, Chen C, Wu X, Yuan H, Gu J, Lu Y, Cai C, Zheng Q. DLX5 promotes osteosarcoma progression via activation of the NOTCH signaling pathway. Am J Cancer Res. 2021;11:3354-3374. [PubMed] |
| 31. | Lee SY, Jeon HM, Ju MK, Jeong EK, Kim CH, Yoo MA, Park HG, Han SI, Kang HS. Dlx-2 is implicated in TGF-β- and Wnt-induced epithelial-mesenchymal, glycolytic switch, and mitochondrial repression by Snail activation. Int J Oncol. 2015;46:1768-1780. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 32. | Procaccio L, Schirripa M, Fassan M, Vecchione L, Bergamo F, Prete AA, Intini R, Manai C, Dadduzio V, Boscolo A, Zagonel V, Lonardi S. Immunotherapy in Gastrointestinal Cancers. Biomed Res Int. 2017;2017:4346576. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |