Published online May 15, 2023. doi: 10.4251/wjgo.v15.i5.787
Peer-review started: November 30, 2022
First decision: December 25, 2022
Revised: January 6, 2023
Accepted: April 7, 2023
Article in press: April 7, 2023
Published online: May 15, 2023
Processing time: 163 Days and 7.8 Hours
Circular RNAs (circRNAs) have attracted extensive attention as therapeutic targets in gastric cancer (GC). Circ_0003356 is known to be downregulated in GC tissues, but its cellular function and mechanisms remain undefined.
To investigate the role of circ_0003356 in GC at the molecular and cellular level.
Circ_0003356, miR-668-3p, and SOCS3 expression were assessed via quantitative real time-polymerase chain reaction (qRT-PCR). Wound healing, EdU, CCK-8, flow cytometry and transwell assays were used to analyze the migration, proliferation, viability, apoptosis and invasion of GC cells. The subcellular localization of circ_0003356 was monitored using fluorescence in situ hybridization. The interaction of circ_0003356 with miR-668-3p was confirmed using RIP-qRT-PCR, RNA pull-down, and dual luciferase reporter assays. We observed protein levels of genes via western blot. We injected AGS cells into the upper back of mice and performed immunohistochemistry staining for examining E-cadherin, N-cadherin, Ki67, and SOCS3 expressions. TUNEL staining was performed for the assessment of apoptosis in mouse tumor tissues.
Circ_0003356 and SOCS3 expression was downregulated in GC cells, whilst miR-668-3p was upregulated. Exogenous circ_0003356 expression and miR-668-3p silencing suppressed the migration, viability, proliferation, epithelial to mese
Circ_0003356 impaired GC development through its interaction with the miR-668-3p/SOCS3 axis.
Core Tip: We observed the low level of circ_0003356 expression in gastric cancer (GC) tissues and cells. Circ_0003356 expression was positively related to GC patient prognosis. Exogenous circ_0003356 overexpression and/or miR-668-3p suppression enhanced apoptosis in GC cells and suppressed GC cell proliferation, migration, invasion, and epithelial to mesenchy-mal transition. The overexpression of circ_0003356 also prevented tumor growth in mice. At the mechansistic level, circ_0003356 was found to interact with the miR-668-3p/SOCS3 axis to impair GC development. Together, we reveal new and important molecular details highlighting circ_0003356 as a novel cancer target.
- Citation: Li WD, Wang HT, Huang YM, Cheng BH, Xiang LJ, Zhou XH, Deng QY, Guo ZG, Yang ZF, Guan ZF, Wang Y. Circ_0003356 suppresses gastric cancer growth through targeting the miR-668-3p/SOCS3 axis. World J Gastrointest Oncol 2023; 15(5): 787-809
- URL: https://www.wjgnet.com/1948-5204/full/v15/i5/787.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i5.787
Gastric cancer (GC) encompasses a range of gastrointestinal malignancies[1,2], and ranks as the 2nd main cause of global cancer-associated deaths[3,4]. Chemotherapy, radiotherapy, and surgical resection have progressed GC treatment[5,6], but five-year survival rates remain low[7,8]. Personalized therapy has shown promise for GC treatment[9], but more effective anti-GC targets are required. Circular RNAs (circRNAs) regulate GC occurrence and development. The downregulation of circ_SKA3 inhibits the colony formation, proliferation, migration, and invasion of GC cells in vitro and suppresses tumorigenesis in vivo[10]. Circ_0005758 up-regulation can inhibit GC cell proliferation, migration and invasion, and suppress tumor growth in xenograft mice[11]. Up-regulation of circ_0021087 impairs proliferation, epithelial to mesenchy-mal transition (EMT), and invasion of GC cells and overpowers tumor development in xenograft mice[12]. CircRNA analysis in GSE184882 indicates that circ_0003356 is lowly expressed in GC tissues, but the molecular effects governing its role in GC suppression have not been investigated at the molecular level.
MicroRNAs (miRNAs) regulate the post-transcriptional expression of a number of genes[13,14] and play a key role in GC progression. The overexpression of miR-548 promotes proliferation, migration and invasion of GC cells in vitro[15]. Silencing of miR-762 reduces GC cell viability and facilitates apoptosis[16]. Inhibition of miR-4742-5p suppresses the proliferation, migration and invasion of GC cells[17]. Especially, miR-668-3p is oncogenic in hepatocellular carcinoma (HCC)[18] and colorectal cancer (CRC) cells[19].
SOCS3 is a negative regulator of hormone and cytokine signaling and plays a key role in cancer development[20,21]. Available studies have highlighted the importance of SOCS3 in the manipulation of cancer development. A study from Yu et al[22] have revealed that the downregulation of SOCS3 can reverse the suppressive effects of miR-3173-5p inhibitor on colony formation, cell proliferation, migration, invasion and EMT in prostate cancer cells. Another study from Li et al[23] have reported that SOCS3 overexpression also suppresses spheroid formation and CRC cell proliferation. In particular, SOCS3 is proven to exert an important role against tumor in GC. For instance, inhibition of miR-340 suppresses cell cycle progression through enhancing the expression of SOCS3 in GC cells[24]. MiR-665 also serves as an oncogene in GC through the downregulation of SOCS3[25]. The regulatory effects of miR-668-3p on SOCS3 in GC cells has not been investigated. Here, we show for the first time that circ_0003356 is downregulated in GC cells and tumor tissues and reveal its association with GC progression.
GC and normal tissues (n = 80 pairs) were collected from surgical specimens of patients with GC at Zhongshan City People’s Hospital. Patients were diagnosed with GC via histopathology. No patients had received prior immunotherapy, adjuvant chemotherapy, or radiation therapy before surgery. GC patients were allocated into circ_0003356 low or high-expression groups based on the median expression value in tumor tissues. The Ethics Committee of Zhongshan City People’s Hospital provided approval (K2017-182). All participants agreed to the study and provided informed written consent.
AGS and HGC-27 cell lines (human) and GES-1 (non-GC) were cultured in Roswell Park Memorial Institute 1640 medium + 10% fetal bovine serum (FBS) + 1% streptomycin-penicillin. Cells were maintained at 37 °C 5% CO2.
Quantitative real time-polymerase chain reaction: Cells were lysed in Trizol and total RNA was quantitated on a NanoDrop 2000c spectrophotometer. For circ_0003356 and SOCS3, PrimeScript™ RT reagent kits were used for complementary DNA (cDNA) synthesis. For miR-668-3p, a TaqMan miRNA Assay kit was used for cDNA synthesis. cDNA was amplified using BeyoFast™ SYBR Green qPCR Mix. Primer sequences as shown in Table 1 were bought from TaKaRa. Relative mRNA expression of circ_0003356 and SOCS3 were normalised to GAPDH and calculated using the 2-ΔΔCt method. miR-668-3p was normalised to U6.
| Genes | Sequences (5’-3’) | |
| Circ_0003356 | Forward | CCAAGCTTGAAGACGGCAAA |
| Reverse | TCATGGGGCTTCACCTTGAC | |
| MiR-668-3p | Forward | TGTCACTCGGCTCGG |
| Reverse | TGCGTGTCGTGGAGTC | |
| SOCS3 | Forward | CCTGCGCCTCAAGACCTTC |
| Reverse | GTCACTGCGCTCCAGTAGAA | |
| U6 | Forward | GCTTCGGCAGCACATATACTAAAAT |
| Reverse | CAGTGCGTGTCGTGGAGT | |
| GAPDH | Forward | GGAGATTGTTGCCATCAACG |
| Reverse | TTGGTGGTGCAGGATGCATT |
The empty pcDNA3.1 vector, circ_0003356 overexpression vector (pcDNA3.1-circ_0003356), miR-668-3p inhibitor, negative control (NC) inhibitor, miR-668-3p mimic, NC mimic, small interfering RNA (siRNA) against SOCS3 (si-SOCS3) and si-NC were bought from GenePharma (Shanghai, China). Above factors were then transfected into AGS and HGC-27 cells using Lipofectamine 3000 (Invitrogen, Carlsbad, California, United States) for 48 h.
CCK-8 assay: The proliferation of HGC-27 and AGS cells was evaluated via a cell counting kit-8 (Dojindo, Osaka, Japan). HGC-27 and AGS cells (1 × 104 cells/well) were plated in 96-well plates for incubating 24 h, 48 h, 72 h and 96 h. After that, each well was treated with CCK-8 reagent (10 μL) for another 1 h. Ultimately, a microplate reader (Bio-Rad, CA, United States) was used for measurement of optical density at 450 nm.
EdU assays: A Cell-Light EdU DNA Cell Proliferation Kit (RiboBio, Guangzhou, China) was used to perform EdU assay. Cells were labeled with 50 μM EdU for 2 h and fixed with 4 % paraformaldehyde (PFA) (4%). Cells were washed in phosphate buffered saline (PBS) containing glycine (2 mg/mL) for 10 min to remove artefacts and permeabilized. Cells were stained with Apollo dye liquor in the dark followed by DAPI. Cells were imaged on a fluorescence microscope.
Flow cytometry analysis: Apoptotic rates were measured by flow cytometry (FACSCalibur, BD, United States). Cells were resuspended in 300 μL binding buffer and incubated with + Annexin V-FITC (5 μL) for 10 min. Cells were labeled with propidium iodide (5 μL) in the dark for 5 min. Apoptotic cells were quantified using FlowJo V10 software.
Wound healing assay: HGC-27 and AGS cells (5 × 104 cells/well) were put to 6-well plates coated with extracellular matrix molecule, which were cultured in RPMI1640 including 10% FBS. When the cell monolayer was formed, a sterile pipette tip was utilized for scratching the monolayer and scratch width was recorded under an inverted microscope (TE2000; Nikon, Tokyo, Japan). After being washed via PBS, FBS-free medium was added to culture cells for 24 h, followed by record of scratch wound. At last, the migration rate was calculated according to the following formula: (1 - 24 h scratch width/0 h scratch width) × 100.
Transwell assay: Matrigel-coated transwell chambers (8 μm; BD Biosciences, Franklin Lakes, NJ, United States) were utilized for evaluating invasion of HGC-27 and AGS cells. In detail, 3 × 103 cells were re-suspended in FBS-free RPMI1640 medium (200 μL), which were appended to the upper chamber. In contrast, RPMI1640 medium containing 10% FBS was appended to bottom chambers. Following 24 h of incubation, cells in upper chambers were removed via a cotton swab. Cells in bottom chambers were fixed via 4% PFA and stained via 0.1% crystal violet. At last, the number of these cells was counted through inverted fluorescence microscopy (Leica Microsystems GmbH, Wetzlar, Germany).
Cytoplasmic and nuclear fractions: AGS and HGC-27 cells were lysed on ice in lysis buffer + protease inhibitors and centrifuged for 3 min. The subsequent pellets and supernatants were harvested as nuclear and cytoplasmic fractions. RNA was extracted from each fraction using Buffer SK. Quantitative real-time polymerase chain reaction (qRT-PCR) analysis was performed for the detection of circ_0003356 expression. U6 and 18S rRNA were used as controls of cytoplasmic and nuclear transcripts, respectively.
Fluorescence in situ hybridization (FISH): Fluorescence in situ hybridization (FISH) was performed using specific probes against circ_0003356 and miR-668-3p. Briefly, cells were fixed in 4% PFA for 15 min and washed with a 70%, 95%, and 100% gradient of ethanol. Cell hybridization was performed at 37 °C in a dark incubator. Cells were blocked in 3% normal goat serum and 1% bovine serum albumin for 1 h. Cells were cultured with HRP-conjugated anti-biotin antibodies at 4 °C for 1 d. Cells were imaged on a fluorescence microscope.
Dual luciferase reporter assay: The online database circinteractome and StarBase v2.0 were used to identify predicted targets of circ_0003356 and miR-668-3p. The interaction of circ_0003356 and miR-668-3p or SOCS3 and miR-668-3p were verified via dual luciferase reporter (DLR) assays. Briefly, the 3’-UTR sequences of WT circ_0003356 or SOCS3 comprising the miR-668-3p binding sites were cloned into pRL-CMV. 3’-UTR sequences of circ_0003356 or SOCS3 containing mutant sites for miR-668-3p were also generated. MiR-668-3p mimic or NC mimic were transfected into HGC-27 and AGS cells and luciferase activity was measured 2 d post-transfection.
RNA binding protein immunoprecipitation assay: Cells were lysed in RNA binding protein immunoprecipitation (RIP) buffer and magnetic beads conjugated with anti-Ago2 and anti-immunoglobulin G (IgG) were added to cell lysates. MiR-668-3p and circ_0003356 levels in the beads were analyzed.
RNA pull-down assays: RNA pull-downs were performed using commercial Magnetic RNA-Protein Pull-Down Kits (Pierce). Cells were transfected with 3’-biotinylated miR-668-3p (Bio-miR-668-3p) or Bio-NC mimics. After 2 d, cells were lysed cells and streptavidin-coupled beads added to pull-down biotin-coupled RNA complexes. Circ_0003356 enrichment in the bound fractions was subsequently assayed.
Western blot analysis: Cells were lysed in RIPA buffer and protein content assessed via BCA assays. A total of 20 μg of protein was resolved on 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to on polyvinylidene difluoride membranes. Membranes were blocked in non-fat milk for 1 h and probed with anti-N-cadherin (1:6000, ab76011), anti-E-cadherin (1:10000, ab40772), anti-SOCS3 (1:1000, ab280884) and β-actin (1:1000, ab8224) primary antibodies at 4 °C for 24 h. Cells were subsequently labeled with anti-rabbit secondary antibodies (1:2000, ab205718) and proteins were visualized using chemiluminescence.
Establishment of xenograft tumor models: Mice (4-5 wk-old; BALB/c nude; male) were housed at room temperature with 50% humidity and 12 h light/dark cycle. All experiments were approved by the Animal Care and Use Committee of Zhongshan City People’s Hospital (K2017-182).
Mice were injected into the upper back with 1 × 107 AGS cells transfected with circ_0003356 or control vector[26]. Mice were assigned into 2 groups (n = 6 mice/group): Thecirc_0003356 group and the vector group. After injection for five weeks, mice were anesthetized with pentobarbital sodium and sacrificed via dislocation of cervical vertebrae. Tumors were dissected and tumor volumes calculated (length × width2)/2.
Immunohistochemistry staining: Mice tumor tissues were fixed in formalin and paraffinized. Tumors were cut into 4 μm sections, dewaxed with xylene, and hydrated in ethanol. Sections were boiled in citrate buffer (10 mmol/L) and cultured using H2O2 solution (3%) to inhibit endogenous peroxidase activity. Sections were probed with anti-Ki67 (1:200, ab16667), anti-SOCS3 (1:1000, ab280884), anti-N-cadherin (1:500, ab76011), and anti-E-cadherin (1:500, ab40772) primary antibodies at 4 °C overnight and labeled with HRP-conjugated secondary antibodies (1:500, ab6112) 30 min. Cells were stained using DAB substrate and counterstained using hematoxylin. Cells were imaged on a light microscope.
TUNEL staining: TUNEL assay was performed using the commercially available in situ Cell Death Detection Kit as per the manufactures recommendations.
GraphPad Prism 8.0 was used for data analysis. Experiments were performed on a minimum of 3 occasions and are shown as the mean ± SD. Survival curves were plotted using the Kaplan-Meier method. Data were compared using a student’s t-test.
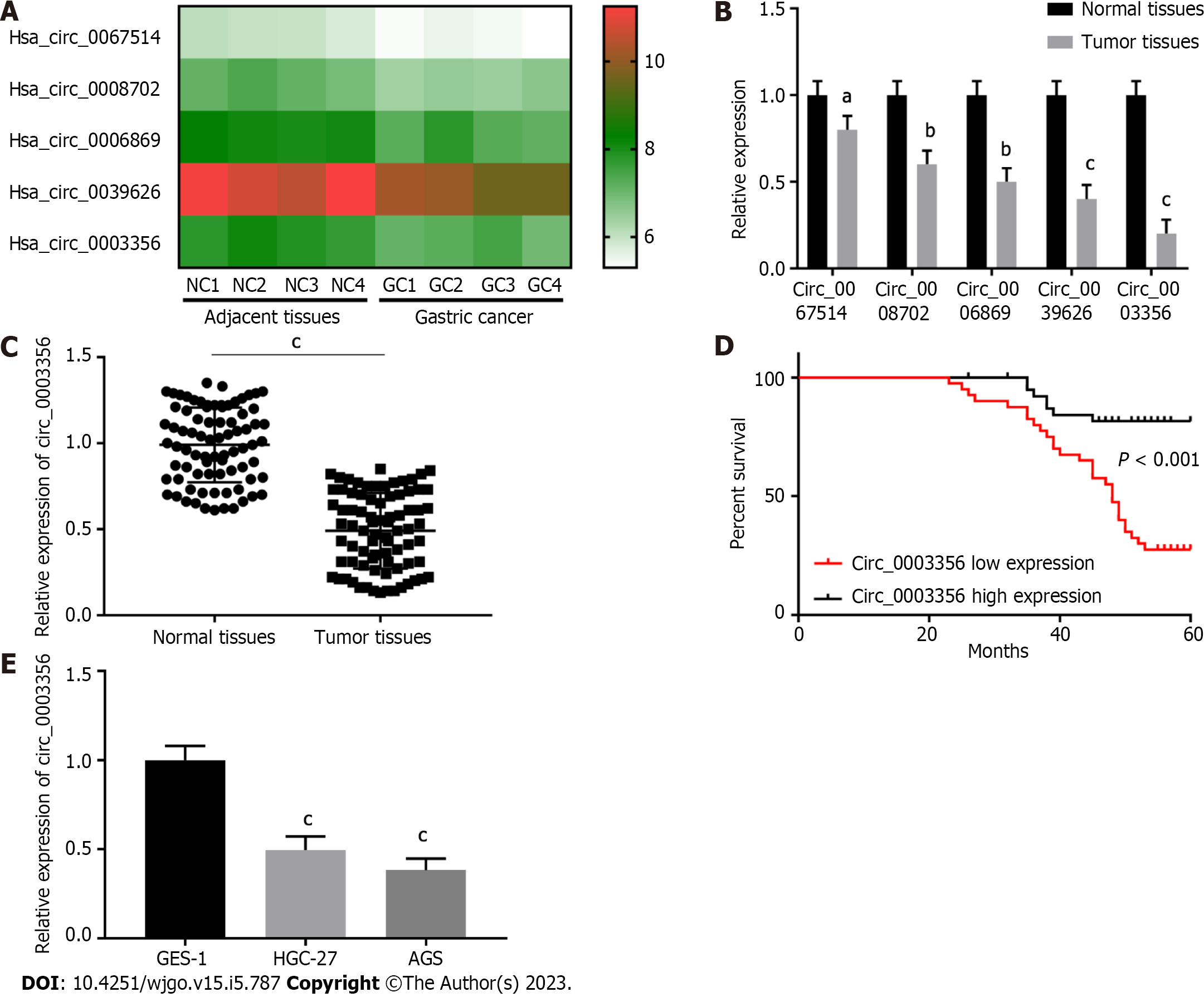
Published GEO and GSE184882 datasets were analyzed[27,28]. Five circRNAs with significant expression differences in GC tissues were identified and selected for heat map analysis (Figure 1A). Then their expression was further determined via qRT-PCR. Five circRNAs with low relative expression in tumor tissues compared to normal tissues were identified (n = 20), amongst which circ_0003356 expression (0.2 ± 0.06532) was the lowest (P = 0.0002, Figure 1B). Circ_0003356 was selected for further analysis.
Circ_0003356 expression was validated in GC normal and tumor tissues (n = 80). As expected, circ_0003356 expression was markedly downregulated in tumor tissues (0.4908 ± 0.02457) compared to normal adjacent tissues (0.99 ± 0.0243; P < 0.0001, Figure 1C). Patients with low circ_0003356 expression correlated with a short survival time (P < 0.0001, Figure 1D). Moreover, circ_0003356 displayed lower expression in AGS (0.3833 ± 0.06012) and HGC-27 cells (0.4967 ± 0.06012) than GES-1 cells (P = 0.0004, P = 0.0001, Figure 1E).
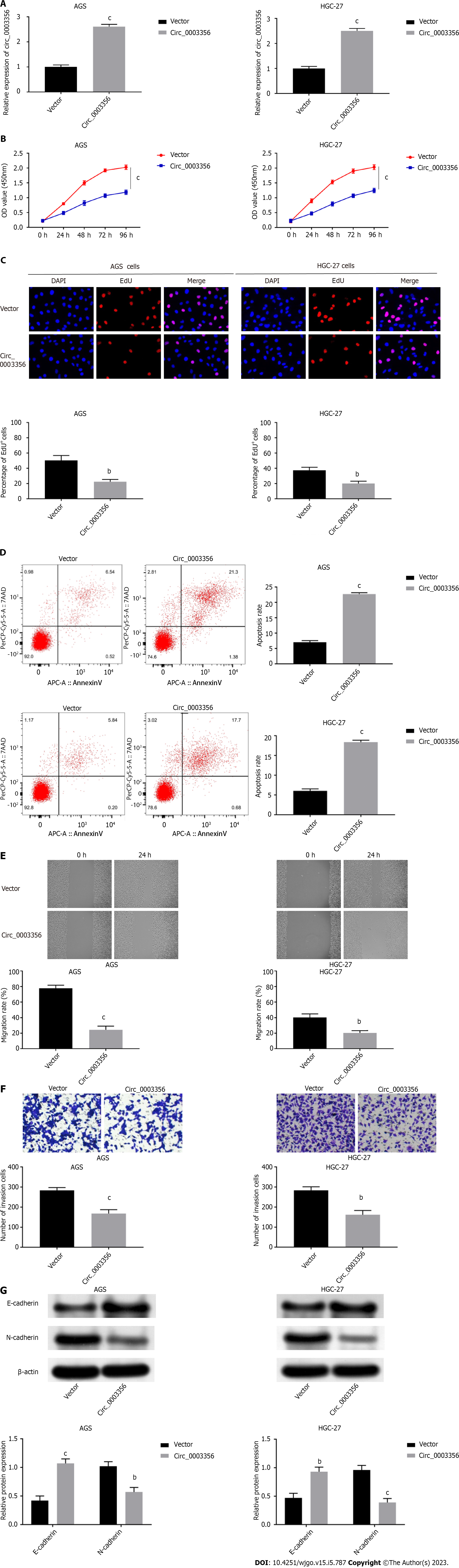
Circ_0003356 was overexpressed in AGS and HGC-27 cells to further explore its influence on GC cell proliferation, apoptosis, migration, invasion, and EMT. Circ_0003356 expression in AGS cells (2.613 ± 0.05207) and HGC-27 cells (2.5 ± 0.05774) was dramatically increased following transfection of pcDNA3.1-circ_0003356 (P < 0.0001, P < 0.0001, Figure 2A). Functional analyses showed that the viability of AGS (1.183 ± 0.06155) and HGC-27 cells (1.247 ± 0.0636) at 96 h were attenuated after circ_0003356 overexpression (P = 0.0002, P = 0.0002, Figure 2B). EdU assays also revealed that the proliferative ability of AGS (22.33 ± 1.764) and HGC-27 cells (20 ± 1.732) was attenuated by circ_0003356 overexpression (P = 0.0025, P = 0.0043, Figure 2C). Apoptotic rates in AGS (22.68 ± 0.2887) and HGC-27 cells’ (18.38 ± 0.2887) were increased by circ_0003356 overexpression (P < 0.0001, P < 0.0001, Figure 2D). In terms of the migration and invasion of AGS (24.33 ± 2.728) and HGC-27 cells (20.33 ± 1.764), decreased numbers of invading AGS (168 ± 11.27) and HGC-27 cells (161.7 ± 12.35) were observed in response to circ_0003356 overexpression (P = 0.0001, P = 0.0031, P = 0.0002, P = 0.0016, Figures 2E and F). Circ_0003356 overexpression also attenuated the EMT of AGS (1.07 ± 0.6531, 0.57 ± 0.5168) and HGC-27 (0.93 ± 0.06533, 0.39 ± 0.06137) cells evidenced by reduced N-cadherin expression and the enhanced expression of E-cadherin (P = 0.0006, P = 0.0023, P = 0.0021, P = 0.0007, Figure 2G).
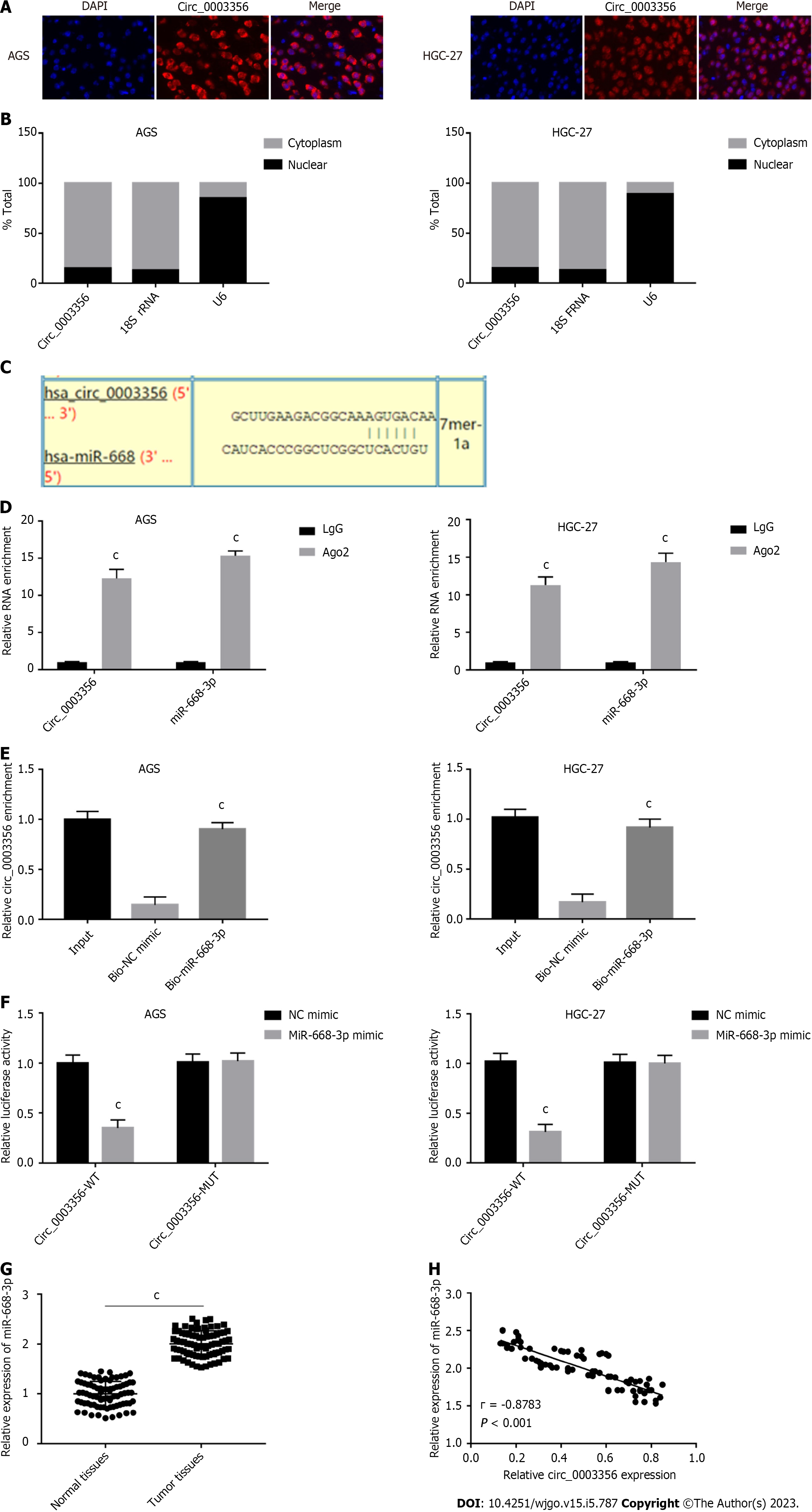
The subvocalization of circ_0003356 in AGS and HGC-27 cells was confirmed through FISH, which showed that circ_0003356 located in AGS and HGC-27 cells’ cytoplasm (Figure 3A). The findings were confirmed by qRT-PCR, showing that the circ_0003356 level was in AGS and HGC-27 cells markedly higher in the cytoplasm (85%) than the nuclear (15%) (Figure 3B).
Target predictions were performed using circinteractome, indentifing binding sites between miR-668-3p and circ_0003356 (Figure 3C). We verified this relationship via RIP-qRT-PCR, RNA pull-down, and DLR assays. The Ago2 group showed enrichment for circ_0003356 and miR-668-3p in AGS (12.3 ± 0.7024, 15.33 ± 0.3801) and HGC-27 cells (11.3 ± 0.6272, 14.33 ± 0.6912) compared to the IgG group (P = 0.000087, P = 0.0000029, P = 0.00008, P = 0.00004, Figure 3D). The biotinylated miR-668-3p probe enriched more circ_0003356 than the biotinylated NC probe in AGS (0.9033 ± 0.03756) and HGC-27 cells (0.9167 ± 0.0491) (P = 0.0002, P = 0.0004, Figure 3E). Relative luciferase activity in AGS (0.35 ± 0.06532) and HGC-27 (0.31 ± 0.06478) were reduced in cells co-transfected with miR-668-3p mimic and circ_0003356 WT relative to miR-668-3p mimic and circ_0003356 MUT (P = 0.00057, P = 0.00041, Figure 3F). High expression of miR-668-3p was observed in tumor tissues (2.006 ± 0.02872) compared to healthy tissues (0.9984 ± 0.02829) (P < 0.0001, Figure 3G). An inverse relationship between the expression of miR-668-3p and circ_0003356 in tumor tissues was also observed (r = -0.8783, P = 0.0007, Figure 3H).
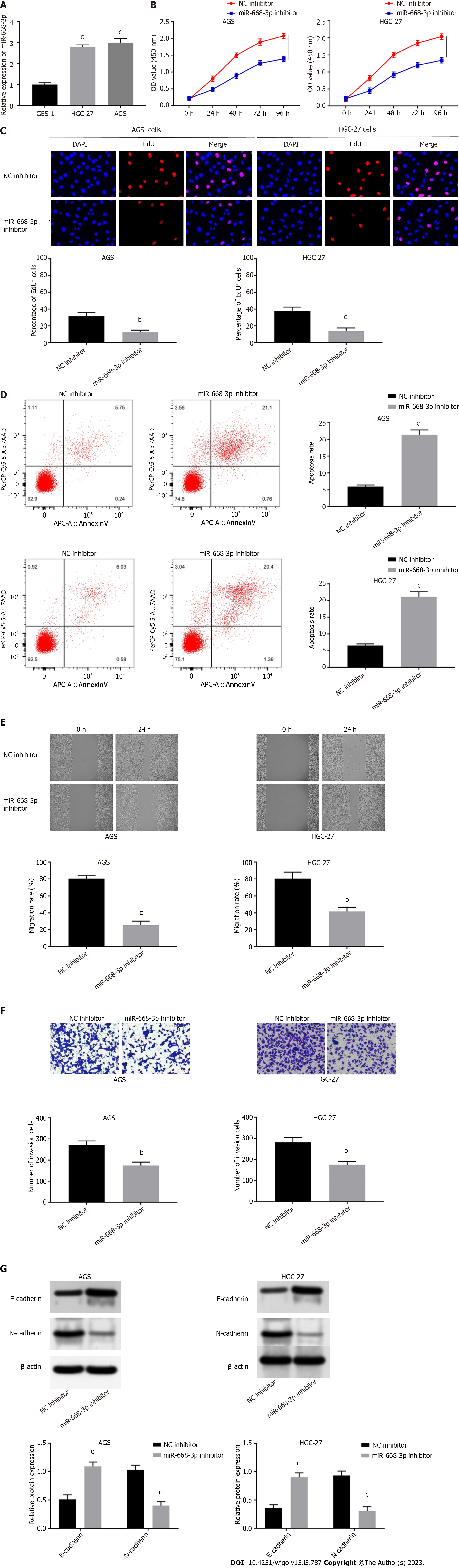
Accordingly, the influences of miR-668-3p on cell migration, apoptosis, proliferation, invasion, and EMT were explored. Firstly, miR-668-3p expression in HGC-27 (2.8 ± 0.05508) and AGS (3 ± 0.1155) cells was found to be remarkably higher than GES-1 cells (1.007 ± 0.05207) (P < 0.0001, P < 0.0001, Figure 4A). When we determined its specific functions, we found that the viability (1.393 ± 0.06128, 1.342 ± 0.0636) and proliferative capacity (12.33 ± 1.453, 14 ± 2.082) of AGS and HGC-27 cells were attenuated by miR-668-3p suppression (P = 0.00037, P = 0.00039, P = 0.0033, P = 0.0020, Figures 4B and C). Apototic rates in AGS (21.29 ± 0.8838) and HGC-27 cells’ (21.09 ± 0.9074) were raised following miR-668-3p suppression (P < 0.0001, P = 0.0001, Figure 4D). The migration (25.67 ± 2.603, 41.67 ± 2.906 ) and invasion (174.7 ± 9.262, 175.7 ± 9.025) of AGS and HGC-27 cells were also repressed following miR-668-3p down-regulation (P = 0.0001, P = 0.0019, P = 0.0026, P = 0.0023, Figures 4E and F). The EMT of AGS (1.09 ± 0.06348, 0.4 ± 0.06137) and HGC-27 cells (0.9 ± 0.05627, 0.31 ± 0.06137) was similarly attenuated by miR-668-3p suppression, evidenced by decreased N-cadherin and increased E-cadherin expression (P = 0.00089, P = 0.00051, P = 0.00066, P = 0.00054, Figure 4G).
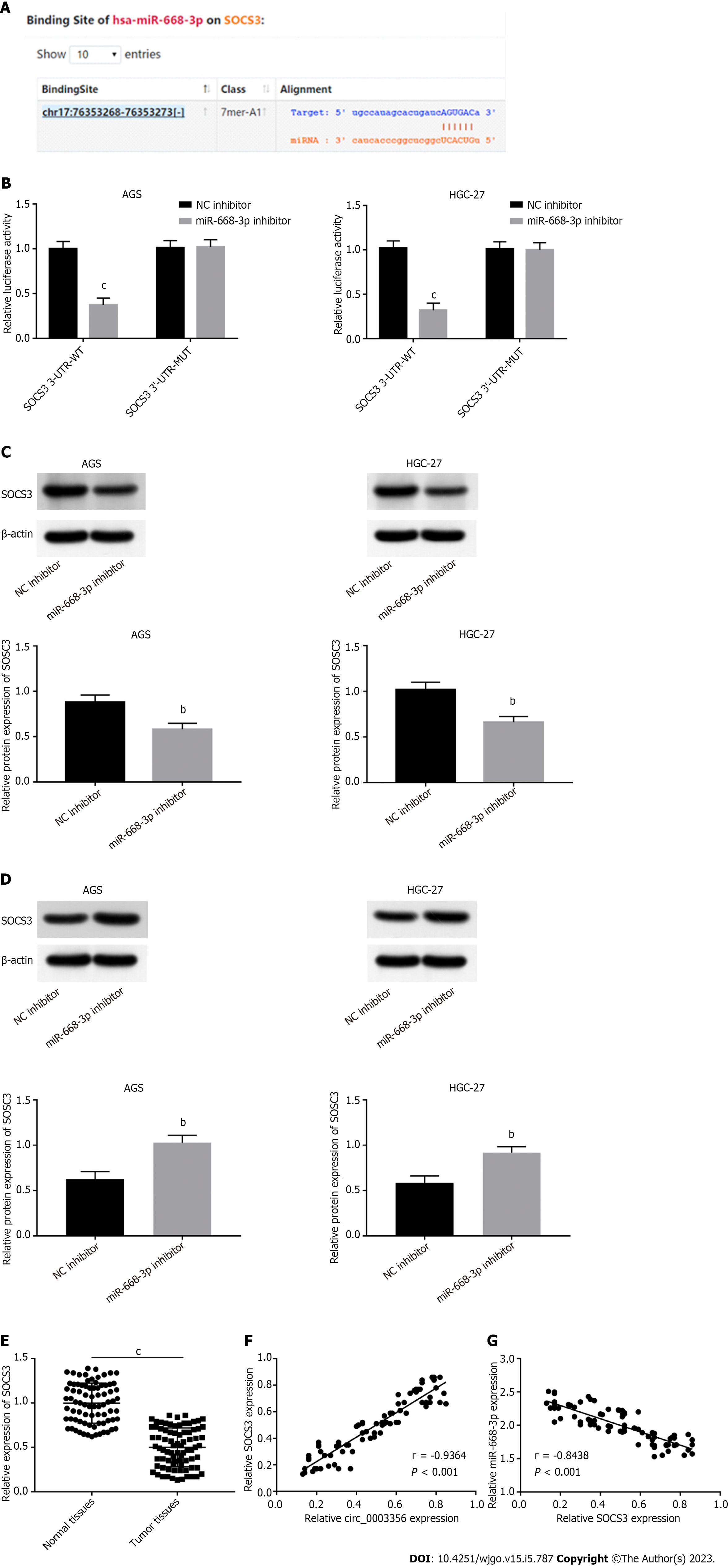
MiR-668-3p targets were assessed using the starbase2.0 database. SOCS3 contained binding sequences (AGUGAC) of miR-668-3p (Figure 5A). DLR assay was performed to validate this interaction. Relative luciferase activity in AGS (0.37 ± 0.06532) and HGC-27 cells (0.32 ± 0.06532) transfected with SOCS3 WT reporter was reduced in response to transfection of miR-668-3p mimic (P = 0.00041, P = 0.00065, Figure 5B). The SOCS3 MUT reporter showed no such changes following miR-668-3p mimic addition (Figure 5B), suggestive of a direct interaction between miR-668-3p and SOCS3. The regulatory relation between miR-668-3p and SOCS3 was assessed through western blotting. SOCS3 expression in AGS and HGC-27 cells was reduced through the addition of miR-668-3p mimic (0.5833 ± 0.03756, 0.6633 ± 0.03528) and boosted by the miR-668-3p inhibitor (1.03 ± 0.04619, 0.92 ± 0.03786), implying an inverse regulatory relationship between miR-668-3p and SOCS3 (P = 0.0076, P = 0.0036; P = 0.0038, P = 0.0047, Figures 5C and D). Lower expression of SOCS3 in tumor tissues (0.5001 ± 0.02447) compared to normal tissues (0.9979 ± 0.02526) (P < 0.0001, Figure 5E) was also observed. Upon correlation analysis between circ_0003356 and SOCS3 expression (P < 0.0001, r = 0.9364; Figure 5F), the expression of SOCS3 was negatively associated with miR-668-3p in tumor tissues (P < 0.0001, r = -0.8438; Figure 5G).
