Published online Jan 15, 2023. doi: 10.4251/wjgo.v15.i1.102
Peer-review started: September 17, 2022
First decision: November 18, 2022
Revised: November 19, 2022
Accepted: December 21, 2022
Article in press: December 21, 2022
Published online: January 15, 2023
Processing time: 114 Days and 20.1 Hours
The multi-target stool DNA test (MT-sDNA) has potential utility in the detection of colorectal cancer (CRC), but validation of its clinical accuracy has been limited in China.
To evaluate the diagnostic performance of MT-sDNA and investigate the combined diagnostic value of alpha-fetoprotein (AFP), carcinoembryonic antigen (CEA), and carbohydrate antigen 199 (CA199) with MT-sDNA in CRC and adenomas.
We evaluated the performance of the MT-sDNA kit based on a hospital clinical trial. In this case-control study, 135 participants from the Affiliated Hospital of Medical School of Ningbo University, including 51 CRC patients, 23 patients with adenomas, and 61 healthy controls were enrolled. We used a risk scoring system to determine the positivity of tests with histological diagnosis or colonoscopy as the reference standard.
The main indices of sensitivity, specificity and accuracy were evaluated. The sensitivity and specificity for CRC detection were 90.2% and 83.3%, respectively, with an accuracy of 89.8%. For adenoma, the sensitivity and specificity were 56.5% and 68.9%, respectively, with an accuracy of 73.1%. The sensitivity and specificity of MT-sDNA combined with CEA in the diagnosis of adenoma were 78.3% and 60.7%, respectively.
The MT-sDNA test showed better performance in the detection of CRC, which was superior to AFP, CEA, and CA199 separately, but not for predicting adenomas. The combination of MT-sDNA with CEA further improved the sensitivity for adenoma diagnosis.
Core Tip: The sensitivity and specificity for colorectal cancer (CRC) detection were 90.2% and 83.3%, respectively, with an accuracy of 89.8%. For adenoma, the sensitivity and specificity were 56.5% and 68.9%, respectively, with an accuracy of 73.1%. The multi-target stool DNA (MT-sDNA) test showed better performance for the detection of CRC, which was superior to alpha-fetoprotein, carcinoembryonic antigen (CEA), and carbohydrate antigen 199 separately, but not for predicting adenomas. The sensitivity and specificity of MT-sDNA combined with CEA in the diagnosis of adenoma were 78.3% and 60.7%, respectively, which suggested that combined detection has certain advantages in adenoma diagnosis. This study can help clinicians select a standardized and optimal management strategy for the treatment of these patients.
- Citation: Gao HL, Lv LB, Zhao WF, Lu QW, Fan JQ. Diagnostic accuracy of the multi-target stool DNA test in detecting colorectal cancer: A hospital-based study. World J Gastrointest Oncol 2023; 15(1): 102-111
- URL: https://www.wjgnet.com/1948-5204/full/v15/i1/102.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i1.102
Colorectal cancer (CRC) is the third most common cancer in terms of incidence rate and the fifth-leading cause of cancer-related deaths in China[1]. In the USA, age-standardized mortality and incidence rates of CRC have recently significantly decreased[2]. Several screening tests, including colonoscopy and the fecal occult blood test (FOBT), are currently used in CRC detection[3]. In addition, tumor markers such as alpha-fetoprotein (AFP), carbohydrate antigen 199 (CA199) and carcinoembryonic antigen (CEA) are common indices used in the diagnosis of CRC[4]. Colonoscopy is unlikely to potentially increase screening rates due to its invasive nature and inconvenience for patients[5]. The FOBT, CA199 and CEA, the most widely used noninvasive tools in CRC screening, lack diagnostic accuracy[6]. In light of this situation, new methods for CRC screening and diagnosis are required[7]. The multi-target stool DNA (MT-sDNA) test was added as a recommended CRC screening option in the 2016 US Preventive Services Task Force and 2018 American Cancer Society guidelines[8,9].
Recently, the MT-sDNA test has arrived in the commercial market and has been optimized in terms of improved sensitivity, sample storage and platform analysis[10]. Cologuard®, the only MT-sDNA kit available in the United States, was approved by the US Food and Drug Administration to evaluate 11 biomarkers, such as KRAS gene mutation, methylation markers and hemoglobin[9]. The commercial kit ColoClear® from New Horizon Health (NHH) Technology combines with N-myc downstream-regulated gene 4 (NDRG4) and Bone Morphogenetic Protein 3 (BMP3) methylation, and KRAS mutation has been proved to have good sensitivity and specificity in Hubei, China[11]. However, another Chinese study showed that the MT-sDNA kit may not be suitable for predicting CRC due to decreased specificity[12]. More evidence is needed for the extensive use of the MT-sDNA test in China. Moreover, combination analyses of tumor markers with the MT-sDNA test are still sparse. The goal of this research was to evaluate the accuracy of the MT-sDNA method in the diagnosis of CRC and to compare the diagnostic performance of different tumor markers combined with MT-sDNA, using histological and colonoscopy confirmation as reference methods.
The study was performed in the Affiliated Hospital of Medical School of Ningbo University and approved by the institutional ethics review committee. The approved identifier number is KY20201111. All subjects signed an informed consent and were told the MT-sDNA results. The primary measures of this research, including sensitivity, specificity, and accuracy, were investigated to evaluate the consistency of the commercial kit ColoClear® (NHH Technology) compared with the reference standards of histopathologic or colonoscopy examination.
A total of 135 participants were recruited from January 2020 to March 2021 in the Affiliated Hospital of Medical School of Ningbo University. Participants who visited inpatient or endoscopy centers were eligible for recruitment. The inclusion criteria were: age > 35 years, and a diagnosis of CRC or adenoma. The exclusion criteria were: A previous diagnosis of CRC, inflammatory bowel disease, familial adenomatous polyposis syndrome, other cancers and cognitive impairment. All participants provided informed consent and the study was approved by the Human Research and Ethics Committee of the Affiliated Hospital of Medical School of Ningbo University.
Fecal samples (4-5 g) were collected prior to bowel preparation for colonoscopy examination in patients with colorectal polyps and before surgical removal of intestinal tumor tissue from CRC patients. All experimental procedures related to the MT-sDNA tests [KRAS mutation, NDRG4, BMP3 methylation and Fecal Immunochemical Test (FIT)] were carried out in the laboratory of NHH Technology (Hangzhou, China). The details regarding probes and primers, as well as the risk prediction algorithm were the same as those in a previously published article[13]. In this risk prediction model, a risk score is provided as a single output. If the risk score value was ≥165, the test was considered “positive”. If the risk score was < 165, the test was regarded as “negative”[11]. Three serum biomarkers, CA199, CEA, and AFP levels were determined by the Department of Testing, Affiliated Hospital of Ningbo University School of Medicine.
Histological diagnosis and colonoscopy were the reference criteria for determining the accuracy of the kit for validating screening performance. All pathological diagnoses were in accordance with the diagnostic criteria of the 2010 World Health Organization Classification of Gastrointestinal Neoplasms.
The sensitivity and specificity were analyzed by receiver operating characteristic (ROC) curves with the area under the ROC curve (AUC) and 95%CI calculated for the MT-sDNA test. Statistical analysis was performed using SPSS software (version 23.0, IBM Corp., USA). The t-test and chi-square test were adopted to compare the differences among different groups. P < 0.05 was considered statistically significant.
One patient who did not meet the inclusion criteria was excluded, and 135 subjects were finally included (Figure 1). The basic demographic characteristics of the 135 enrolled patients are summarized in Table 1. The group of patients with CRC, adenoma and normal controls comprised 51, 23 and 61 participants, with an average age and standard deviation of 66.14 ± 9.47, 60.13 ± 12.40 and 54.18 ± 10.30, and a female-to-male ratio of 2.4, 1.88 and 1.03, respectively. The rectum was the most common tumor site (52.94%) in CRC patients. Ulcerative type, medium differentiation and Dukes stage A accounted for 78.43%, 72.55% and 78.43% of CRC, respectively. 95.74% of CRC patients had adenocarcinoma.
| Variable | Case group | Control group | |
| Colorectal cancer | Adenomas | Healthy subjects | |
| Gender | |||
| Female | 36 (70.60) | 15 (65.20) | 31 (51.81) |
| Male | 15 (29.49) | 8 (34.80) | 30 (49.18) |
| Age | |||
| mean ± SD | 66.14 ± 9.47 | 60.13 ± 12.40 | 54.18 ± 10.30 |
| < 60 yr | 16 (31.40) | 10 (43.50) | 42 (68.90) |
| ≥ 60 yr | 35 (68.60) | 13 (56.50) | 19 (31.10) |
| Education level | |||
| Junior high school and below | 42 (82.40) | 14 (60.90) | 16 (26.20) |
| Senior high school and above | 9 (17.60) | 9 (39.10) | 45 (73.80) |
| BMI | |||
| < 23.00 | 31 (60.80) | 12 (52.20) | 21 (34.42) |
| ≥ 23.00 | 20 (39.20) | 11 (47.80) | 40 (65.57) |
| Tumor location | |||
| Colon | 24 (47.06) | - | - |
| Rectum | 27 (52.94) | - | - |
| Pathogenic type | - | - | |
| Protruding type | 7 (13.73) | - | - |
| Infiltrating type | 4 (7.84) | - | - |
| Ulcerative type | 40 (78.43) | - | - |
| Differentiation | - | - | |
| High | 4 (7.84) | - | - |
| Medium | 37 (72.55) | - | - |
| Low | 10 (19.61) | - | - |
| Histological type | - | - | |
| Adenocarcinoma | 49 (95.74) | - | - |
| Other types | 2 (4.26) | - | - |
| Dukes stage | - | - | |
| A | 40 (78.43) | - | - |
| B | 10 (19.61) | - | - |
| C | 1 (1.06) | - | - |
| D | 0 (0.0) | - | - |
As shown in Table 2, the levels of tumor biomarkers AFP, CEA, CA199 and the risk score were elevated in CRC patients compared with healthy controls (P < 0.05). Regarding the tumor biomarkers, the value of CEA was higher in adenoma patients compared with healthy controls, and the risk score was obviously increased in adenoma patients compared with healthy controls, but no significant differences between adenoma patients and healthy controls were observed in terms of AFP and CA199 (P > 0.05).
| Variable | Case group | Control group | P value | ||
| Colorectal cancer | Adenomas | Healthy subjects | CRC patients vs healthy controls | Adenoma patients vs healthy controls | |
| AFP | |||||
| mean ± SD | 5.87 ± 17.59 | 3.03 ± 1.57 | 3.32 ± 1.36 | 0.03 | 0.43 |
| ≤ 7 µg/L | 47 (92.20) | 22 (95.70) | 61 (100.00) | ||
| > 7.1 µg/L | 4 (7.80) | 1 (4.30) | 0 (0.00) | ||
| CEA | |||||
| mean ± SD | 37.12 ± 149.74 | 5.21 ± 3.58 | 2.20 ± 1.58 | 0.00 | 0.00 |
| ≤ 5 µg/L | 34 (66.70) | 11 (47.80) | 56 (91.80) | ||
| > 5.1 µg/L | 17 (33.30) | 12 (52.20) | 5 (8.20) | ||
| CA199 | |||||
| mean ± SD | 63.05 ± 276.78 | 13.39 ± 10.19 | 9.77±8.89 | 0.01 | 0.15 |
| ≤ 25 µ/mL | 41 (80.40) | 18 (78.30) | 59 (96.70) | ||
| > 25.1 µ/mL | 10 (19.60) | 5 (21.70) | 2 (3.30) | ||
| Complex value | |||||
| mean ± SD | 806.54 ± 289.28 | 351.61 ± 369.85 | 105.11 ± 90.95 | 0.00 | 0.00 |
| < 165 | 5 (9.80) | 13 (56.52) | 59 (96.72) | ||
| ≥ 165 | 46 (90.20) | 10 (43.48) | 2 (3.28) | ||
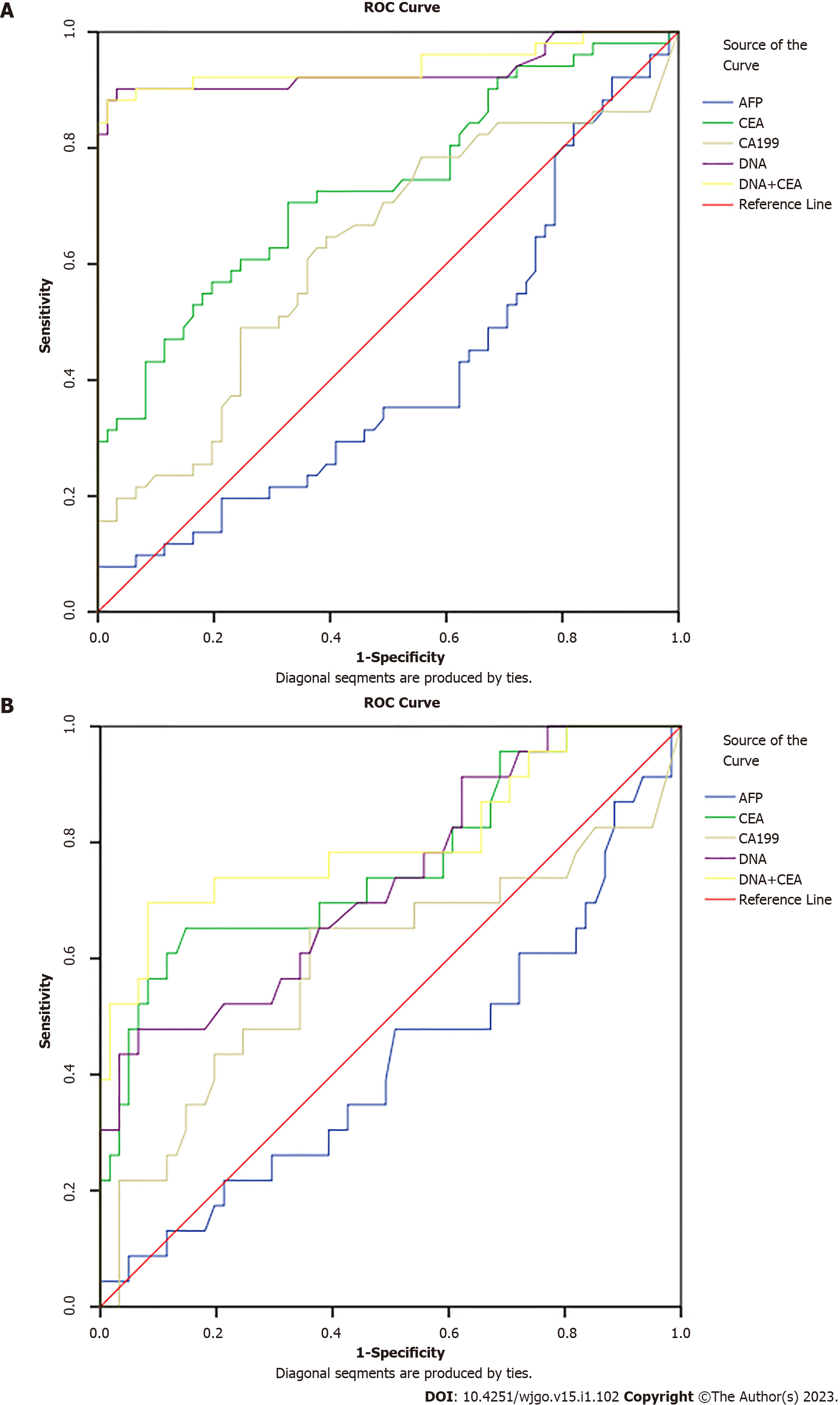
We tested the diagnostic value of MT-sDNA and tumor markers in healthy controls. We found that in CRC, the AUC value, the sensitivity and specificity of MT-sDNA was similar to the combined detection results of MT-sDNA and CEA, and the AUC value was 89.8% (Table 3 and Figure 2A), indicating that there was no significant difference in diagnostic value between the MT-sDNA test and combined test in CRC.