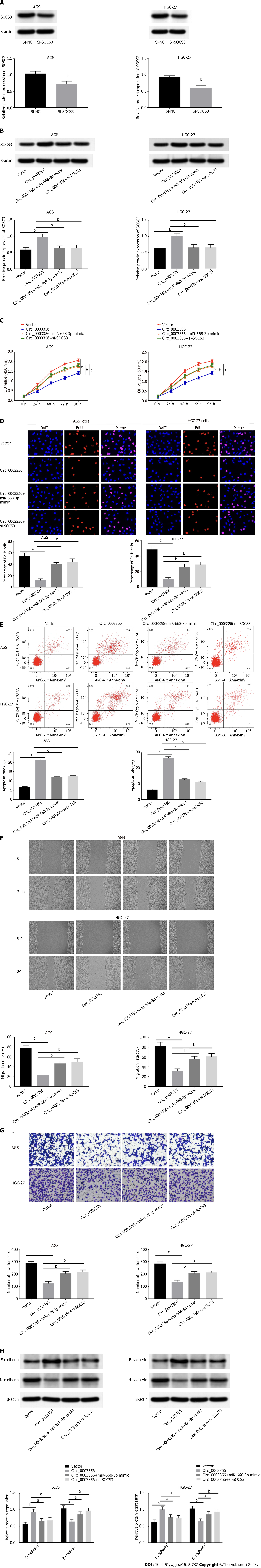
SOCS3 was silenced via transfection of si-SOCS3 and its reduced expression confirmed in AGS (0.7267 ± 0.05044) and HGC-27 cells (0.5933 ± 0.05207) (P = 0.0086, P = 0.0065, Figure 6A). miR-668-3p overexpression (0.6467 ± 0.06394, 0.6633 ± 0.04322 ) or SOCS3 downregulation (0.6433 ± 0.06394, 0.6267 ± 0.04527) reversed the promoting effect of circ_0003356 on SOCS3 expression level in AGS and HGC-27 cells (P = 0.0012, P = 0.0032, P = 0.0030, P = 0.0022, P = 0.0031, P = 0.0031, Figure 6B). The viability and proliferative capacity of AGS and HGC-27 cells were attenuated by circ_0003356 overexpression, whilst miR-668-3p upregulation or SOCS3 downregulation recovered the circ_0003356-mediated decrease in cell viability and proliferative capacity (P = 0.00039, P = 0.0025, P = 0.0037, P = 0.00042, P = 0.0035, P = 0.0051; P < 0.0001, P = 0.0002, P < 0.0001, P < 0.0001, P = 0.0044, P = 0.0012; Figures 6C and D). The effects of circ_0003356 on the apoptotic rates of AGS and HGC-27 cells were similarly reversed by miR-668-3p upregulation or SOCS3 downregulation (P < 0.0001, Figure 6E). When it came to circ_0003356-mediated inhibition of migration, invasion, and EMT, miR-668-3p up-regulation or SOCS3 down-regulation could partially eliminate them (P < 0.0001, P = 0.0017, P = 0.0007, P < 0.0001, P = 0.0039, P = 0.0011; P < 0.0001, P = 0.0009, P = 0.0004, P < 0.0001, P = 0.0006, P = 0.0003; P = 0.0028, P = 0.010, P = 0.0164, P = 0.0029, P = 0.023, P = 0.0278; P = 0.00314, P = 0.0213, P = 0.0144, P = 0.00285, P = 0.0267, P = 0.00914; Figures 6F-H).
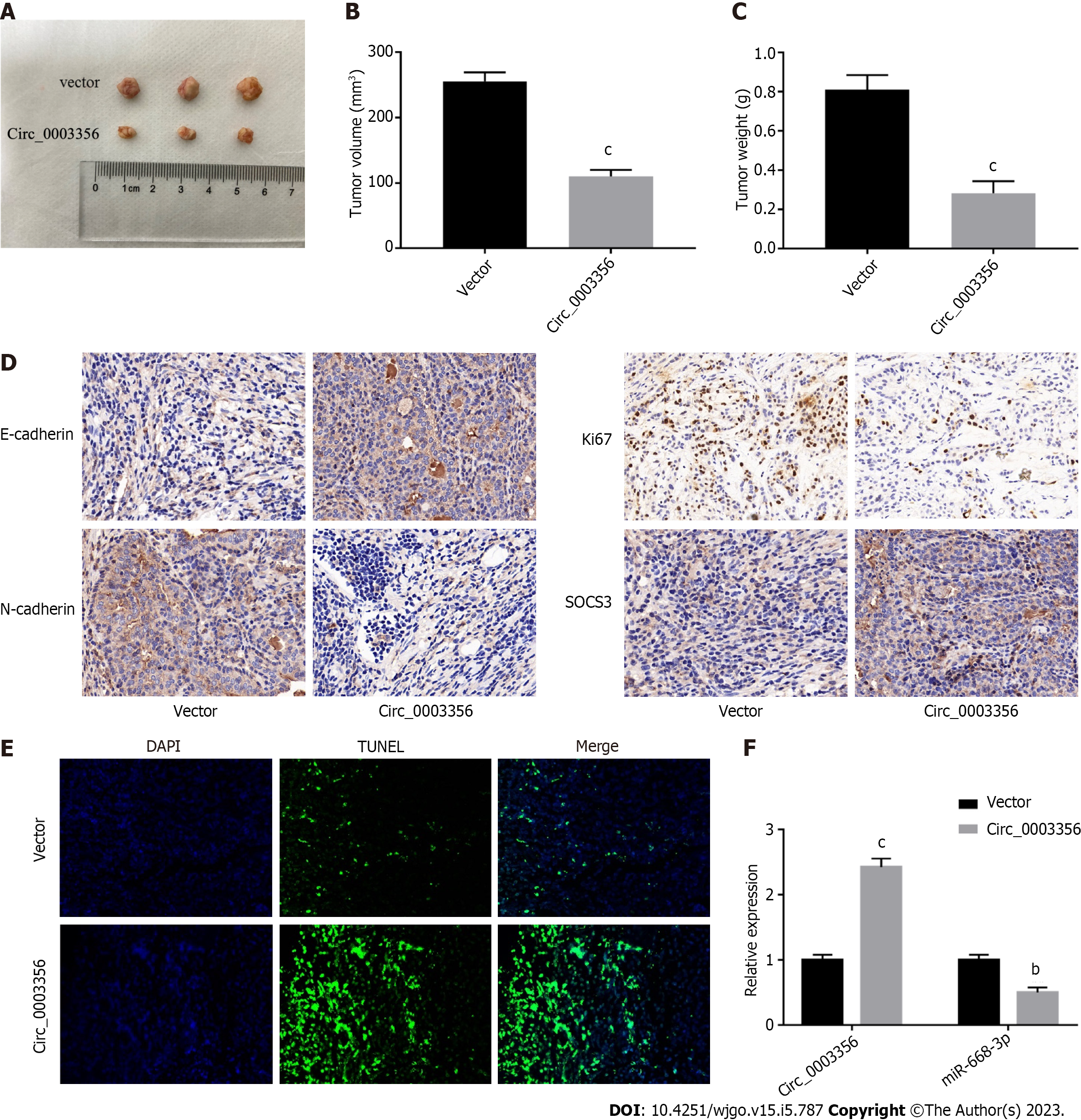
A xenograft mouse model was constructed to confirm the anti-tumor role of circ_0003356 in GC. As depicted in Figures 7A-C, tumor size, volume (110 ± 5.859, P = 0.0001), and weight (0.2833 ± 0.03528, P = 0.0007) were reduced when circ_0003356 was overexpressed. Decreased expressions of Ki67 (proliferative marker protein) and N-cadherin were observed following circ_0003356 overexpression, while SOCS3 and E-cadherin expression increased, suggestive of inhibitory effects on cell proliferation and EMT (Figure 7D). TUNEL staining indicated that circ_0003356 upregulation increased apoptosis in tumor tissues of mice (Figure 7E). Circ_0003356 expression was found to be boosted (2.423 ± 0.09135) and expression of miR-668-3p (0.5 ± 0.06531) was diminished after circ_0003356 was overexpressed in mouse tumor tissues (P = 0.000099, P = 0.0016, Figure 7F).
Due to a lack of effective treatment strategies and prognostic indicators, GC remains a threat to human health[29,30]. Emerging evidence has implicated circRNAs during GC development[31,32]. Thence, great efforts should be made to comprehensively understand circRNAs. To date, many circRNAs show low expression in GC, such as circ_0021087[12], hsa_circ_0005556[33], and circPSMC3[34]. Here, we observed low expression levels of circ_0003356 in GC cells and tissues. This is the first report to show an association of circ_0003356 with GC. Moreover, increasing findings have indicated that several circRNAs act as prognostic indicators for GC, including hsa_circ_0005556[33] and circ_0000260[35]. Here, based on Kaplan-Meier analysis, patients with low circ_0003356 had poor survival rates.
Accumulating data reveal that circRNAs have an anti-tumor function in GC. For instance, circ_0021087 overexpression suppresses the proliferation, migration, invasion, and EMT of GC cells and impairs tumorigenesis in xenograft mice[12]. CircPSMC3 up-regulation prevents GC cells from migrating, proliferating, and invading, thereby suppressing tumorigenesis in xenograft mice[36]. Similarly, we found that circ_0003356 overexpression repressed proliferation, migration, invasion, and EMT of GC cells and enhanced GC cell apoptosis. In vivo, circ_0003356 overexpression suppressed tumorigenesis. These results suggest an anti-tumor role for circ_0003356’s in GC, highlighting its potential in the clinical management of GC.
To our knowledge, circRNAs affect gene expression via sponging downstream miRNAs in cancer cell[37,38]. Inspired by this, we predicted downstream miRNAs of circ_0003356 using circinteractome. MiR-668-3p was identified as one such target. We performed RIP-qRT-PCR, RNA pull-downs, and DLR assays to verify our prediction. In previous reports, miR-668-3p has shown high expression and carcinogenesis in HCC and CRC[18,19]. Consistent with these studies, we discovered high miR-668-3p expression in GC tissues and cells. The down-regulation of miR-668-3p enhanced cell apoptosis and impaired the proliferation, migration, invasion, and EMT of cells (AGS and HGC-27). These outcomes suggest that miR-668-3p is oncogenic in GC. Several cirRNAs, including circ_0014717 and circTMEM59, target miR-668-3p to suppress tumor development[18,19]. Here, an inverse relationship was observed between miR-668-3p and circ_0003356 expression in tumor tissues of GC patients. MiR-668-3p was negatively regulated by circ_0003356 in mouse tumor tissues. In particular, the inhibitory influence of circ_0003356 on the proliferation, migration, invasion. EMT of GC cells were reversed by miR-3619-5p overexpression. We deduced that circ_0003356 prevents GC malignancy through sponging miR-668-3p.
It is acknowledged that the miRNA-circRNA-mRNA network is closely related to the regulation of GC progression[39]. We therefore predicted target genes of miR-668-3p and found that SOCS3 was targeted by miR-668-3p. SOCS3 has been shown to display low expression in GC tissues and to participate in tumor suppression in GC[24,40]. In this study, we observed that the downregulation of SOCS3 expression in GC tissues was regulated by miR-668-3p. An inverse correlation between SOCS3 and miR-668-3p expression levels in tumor tissues of GC patients was observed. Thus, we deduced that miR-668-3p acts as a tumor promoter, targeting SOCS3 in GC. Additionally, we identified a positive relationship between SOCS3 and circ_0003356 and found that the suppressive effects of circ_0003356 on cell proliferation, migration, invasion, and EMT were reversed by SOCS3 upregulation in GC cells. These findings strongly support the notion that circ_0003356 has an anti-tumor role through sponging miR-668-3p to target SOCS3 in GC.
In summary, we show that circ_0003356 expression is low in GC tissues and cells and positively related to GC patient prognosis. Circ_0003356 up-regulation or miR-668-3p inhibition facilitated cell apoptosis and suppressed proliferation, migration, invasion and EMT in cells (AGS and HGC-27). Circ_0003356 overexpression also prevented tumor growth in mice. Mechanistically, circ_0003356 was found to interact with the miR-668-3p/SOCS3 axis to impair GC development. Overall, this investigation provides a new perspective on the therapeutic targets of GC.
Gastric cancer (GC) is a common malignant tumor with high prevalence and mortality. Circular RNA (circRNA) analysis in GSE184882 has indicated that circ_0003356 shows abnormal expression in GC tissues, but the function of circ_0003356 remains unclear in GC.
To seek the prognostic biomarker and therapeutic target for GC.
In order to explore the function and mechanism of circ_0003356 in GC.
Quantitative real-time polymerase chain reaction was used to detect expression of circ_0003356. The clinical value of circ_0003356 for GC prognosis was evaluated. The role of circ_0003356 in GC was assessed via in vitro and in vivo experiments. The molecular mechanism of circ_0003356 on miR-668-3p/SOCS3 axis was validated.
Circ_0003356 expression was markedly decreased in GC tissues and cells, and it was positively associated with survival time of GC patients. Moreover, functional analyses showed that circ_0003356 overexpression inhibited malignant behaviors of GC cells via targeting the miR-668-3p/SOCS3 axis.
Circ_0003356 is lowly expressed in GC tissues and cells, displaying potential as a prognostic biomarker for GC. Circ_0003356 up-regulation represses the malignancy of GC via targeting the miR-668-3p/SOCS3 axis.
Other mechanisms of circ_0003356 in GC may be probed in future researches, and its application in treatment of GC will be extended.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Shu C, China; Tanabe S, Japan S-Editor: Wang JJ L-Editor: A P-Editor: Zhang XD
| 1. | Curea FG, Hebbar M, Ilie SM, Bacinschi XE, Trifanescu OG, Botnariuc I, Anghel RM. Current Targeted Therapies in HER2-Positive Gastric Adenocarcinoma. Cancer Biother Radiopharm. 2017;32:351-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 2. | Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1150] [Cited by in RCA: 2858] [Article Influence: 571.6] [Reference Citation Analysis (5)] |
| 3. | Amiri M, Janssen F, Kunst AE. The decline in stomach cancer mortality: exploration of future trends in seven European countries. Eur J Epidemiol. 2011;26:23-28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 24] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 4. | Ruiz EF, Torres-Roman JS, Servan SA, Martinez-Herrera JF, Arce-Huamani MA, Carioli G, La Vecchia C. Trends and geographic pattern of stomach cancer mortality in Peru. Cancer Epidemiol. 2019;58:193-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 5. | Sexton RE, Al Hallak MN, Diab M, Azmi AS. Gastric cancer: a comprehensive review of current and future treatment strategies. Cancer Metastasis Rev. 2020;39:1179-1203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 459] [Cited by in RCA: 460] [Article Influence: 92.0] [Reference Citation Analysis (0)] |
| 6. | Hoshi H. Management of Gastric Adenocarcinoma for General Surgeons. Surg Clin North Am. 2020;100:523-534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 7. | Allemani C, Weir HK, Carreira H, Harewood R, Spika D, Wang XS, Bannon F, Ahn JV, Johnson CJ, Bonaventure A, Marcos-Gragera R, Stiller C, Azevedo e Silva G, Chen WQ, Ogunbiyi OJ, Rachet B, Soeberg MJ, You H, Matsuda T, Bielska-Lasota M, Storm H, Tucker TC, Coleman MP; CONCORD Working Group. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet. 2015;385:977-1010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1738] [Cited by in RCA: 1735] [Article Influence: 173.5] [Reference Citation Analysis (0)] |
| 8. | Ang TL, Fock KM. Clinical epidemiology of gastric cancer. Singapore Med J. 2014;55:621-628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 274] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 9. | Fontana E, Smyth EC. Novel targets in the treatment of advanced gastric cancer: a perspective review. Ther Adv Med Oncol. 2016;8:113-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 10. | Wang C, Jiang H, Peng J, Weng D, Zhang Y, Zhou Y, Zhang Q. Circular RNA circ_SKA3 enhances gastric cancer development by targeting miR-520h. Histol Histopathol. 2023;38:317-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 11. | Li P, Xiao W. Circ_0005758 impedes gastric cancer progression through miR-1229-3p/GCNT4 feedback loop. Toxicol In Vitro. 2022;85:105454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 12. | Yu Y, Li H, Wu C, Li J. Circ_0021087 acts as a miR-184 sponge and represses gastric cancer progression by adsorbing miR-184 and elevating FOSB expression. Eur J Clin Invest. 2021;51:e13605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 13. | Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J Cell Physiol. 2019;234:5451-5465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1431] [Cited by in RCA: 1335] [Article Influence: 222.5] [Reference Citation Analysis (0)] |
| 14. | Bartel DP. Metazoan MicroRNAs. Cell. 2018;173:20-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2329] [Cited by in RCA: 2660] [Article Influence: 380.0] [Reference Citation Analysis (0)] |
| 15. | Chen G, Shang J, Li M, Zhang H, Xu H. miR-548 predicts clinical prognosis and functions as a tumor promoter in gastric cancer. Clin Exp Med. 2022;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 16. | Yu K, Zhu H. MiR-762 regulates the activation of PI3K/AKT and Hippo pathways involved in the development of gastric cancer by targeting LZTS1. Am J Transl Res. 2022;14:5050-5058. [PubMed] |
| 17. | Bae WJ, Woo KJ, Ahn JM, Yang CM, Kim YS, Kim S, Lee D. miR-4742-5p promotes invasiveness of gastric cancer via targeting Rab43: An in vitro study. Biochem Biophys Res Commun. 2022;613:180-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 18. | Ma H, Huang C, Huang Q, Li G, Li J, Huang B, Zhong Q, Cao C. Circular RNA circ_0014717 Suppresses Hepatocellular Carcinoma Tumorigenesis Through Regulating miR-668-3p/BTG2 Axis. Front Oncol. 2020;10:592884. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 19. | Feng Y, Wang X, Huang C, Zhang D, Liu T, Zhang C, Zhang Y, Ji D, Tang J, Sun Y. Upregulated circTMEM59 Inhibits Cell Growth and Metastasis by miR-668-3p/ID4 Axis in Colorectal Cancer. Oxid Med Cell Longev. 2022;2022:7242124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 20. | Gao Y, Zhao H, Wang P, Wang J, Zou L. The roles of SOCS3 and STAT3 in bacterial infection and inflammatory diseases. Scand J Immunol. 2018;88:e12727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 94] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 21. | Qin H, Holdbrooks AT, Liu Y, Reynolds SL, Yanagisawa LL, Benveniste EN. SOCS3 deficiency promotes M1 macrophage polarization and inflammation. J Immunol. 2012;189:3439-3448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 366] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 22. | Yu C, Fan Y, Zhang Y, Liu L, Guo G. LINC00893 inhibits the progression of prostate cancer through miR-3173-5p/SOCS3/JAK2/STAT3 pathway. Cancer Cell Int. 2022;22:228. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 23. | Li L, Zhang J, Peng H, Jiang X, Liu Z, Tian H, Hou S, Xie X, Peng Q, Zhou T. Knockdown of miR-92a suppresses the stemness of colorectal cancer cells via mediating SOCS3. Bioengineered. 2022;13:5613-5624. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 24. | Xiao C, Hong H, Yu H, Yuan J, Guo C, Cao H, Li W. MiR-340 affects gastric cancer cell proliferation, cycle, and apoptosis through regulating SOCS3/JAK-STAT signaling pathway. Immunopharmacol Immunotoxicol. 2018;40:278-283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 25. | Tang H, Long Q, Zhuang K, Yan Y, Han K, Guo H, Lu X. miR-665 promotes the progression of gastric adenocarcinoma via elevating FAK activation through targeting SOCS3 and is negatively regulated by lncRNA MEG3. J Cell Physiol. 2020;235:4709-4719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 26. | Luo Z, Rong Z, Zhang J, Zhu Z, Yu Z, Li T, Fu Z, Qiu Z, Huang C. Circular RNA circCCDC9 acts as a miR-6792-3p sponge to suppress the progression of gastric cancer through regulating CAV1 expression. Mol Cancer. 2020;19:86. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 140] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 27. | Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991-D995. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4527] [Cited by in RCA: 6786] [Article Influence: 522.0] [Reference Citation Analysis (0)] |
| 28. | Song J, Yu S, Zhong D, Yang W, Jia Z, Yuan G, Li P, Zhang R, Li Y, Zhong G, Chen Z. The circular RNA hsa_circ_000780 as a potential molecular diagnostic target for gastric cancer. BMC Med Genomics. 2021;14:282. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 29. | Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23:700-713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1159] [Cited by in RCA: 1327] [Article Influence: 120.6] [Reference Citation Analysis (0)] |
| 30. | Johnston FM, Beckman M. Updates on Management of Gastric Cancer. Curr Oncol Rep. 2019;21:67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 316] [Article Influence: 52.7] [Reference Citation Analysis (1)] |
| 31. | Li R, Jiang J, Shi H, Qian H, Zhang X, Xu W. CircRNA: a rising star in gastric cancer. Cell Mol Life Sci. 2020;77:1661-1680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 279] [Article Influence: 55.8] [Reference Citation Analysis (0)] |
| 32. | Wei L, Sun J, Zhang N, Zheng Y, Wang X, Lv L, Liu J, Xu Y, Shen Y, Yang M. Noncoding RNAs in gastric cancer: implications for drug resistance. Mol Cancer. 2020;19:62. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 279] [Cited by in RCA: 336] [Article Influence: 67.2] [Reference Citation Analysis (0)] |
| 33. | Yang L, Yu Y, Yu X, Zhou J, Zhang Z, Ying S, Guo J, Yan Z. Downregulated Expression of hsa_circ_0005556 in Gastric Cancer and Its Clinical Significance. Dis Markers. 2019;2019:2624586. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 34. | Rong D, Lu C, Zhang B, Fu K, Zhao S, Tang W, Cao H. CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR-296-5p. Mol Cancer. 2019;18:25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 147] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 35. | Liu S, Wu M, Peng M. Circ_0000260 Regulates the Development and Deterioration of Gastric Adenocarcinoma with Cisplatin Resistance by Upregulating MMP11 via Targeting MiR-129-5p. Cancer Manag Res. 2020;12:10505-10519. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 36. | Rong D, Lu C, Zhang B, Fu K, Zhao S, Tang W, Cao H. Correction to: CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR-296-5p. Mol Cancer. 2020;19:140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 37. | Li J, Sun D, Pu W, Wang J, Peng Y. Circular RNAs in Cancer: Biogenesis, Function, and Clinical Significance. Trends Cancer. 2020;6:319-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 524] [Cited by in RCA: 475] [Article Influence: 95.0] [Reference Citation Analysis (0)] |
| 38. | Wang KW, Dong M. Role of circular RNAs in gastric cancer: Recent advances and prospects. World J Gastrointest Oncol. 2019;11:459-469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 32] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 39. | Cheng J, Zhuo H, Xu M, Wang L, Xu H, Peng J, Hou J, Lin L, Cai J. Regulatory network of circRNA-miRNA-mRNA contributes to the histological classification and disease progression in gastric cancer. J Transl Med. 2018;16:216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 139] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 40. | Shi Y, Zheng C, Jin Y, Bao B, Wang D, Hou K, Feng J, Tang S, Qu X, Liu Y, Che X, Teng Y. Reduced Expression of METTL3 Promotes Metastasis of Triple-Negative Breast Cancer by m6A Methylation-Mediated COL3A1 Up-Regulation. Front Oncol. 2020;10:1126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 98] [Article Influence: 19.6] [Reference Citation Analysis (0)] |