| Detection method | AUC (%) | Sensitivity (%) | Specificity (%) | P value |
| AFP | 41.3 | 35.3 | 48.8 | 0.264 |
| CEA | 73.2 | 60.8 | 63.1 | 0.001 |
| CA199 | 62.5 | 49.0 | 69.0 | 0.069 |
| DNA | 93.3 | 90.2 | 83.3 | 0.000 |
| DNA+CEA | 94.7 | 90.2 | 75.0 | 0.000 |
As shown in Table 4 and Figure 2B, the sensitivity and specificity of MT-sDNA combined with CEA in the diagnosis of adenoma were 78.3% and 60.7%, and the diagnostic accuracy was 80.4%, which was higher than the MT-sDNA and CEA test alone, with an accuracy of 73.1% and 76.1%, respectively (Table 4 and Figure 2B).
| Detection method | AUC (%) | Sensitivity (%) | Specificity (%) | P value |
| AFP | 42.1 | 34.8 | 54.1 | 0.263 |
| CEA | 76.1 | 69.6 | 62.3 | 0.000 |
| CA199 | 59.2 | 56.5 | 65.6 | 0.196 |
| DNA | 73.1 | 56.5 | 68.9 | 0.001 |
| DNA+CEA | 80.4 | 78.3 | 60.7 | 0.000 |
Screening for CRC is crucial as it can improve patient outcome when diagnosed at an early stage[14]. The MT-sDNA test was developed for colorectal screening in recent years[15]. In the present study, we recruited 135 participants who all underwent histological or colonoscopy examination, the MT-sDNA test and tumor biomarker detection. We found that the risk score of MT-sDNA was significantly increased in CRC and adenoma patients compared with healthy controls which potentially makes it a promising non-invasive tumor biomarker for CRC detection[16].
We also found that the diagnostic accuracy, sensitivity and specificity of the risk score were 89.8%, 90.2% and 83.3% for CRC, respectively. The diagnostic sensitivity of MT-sDNA was lower in the present study compared with 92.3% in the United States study[17], possibly due to the younger age of the participants[18].
Similar to other studies, our study demonstrated that the sensitivity of MT-sDNA in the diagnosis of adenoma was low[19], indicating that MT-sDNA is not suitable for the diagnosis of adenoma, although previous studies have shown that the sensitivity of the MT-sDNA test was relatively high for advanced adenomas[20-22]. Previous studies mostly focused on comparing the accuracy of MT-sDNA and FIT detection, whereas no studies have focused on the combination of tumor markers and the MT-sDNA test in the diagnosis of adenoma. We confirmed the diagnostic accuracy of the risk score and tumor biomarkers for adenoma. We noted that, in the detection of adenoma, the accuracy and sensitivity of CEA combined with MT-sDNA increased which suggested that this combination has certain advantages in the diagnosis of adenoma. In addition, compared with MT-sDNA alone, the diagnostic accuracy of CEA combined with MT-sDNA tended to be superior for CRC detection, but there was no increase in sensitivity. This indicated that the combination had little effect on the diagnosis of CRC.
This study has several limitations. One limitation is the small sample size; thus, we did not subdivide adenomas and the accuracy of the results requires further verification. In addition, the relationship between overall survival and the risk score could not be determined due to the limited follow-up time. Therefore, analyses with longer follow-up duration should be conducted.
In summary, the present research found that the risk score of fecal MT-sDNA was increased in CRC and adenoma patients. MT-sDNA has high diagnostic value in the diagnosis of CRC. The combination of MT-sDNA and CEA could improve sensitivity, although the specificity decreased in adenoma detection. Fecal MT-sDNA together with CEA is helpful in diagnosing patients at high-risk of adenoma. This can help clinicians to select a standardized and optimal management strategy for the treatment of these patients.
The multi-target stool DNA test (MT-sDNA) has potential utility in the detection of colorectal cancer (CRC), but validation of its clinical accuracy has been limited in China.
More evidence is needed for the extensive use of the MT-sDNA test in China. Moreover, combination analyses of tumor markers with the MT-sDNA test are still sparse.
The goal of this research was to evaluate the accuracy of the MT-sDNA method in the diagnosis of CRC and to compare the diagnostic performance of different tumor markers combined with MT-sDNA.
In this study, routine clinical test results [alpha-fetoprotein, carcinoembryonic antigen (CEA), and carbohydrate antigen 199] and MT-sDNA test were used as evaluation indexes for the combined diagnosis of colorectal cancer and adenoma, so as to improve the diagnostic performance of this research.
The sensitivity and specificity for CRC detection were 90.2% and 83.3%, respectively, with an accuracy of 89.8%. For adenoma, the sensitivity and specificity were 56.5% and 68.9%, respectively, with an accuracy of 73.1%.
The MT-sDNA test showed better performance for the detection of CRC, which was superior to alpha-fetoprotein, CEA, and carbohydrate antigen 199 separately, but not for predicting adenomas. The sensitivity and specificity of MT-sDNA combined with CEA in the diagnosis of adenoma were 78.3% and 60.7%, respectively, which suggested that combined detection has certain advantages in adenoma diagnosis.
This study can help clinicians select a standardized and optimal management strategy for the treatment of these patients.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Mo X China; Sipahi A, Brazil S-Editor: Liu GL L-Editor: A P-Editor: Liu GL
| 1. | Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4235] [Cited by in RCA: 11419] [Article Influence: 3806.3] [Reference Citation Analysis (4)] |
| 2. | Cardoso R, Guo F, Heisser T, Hackl M, Ihle P, De Schutter H, Van Damme N, Valerianova Z, Atanasov T, Májek O, Mužík J, Nilbert MC, Tybjerg AJ, Innos K, Mägi M, Malila N, Bouvier AM, Bouvier V, Launoy G, Woronoff AS, Cariou M, Robaszkiewicz M, Delafosse P, Poncet F, Katalinic A, Walsh PM, Senore C, Rosso S, Vincerževskienė I, Lemmens VEPP, Elferink MAG, Johannesen TB, Kørner H, Pfeffer F, Bento MJ, Rodrigues J, Alves da Costa F, Miranda A, Zadnik V, Žagar T, Lopez de Munain Marques A, Marcos-Gragera R, Puigdemont M, Galceran J, Carulla M, Chirlaque MD, Ballesta M, Sundquist K, Sundquist J, Weber M, Jordan A, Herrmann C, Mousavi M, Ryzhov A, Hoffmeister M, Brenner H. Colorectal cancer incidence, mortality, and stage distribution in European countries in the colorectal cancer screening era: an international population-based study. Lancet Oncol. 2021;22:1002-1013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 257] [Article Influence: 64.3] [Reference Citation Analysis (0)] |
| 3. | Lu DC, Zhang QF, Li L, Luo XK, Liang B, Lu YH, Hu BL, Jiang HX. Methylated Septin9 has moderate diagnostic value in colorectal cancer detection in Chinese population: a multicenter study. BMC Gastroenterol. 2022;22:232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 4. | Liu T, Li X, Liu D, Liu S, Dong M. Increased serum CA125 II, but not CEA,CA19-9,AFP or CA72-4 in colon cancer compared to rectal cancer. Br J Biomed Sci. 2021;78:218-220. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (1)] |
| 5. | Kanth P, Inadomi JM. Screening and prevention of colorectal cancer. BMJ. 2021;374:n1855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 165] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 6. | Xia C, Dong X, Li H, Cao M, Sun D, He S, Yang F, Yan X, Zhang S, Li N, Chen W. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J (Engl). 2022;135:584-590. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1790] [Cited by in RCA: 2198] [Article Influence: 732.7] [Reference Citation Analysis (1)] |
| 7. | Chan SCH, Liang JQ. Advances in tests for colorectal cancer screening and diagnosis. Expert Rev Mol Diagn. 2022;22:449-460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 42] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 8. | Benson M, Johannes A, Weiss JM, Lucey M, Pier J, Pfau P. Colorectal Cancer Screening After Changes in US Preventive Services Task Force Guidelines With Increased Screening Options. WMJ. 2021;120:127-130. [PubMed] |
| 9. | Anand S, Liang PS. A Practical Overview of the Stool DNA Test for Colorectal Cancer Screening. Clin Transl Gastroenterol. 2022;13:e00464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 17] [Article Influence: 5.7] [Reference Citation Analysis (1)] |
| 10. | Carethers JM. Fecal DNA Testing for Colorectal Cancer Screening. Annu Rev Med. 2020;71:59-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 11. | Xu H, Chen H, Hu J, Xiong Z, Li D, Wang S, Yu J. Feasibility of quantification based on novel evaluation with stool DNA and fecal immunochemical test for colorectal cancer detection. BMC Gastroenterol. 2022;22:384. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 12. | Mu J, Huang Y, Cai S, Li Q, Song Y, Yuan Y, Zhang S, Zheng S. Plausibility of an extensive use of stool DNA test for screening advanced colorectal neoplasia. Clin Chim Acta. 2020;501:42-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 13. | Jin P, You P, Fang J, Kang Q, Gu F, Cai Y, Zhai H, Wang B, Li Y, Xu J, Wang J, He Y, Wang Y, Dai M, Sheng J. Comparison of Performance of Two Stool DNA Tests and a Fecal Immunochemical Test in Detecting Colorectal Neoplasm: A Multicenter Diagnostic Study. Cancer Epidemiol Biomarkers Prev. 2022;31:654-661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (1)] |
| 14. | Ladabaum U, Dominitz JA, Kahi C, Schoen RE. Strategies for Colorectal Cancer Screening. Gastroenterology. 2020;158:418-432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 411] [Article Influence: 82.2] [Reference Citation Analysis (0)] |
| 15. | Bosch LJW, Melotte V, Mongera S, Daenen KLJ, Coupé VMH, van Turenhout ST, Stoop EM, de Wijkerslooth TR, Mulder CJJ, Rausch C, Kuipers EJ, Dekker E, Domanico MJ, Lidgard GP, Berger BM, van Engeland M, Carvalho B, Meijer GA. Multitarget Stool DNA Test Performance in an Average-Risk Colorectal Cancer Screening Population. Am J Gastroenterol. 2019;114:1909-1918. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 69] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 16. | Anderson JC, Robinson CM, Hisey W, Limburg PJ, Butterly LF. Colonoscopy Findings in FIT+ and mt-sDNA+ Patients vs in Colonoscopy-only Patients: New Hampshire Colonoscopy Registry Data. Cancer Prev Res (Phila). 2022;15:455-464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 17. | Finney Rutten LJ, Jacobson RM, Wilson PM, Jacobson DJ, Fan C, Kisiel JB, Sweetser S, Tulledge-Scheitel SM, St Sauver JL. Early Adoption of a Multitarget Stool DNA Test for Colorectal Cancer Screening. Mayo Clin Proc. 2017;92:726-733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 18. | Redwood DG, Asay ED, Blake ID, Sacco PE, Christensen CM, Sacco FD, Tiesinga JJ, Devens ME, Alberts SR, Mahoney DW, Yab TC, Foote PH, Smyrk TC, Provost EM, Ahlquist DA. Stool DNA Testing for Screening Detection of Colorectal Neoplasia in Alaska Native People. Mayo Clin Proc. 2016;91:61-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 79] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 19. | Sun M, Liu J, Hu H, Guo P, Shan Z, Yang H, Wang J, Xiao W, Zhou X. A novel panel of stool-based DNA biomarkers for early screening of colorectal neoplasms in a Chinese population. J Cancer Res Clin Oncol. 2019;145:2423-2432. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 20. | Dolatkhah R, Dastgiri S, Jafarabadi MA, Abdolahi HM, Somi MH. Diagnostic accuracy of multitarget stool DNA testing for colorectal cancer screening: A systematic review and meta-analysis. Gastroenterol Hepatol. 2022;45:753-766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 21. | Zou J, Xiao Z, Wu Y, Yang J, Cui N. Noninvasive fecal testing for colorectal cancer. Clin Chim Acta. 2022;524:123-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 22. | Yang C, Wu W, Yang Y, Yang X, Sun J, Zhang W, Liu K, Ying H, Jiang S, Yu X, Shi Y, Zhou Y, Zhu S, Xu Y, Ding Y, Xie L, Cai B, Xin X, Chen P, Zhao R, Wu Y. Multitarget stool DNA test compared with fecal occult blood test for colorectal cancer screening. Oncol Lett. 2020;20:1193-1200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |