Published online Dec 15, 2021. doi: 10.4251/wjgo.v13.i12.2088
Peer-review started: March 22, 2021
First decision: July 3, 2021
Revised: July 10, 2021
Accepted: September 16, 2021
Article in press: September 16, 2021
Published online: December 15, 2021
Processing time: 267 Days and 17.9 Hours
Pancreaticobiliary cancer (PB Ca) is a lethal disease, and a useful diagnostic marker is urgently needed. A correlation between the human microbiota and malignant gastrointestinal diseases was recently reported.
To investigate the efficacy of the duodenal microbiota for diagnosing PB Ca.
We recruited 22 patients with benign pancreaticobiliary diseases (benign group) and 12 patients with PB Ca (malignant group). The duodenal microbiota of each patient was analyzed by the 16S rDNA terminal restriction fragment length polymorphism method. Patient characteristics, tumor markers, and relative abundances of the duodenal microbiota were compared between the benign and malignant groups.
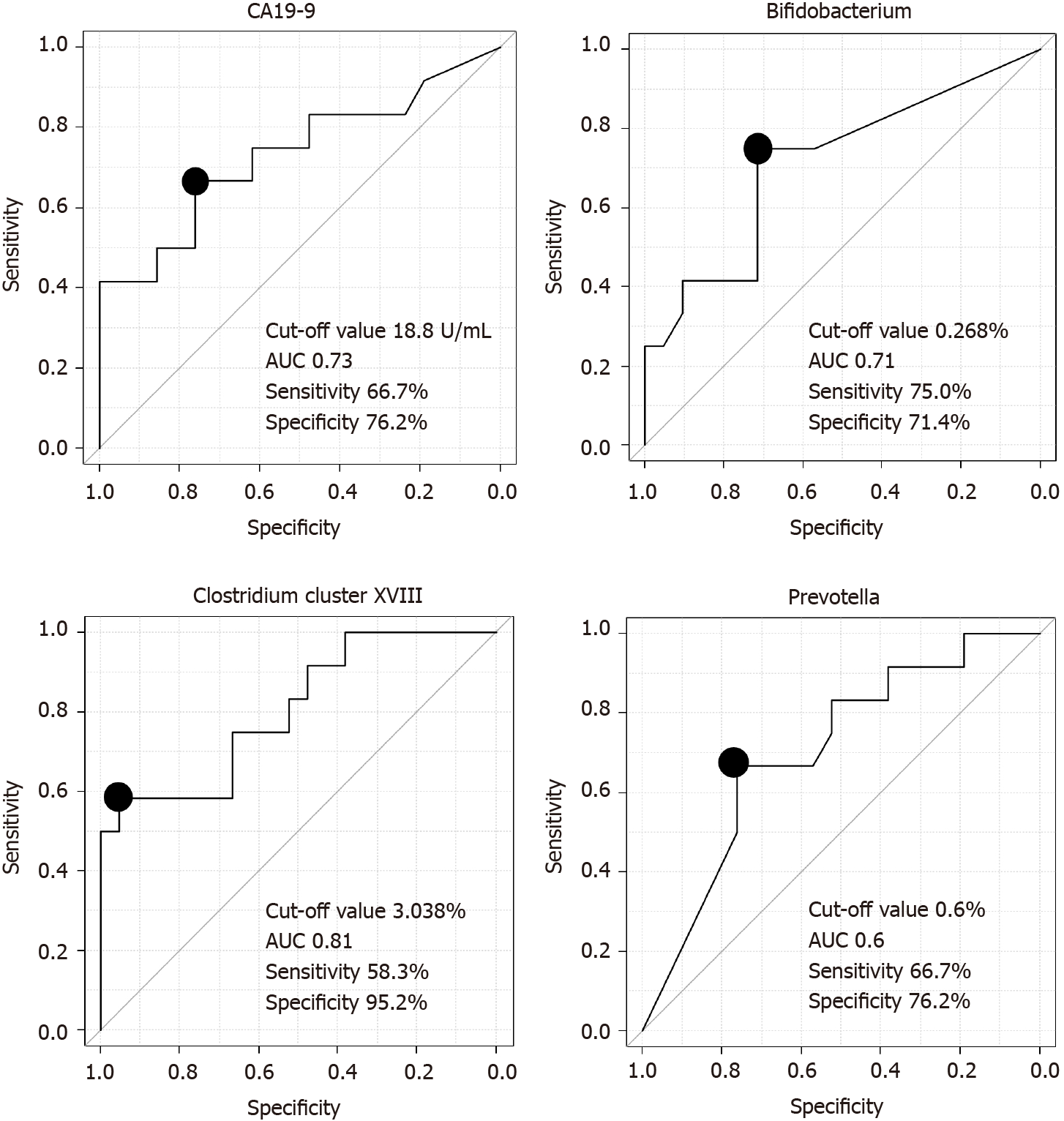
Cancer antigen 19-9 (CA19-9), Bifidobacterium, Clostridium cluster XVIII, and Prevotella levels differed significantly between the benign and malignant groups. Clostridium cluster XVIII had the greatest area under the receiver operating characteristic curve (AUC) among the four factors with respect to diagnosing PB Ca (cutoff value: 3.038%; sensitivity: 58.3%; specificity: 95.2%; AUC: 0.81). The combination of Clostridium cluster XVIII (cutoff value: 3.038%) and CA19-9 Levels (cutoff value: 18.8 U/mL) showed 91.7% sensitivity and 71.4% specificity for diagnosing PB Ca.
The duodenal microbiota may be useful for PB Ca screening.
Core Tip: Recently, a correlation between the human microbiota and malignant gastro
- Citation: Sugimoto M, Abe K, Takagi T, Suzuki R, Konno N, Asama H, Sato Y, Irie H, Watanabe K, Nakamura J, Kikuchi H, Takasumi M, Hashimoto M, Kato T, Kobashi R, Hikichi T, Ohira H. Dysbiosis of the duodenal microbiota as a diagnostic marker for pancreaticobiliary cancer. World J Gastrointest Oncol 2021; 13(12): 2088-2100
- URL: https://www.wjgnet.com/1948-5204/full/v13/i12/2088.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v13.i12.2088
Pancreaticobiliary cancer (PB Ca) is a lethal disease[1]. Surgery is the only radical treatment for pancreatic cancer, but unfortunately, many pancreatic cancer patients have advanced-stage lesions or other organ metastases, and they thus are not candidates for surgery[2]. For those who can undergo surgical treatment, the 5-year survival rate is reported to be 20%-30%[3,4].
In general, biliary tract cancer is difficult to diagnose. Conventional diagnostic methods include evaluating tumor markers [cancer antigen 19-9 (CA19-9) or carcinoembryonic antigen (CEA)], biliary biopsy, biliary juice cytology, and brush cytology, but the diagnostic power of these methods is not sufficient[5-19]. It has been reported that serum CA19-9 elevation is observed in 85% of patients with cholangiocarcinoma, though elevation of this marker can also be found in benign obstructive jaundice. Similarly, elevated serum CEA, which is not seen in obstructive jaundice, occurs in only 30% of patients with cholangiocarcinoma[20]. Therefore, effective diagnostic methods for the early diagnosis of PB Ca are urgently needed. Recently, a correlation between the human microbiota and malignant gastrointestinal diseases was reported[21-26]. In addition, oral and salivary microbiota communities have been reported to be effective in diagnosing pancreatic cancer or predicting the onset of pancreatic cancer[27-29], and the risk of pancreatic cancer is reportedly increased in patients with a history of periodontal disease[30]. Furthermore, serum antibodies against oral microbiota are reported to be a risk factor for the onset of pancreatic cancer[31]. However, the mechanism by which this dysbiosis leads to pancreatic cancer is unknown, especially as the pancreas is relatively distant from the mouth.
Thus, we hypothesized that the duodenal microbiota would be more efficient than the oral microbiota for diagnosing PB Ca because the duodenum is closer to the bile duct and pancreas than the oral cavity. The aim of this study was to determine the efficacy of the duodenal microbiota for diagnosing PB Ca.
This study was approved by the Institutional Review Board of Fukushima Medical University.
We assessed 34 patients with pancreaticobiliary disease who visited our hospital over two years. Twenty-two patients were diagnosed with benign pancreaticobiliary diseases (benign group) [chronic pancreatitis: 6; intraductal papillary mucinous neoplasm (IPMN): 5; gallbladder adenomyomatosis: 3; autoimmune pancreatitis: 3; benign common bile duct (CBD) stricture of unknown origin: 2; serous cystic neoplasm: 2; and CBD stone: 1] (Table 1). The other 12 patients were diagnosed with PB Ca (malignant group) (pancreatic cancer: 9; bile duct cancer: 3). The patients provided written informed consent to participate in this study. For all pancreatic cancer cases, the lesion was located in the head. Eight pancreatic cancer patients were diagnosed by endoscopic ultrasound-guided fine needle aspiration. One pancreatic cancer patient was diagnosed with intraductal papillary mucinous carcinoma with evident worsening of the lesion by imaging. Benign diseases were diagnosed by no histological malignancy or unchanging lesions after a clinical course of at least six months. Furthermore, the IPMN patients in the benign group did not have high-risk stigmata or worrisome features[32]. The cases of bile duct cancer were diagnosed by biliary biopsy or surgery. According to the cytology grade, classes IV and V were diagnosed as malignancies. The stage of PB Ca was determined based on the UICC classification, ver. 8.
| Benign group (n = 22) | Malignant group (n = 12) | ||
| Chronic pancreatitis | 6 | Pancreatic cancer, stage (I/II/III/IV) | 9 (2/5/1/1/) |
| IPMN | 5 | Biliary ductal cancer, stage (I/II/III/IV) | 3 (1/2/0/0) |
| GB ADM | 3 | ||
| Autoimmune pancreatitis | 3 | ||
| CBD stricture of unknown origin | 2 | ||
| Serous cystic neoplasm | 2 | ||
| CBD stone | 1 | ||
The patients did not receive antibiotic agents for at least a week prior to duodenal juice collection, and they did not receive steroids at all.
An endoscope was used under sedation with midazolam. The endoscope was advanced to the duodenum, and 0.5-1.0 mL of duodenal juice was collected through a catheter and stored at -20°C. The endoscope used was Q260 and Q260H, and the catheter was a PR-109Q-1 or PR-104Q-1 (Olympus, Tokyo, Japan).
Bacterial DNA was extracted from duodenal juice samples in accordance with a previous report by Takahashi et al[33].
Terminal restriction fragment (T-RF) length polymorphism (T-RFLP) was performed by TechnoSuruga Laboratory (Shizuoka, Japan) according to Nagashima’s methods[34,35]. The 16S rRNA gene was amplified from the extracted DNA using the primers 5’ FAM-labeled 516F (5’-TGCCAGCAGCCGCGGTA-3’) and 1510R (5’- GGTTACCTTGTTACGACTT-3’) and HotStarTaq DNA Polymerase (Qiagen, Hilden, Germany) with a Thermal Cycler Dice (Takara, Shiga, Japan). The amplification program used was as follows: preheating at 94°C for 15 min; 35 cycles of denaturation at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 2 min; and a terminal extension at 72°C for 10 min. DNA amplification was verified by electrophoresis of the polymerase chain reaction (PCR) products (2 μL) through a 1.0% agarose gel with Tris-acetate-EDTA buffer. The amplified DNA was purified by a MultiScreen PCR96 Filter Plate (Millipore, Billerica, MA, United States).
The purified PCR product (3 μL) was digested with 10 U of Fast Digest BseLI (BslI) (Thermo Fisher Scientific) in a total volume of 15 μL at 37°C for 10 min. The restriction digestion products (0.5 μL) were mixed with 0.1 μL of a DNA fragment-length standard size marker and 10 μL of deionized formamide. The standard size marker was MapMarker X-Rhodamine Labeled 50-1000 bp (Bio Ventures, Murfreesboro, TN, United States). The samples were denatured at 95°C for 2 min and then placed immediately on ice. The T-RF length was established using an ABI PRISM 3130xl genetic analyzer (Thermo Fisher Scientific), and the length and peak area were determined using the genotyping software GeneMapper (Thermo Fisher Scientific). The fragment sizes were estimated using the Local Southern method in GeneMapper software (Thermo Fisher Scientific). If the peak height was less than 50 fluorescence units, the T-RF was excluded from the analysis. The fragments were resolved to one base pair by manual alignment of the size standard peaks from different electro
Patient characteristics and tumor markers (age, sex, reduction in body weight ≥ 5 kg within 6 mo prior to duodenal juice sampling, intake of proton pump inhibitors, CA19-9) were compared between the two groups. The body weight marker was selected for the following reasons. The composition ratio of the microbiota has been reported to be different between subjects with obesity and those with a normal body mass index[36]. Because the intake of high-fat foods influences the quantity and composition of bile acid, the intestinal bacterial flora might change[37]. The relative abundances of duodenal microbiota members (Bacteroides, Bifidobacterium, Lactobacillales, Prevotella, Clostridium cluster IV, Clostridium subcluster XIVa, Clostridium cluster IX, Clostridium cluster XI, Clostridium cluster XVIII, and others) were compared between the benign and malignant groups.
Normally distributed continuous variables were compared using Student’s t test and nonnormally distributed continuous variables using the Mann-Whitney U test. Nominal variables were compared with Fisher’s exact test. A receiver operating characteristic (ROC) curve was employed to compare the accuracy of the biomarkers. The P value < 0.05 was considered statistically significant.
These statistical analyses were performed using the EZR platform (Saitama Medical Center, Jichi Medical University, Saitama, Japan), which is a graphical user interface for R (The R Foundation for Statistical Computing, Vienna, Austria). EZR is a modified version of R-commander that was designed to perform functions that are frequently used in biostatistics[38].
Among the patient characteristics and tumor markers, age and CA19-9 Levels were significantly different between the benign and malignant groups (mean ± SD, age: 63.3 ± 12.2 vs 73.0 ± 8.3 years, P value = 0.016; median (range), CA19-9: 5.4 (2.0-54.8) vs 22.8 (2.0-9893.2), P value = 0.03) (Table 2).
| Benign group (n = 22) | Malignant group (n = 12) | P value | |
| Age, yr (mean ± SD) | 63.0 ± 12.2 | 73.0 ± 8.3 | 0.016 |
| Sex (male/female) | 8/14 | 4/8 | 1.0 |
| Reduction in body weight ≥ 5 kg within 6 mo before duodenal juice sampling, n (%) | 1 (4.5) | 2 (16.7) | 0.28 |
| Intake of proton pump inhibitors, n (%) | 4 (18.2) | 2 (16.7) | 1.0 |
| CA19-9, U/mL, median (range) | 5.4 (2.0-54.8) | 22.8 (2.0-9893.2) | 0.03 |
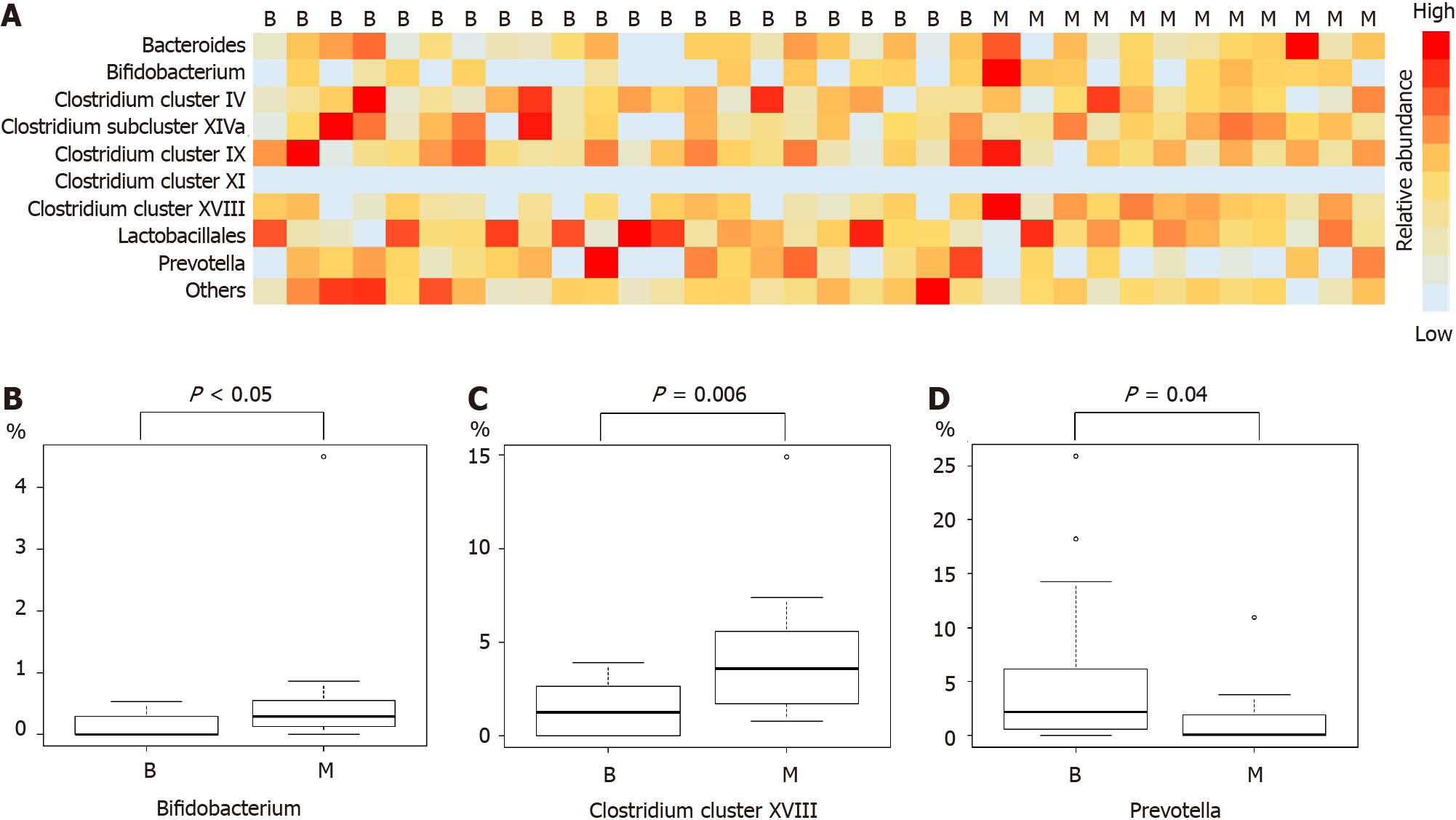
Comparison of microbiome components revealed Bifidobacterium, Clostridium cluster XVIII, and Prevotella to be significantly different between the benign and malignant groups (median (range), Bifidobacterium: 0 (0-0.5)% vs 0.3 (0-4.5)%, P value < 0.05; Clostridium cluster XVIII: 1.3 (0-3.9)% vs 3.6 (0.8-14.9)%, P value = 0.006; Prevotella: 2.2 (0-25.9)% vs 0.1 (0-10.9)%, P value = 0.04) (Figure 1 and Table 3).
| Benign group (n = 22) | Malignant group (n = 12) | P value | |
| Bacteroides, %, median (range) | 4.3 (0-26.1) | 5.6 (0-46.4) | 0.55 |
| Bifidobacterium, %, median (range) | 0 (0-0.5) | 0.3 (0-4.5) | < 0.05 |
| Clostridium cluster IV, %, median (range) | 2.9 (0-10.8) | 3.4 (0-8.8) | 0.80 |
| Clostridium cluster IX, %, median (range) | 4.7 (0.6-19.5) | 4.9 (0-17.8) | 0.68 |
| Clostridium cluster XI, %, median (range) | 0 (0-0) | 0 (0-0) | |
| Clostridium cluster XVIII, %, median (range) | 1.3 (0-3.9) | 3.6 (0.8-14.9) | 0.006 |
| Clostridium subcluster XIVa, %, median (range) | 5.1 (0-23.1) | 6.4 (2.9-13.7) | 0.38 |
| Lactobacillales, %, mean ± SD | 63.0 ± 19.7 | 62.6 ± 18.3 | 0.95 |
| Prevotella, %, median (range) | 2.2 (0-25.9) | 0.1 (0-10.9) | 0.04 |
| Others, %, median (range) | 4.9 (2.5-20.4) | 4.1 (1.5-6.3) | 0.14 |
To determine the influence of age on CA19-9 Levels and microbiome composition, these factors were compared between the subgroup of patients < 69 years and the subgroup of those ≥ 69 years (the median age of all patients was 69 years). According to the results, CA19-9 Levels, Bifidobacterium, Clostridium cluster XVIII, and Prevotella were not influenced by age (Table 4).
| Age < 69 yr (n = 17) | Age ≥ 69 yr (n = 17) | P value | |
| CA19-9, U/mL, median (range) | 7.1 (2-129.3) | 4.9 (2-9893.2) | 0.77 |
| Bifidobacterium, %, median (range) | 0.3 (0-0.5) | 0 (0-4.5) | 0.3 |
| Clostridium cluster XVIII, %, median (range) | 2.6 (0-5.7) | 1.8 (0-14.9) | 0.82 |
| Prevotella, %, median (range) | 1.3 (0-18.2) | 1.9 (0-25.9) | 0.56 |
We assessed the ability of the microbiota to diagnose PB Ca by calculating the area under the ROC curve (AUC) and found that Clostridium cluster XVIII had the highest AUC (cutoff value: 3.038%, sensitivity: 58.3%, specificity: 95.2%, AUC: 0.81) among the three microbiome components (Bifidobacterium, Clostridium cluster XVIII, Prevotella) and CA19-9 Levels (Figure 2).
The combination of Clostridium cluster XVIII (cutoff value: 3.038%) and CA19-9 Levels (cutoff value: 18.8 U/mL) was also examined as a marker to diagnose PB Ca; the sensitivity of this combination was 91.7% (11/12), and the specificity was 71.4% (15/21) (Table 5). CA19-9 data were missing for one patient in the benign group.
In this study, we investigated which members of the duodenal microbiota could aid in diagnosing PB Ca and found Clostridium cluster XVIII to be more useful than CA19-9 Levels and other bacteria for diagnosing PB Ca. Notably, the combination of Clostridium cluster XVIII and CA19-9 Levels showed high sensitivity, indicating that this combination is valuable for screening patients for PB Ca.
As mentioned above, the oral microbiota has been considered to be a biomarker in pancreatic cancer. First, Michaud et al[30] reported that a history of periodontal disease was a risk factor for pancreatic cancer. After that, several oral microbes were reported to be more abundant and possible predictors and risk factors for survival in pancreatic cancer (Table 6)[27-29,39,40]. In addition, antibodies against Porphyromonas gingivalis can serve as a risk factor for pancreatic cancer onset[27].
| Disease | Ref. | Microbes | Sample | Role |
| Pancreatic cancer | Michaud et al[30] | A history of periodontal diseases | Risk factor | |
| Farrell et al[28] | A combination of Neisseria elongate and Streptococcus mitis | Oral | Distinguishing from healthy controls | |
| Torres et al[29] | Ratio of Leptotrichia to Porphyromonas | Saliva | Higher in pancreatic cancer patients | |
| Fan et al[27] | Porphyromonas gingivalis | Oral, antibody | Risk factor | |
| Olson et al[39] | Firmicutes | Oral | More abundant | |
| Lu et al[40] | Leptotrichia, Fusobacterium, Rothia, Actinomyces, Corynebacterium, Atopobium, Peptostreptococcus, Catonella, Oribacterium, Filifactor, Campylobacter, Moraxella, Tannerella | Tongue coating | More prevalent | |
| Mei et al[46] | Acinetobactor, Aquabacterium, Oceanobacillus, Rahnella, Massilia, Delftia, Deinococcus, Sphingobium | Duodenal mucosa | More abundant | |
| Mitsuhashi et al[43] | Fusobacterium species | Cancer tissue | Poor prognosis | |
| Riquelme et al[44] | Pseudoxanthomonas, Streptomyces, Saccharopolyspora, Bacillus clausii | Cancer tissue | Long-term survival | |
| Pancreatic and ampullary cancer | Di Calro et al[45] | Escherichia coli, Klebsiella pneumoniae | Bile juice | Predictor for survival |
| IPMN with high-grade dysplasia | Gaiser et al[47] | Granulicatella adiacens, Fusobacterium nucleatum | Cyst fluid | More abundant |
Although the salivary microbiome may be useful for the medical care of patients with pancreatic cancer, it is influenced by differences in oral hygiene, mastication, and swallowing among individuals[27,41,42]. In contrast, the duodenal microbiota is more relevant to the pancreas and bile duct than is the salivary microbiota. Therefore, the duodenal microbiota was hypothesized to directly reflect the dysbiosis associated with PB Ca, and in fact, the results of this study reveal that the duodenal microbiota might be beneficial for screening PB Ca. The microbiota around the pancreas and biliary duct have been reported. Microbes of the duodenal mucosa, bile juice, cancer tissue, and cyst fluid of IPMN in pancreaticobiliary tumor patients have been investigated (Table 6)[30,43-47]. However, the methods used to analyze the microbiota around the pancreas and bile duct in these reports were more invasive than duodenal juice sampling.
The mechanism by which microbes lead to PB Ca remains unknown. The etiology with respect to the salivary microbiota has been considered in past reports. P. gingivalis can interrupt signaling pathways by modulating receptor expression and cytokine secretion to evade the host’s immune system[48-52]. Moreover, P. gingivalis activates the Toll-like receptor signaling pathway[53,54], which has been reported to be related to pancreatic carcinogenesis[55,56]. In other reports, oral bacteria were found outside of the oral cavity in the gastrointestinal tract. Immune responses against these bacteria can cause inflammation and carcinogenesis in the pancreas. Lipopolysaccharide has also been reported to drive pancreatic carcinogenesis by blocking the MyD88-dependent, Toll-like receptor 4 and MyD88-independent pathways[57]. In another report, the mechanism was described as follows. Bacterial ligands detected by Toll-like receptors cause a Th1/Th2/Th17 imbalance in the tumor microenvironment, promoting tumorigenesis in combination with Kras mutation. In the duodenum, microbes may reach the pancreatic duct or biliary duct through the Vater papilla. In the pancreas or bile duct, pattern recognition receptors (such as Toll-like receptors) are stimulated by the pathogenic molecular patterns of bacterial ligands and induce lower levels of immune suppression, leading to the development of PB Ca[58]. These results from past reports suggest that some type of immune system response is the link between the duodenal microbiota and PB Ca.
On the one hand, the relationship between Clostridium cluster XVIII and carcinogenesis has not been reported, even though Clostridium cluster XVIII is reported to have the potential to enhance regulatory T (Treg) cells[59]. Many Treg cells exist in tumor tissue and prevent the immune response to tumors. Therefore, Tregs contribute to tumor progression and poor prognosis[60-64]. Thus, Clostridium cluster XVIII may increase in response to cancer and activate Tregs. Alternatively, Clostridium cluster XVIII may activate Tregs, with oncogenesis advancing.
This report has some limitations. First, this study was small and performed at a single institution. However, based on the data from Clostridium cluster XVIII, the average value of the malignant group was 4.5%, and that of the benign group was 1.5%. Total thirty patients were needed to achieve an α error of 5% and a β value of 0.2. When Clostridium cluster XVIII was the main outcome, the minimum necessary sample size was secured. Although this is the first report to describe the relationship between the duodenal juice microbiota and PB Ca, the diseases in the malignant group were not uniform. If subgroup analyses of pancreatic diseases were performed, the abundance of some duodenal microbes would be significantly different between the benign and malignant groups (Table 7). We hope that a future study with a larger number of patients will confirm our results for both pancreatic cancer and biliary cancer. Second, healthy control subjects were not enrolled in this study. However, as esophagogastroduodenoscopy under sedation is rarely performed in healthy patients, this limitation was unavoidable in the study design. Third, T-RFLP was applied. Investigations into the duodenal microbiota have been limited because duodenal juice cannot be collected in large volumes (less than 0.5 mL is typically collected). However, the measurement of the duodenal microbiota was demonstrated to be possible. Follow-up studies using next-generation sequencing are warranted[65]. Fourth, examining the duodenal microbiota requires a somewhat invasive technique. In the future, the development of serum antibody testing for the duodenal microbiota should be pursued.
| Benign pancreatic diseases (n = 16) | Pancreatic cancer (n = 9) | P value | |
| Bacteroides, %, median (range) | 2.1 (0-26.1) | 5.8 (0-46.4) | 0.17 |
| Bifidobacterium, %, median (range) | 0 (0-0.5) | 0.48 (0-4.5) | 0.03 |
| Clostridium cluster IV, %, mean ± SD | 4.0 ± 3.3 | 3.6 ± 2.9 | 0.76 |
| Clostridium cluster IX, %, mean ± SD | 5.3 ± 3.6 | 6.4 ± 5.2 | 0.57 |
| Clostridium cluster XI, %, median (range) | 0 (0-0) | 0 (0-0) | |
| Clostridium cluster XVIII, %, median (range) | 1.4 (0-3.9) | 3.0 (0.8-14.9) | 0.04 |
| Clostridium subcluster XIVa, %, median (range) | 3.8 (0-21.4) | 6.0 (2.9-13.7) | 0.32 |
| Lactobacillales, %, mean ± SD | 68.4 ± 19.3 | 59.7 ± 20.4 | 0.3 |
| Prevotella, %, median (range) | 4.2 (2.5-20.4) | 4.0 (1.5-6.3) | 0.3 |
| Others, %, median (range) | 2.0 (0-18.3) | 0.3 (0-11.0) | 0.3 |
In conclusion, the duodenal microbiota may contribute to PB Ca screening.
Pancreaticobiliary cancer (PB Ca) is a lethal disease; however, there are currently no appropriate diagnostic and prognostic markers. Recently, the human microbiota was reported to be a causative factor, diagnostic marker, and prognostic marker for gastrointestinal malignant diseases.
The oral and fecal microbiota have been reported to be useful diagnostic markers for gastrointestinal cancer. The duodenum is located closer to the pancreas and bile duct than the oral cavity and colon. Therefore, we hypothesized that assessment of the duodenal microbiota might improve the diagnostic accuracy for PB Ca.
To investigate the diagnostic accuracy of duodenal microbiota evaluation for PB Ca.
Thirty-four PB Ca and benign pancreaticobiliary disease patients were recruited for this study, and their duodenal juice was aseptically collected by endoscopy. The duodenal microbiota was analyzed, and the relative abundances of species in the duodenal microbiota were compared between PB Ca patients and benign pancreaticobiliary disease patients. The PB Ca diagnosability was compared between a conventional tumor marker and species in the duodenal microbiota with significantly different abundances in PB Ca patients vs benign pancreaticobiliary disease patients.
The abundances of cancer antigen 19-9 (CA19-9), Bifidobacterium, Clostridium cluster XVIII, and Prevotella were significantly different between PB Ca patients and benign pancreaticobiliary disease patients. The diagnostic capacity of Clostridium cluster XVIII was the highest among the four markers (CA19-9, Bifidobacterium, Clostridium cluster XVIII, and Prevotella). The combined assessment of Clostridium cluster XVIII and CA19-9 Levels was useful for PB Ca diagnosis.
It was possible to investigate the microbiota of duodenal juice. Duodenal microbiota evaluation may contribute to the diagnosis of PB Ca.
In the future, novel diagnostic and prognostic markers and treatments could be developed by investigating the relationship between the duodenal microbiota and PB Ca.
We thank all the staff at the Department of Gastroenterology of Fukushima Medical University, the Department of Endoscopy of Fukushima Medical University Hospital, and the Gastroenterology Ward of Fukushima Medical University Hospital. We also thank TechnoSuruga Laboratory for T-RFLP.
Provenance and peer review: Invited article; Externally peer reviewed.
Corresponding Author's Membership in Professional Societies: Fukushima Medical University, No. 03727100.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Japan
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Yao D S-Editor: Ma YJ L-Editor: A P-Editor: Liu JH
| 1. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11573] [Cited by in RCA: 13161] [Article Influence: 1880.1] [Reference Citation Analysis (4)] |
| 2. | Geer RJ, Brennan MF. Prognostic indicators for survival after resection of pancreatic adenocarcinoma. Am J Surg. 1993;165:68-72; discussion 72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 548] [Cited by in RCA: 548] [Article Influence: 17.1] [Reference Citation Analysis (1)] |
| 3. | Picozzi VJ, Oh SY, Edwards A, Mandelson MT, Dorer R, Rocha FG, Alseidi A, Biehl T, Traverso LW, Helton WS, Kozarek RA. Five-Year Actual Overall Survival in Resected Pancreatic Cancer: A Contemporary Single-Institution Experience from a Multidisciplinary Perspective. Ann Surg Oncol. 2017;24:1722-1730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 4. | White RJ, Hasan S, Monga D, Finley G, Islam M, Schiffman S, Williams HK, Kulkarni A, Thakkar S, Kirichenko AV, Wegner RE. Time to Adjuvant Systemic Therapy Following Pancreatic Cancer Resection and Effect on Outcome. Pancreas. 2019;48:1086-1091. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 5. | Uchida N, Kamada H, Ono M, Aritomo Y, Masaki T, Nakatsu T, Kuriyama S. How many cytological examinations should be performed for the diagnosis of malignant biliary stricture via an endoscopic nasobiliary drainage tube? J Gastroenterol Hepatol. 2008;23:1501-1504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 18] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 6. | Rösch T, Hofrichter K, Frimberger E, Meining A, Born P, Weigert N, Allescher HD, Classen M, Barbur M, Schenck U, Werner M. ERCP or EUS for tissue diagnosis of biliary strictures? Gastrointest Endosc. 2004;60:390-396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 232] [Cited by in RCA: 215] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 7. | Foutch PG, Kerr DM, Harlan JR, Kummet TD. A prospective, controlled analysis of endoscopic cytotechniques for diagnosis of malignant biliary strictures. Am J Gastroenterol. 1991;86:577-580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 8. | Kubota Y, Takaoka M, Tani K, Ogura M, Kin H, Fujimura K, Mizuno T, Inoue K. Endoscopic transpapillary biopsy for diagnosis of patients with pancreaticobiliary ductal strictures. Am J Gastroenterol. 1993;88:1700-1704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 9. | Lee JG, Leung JW, Baillie J, Layfield LJ, Cotton PB. Benign, dysplastic, or malignant--making sense of endoscopic bile duct brush cytology: results in 149 consecutive patients. Am J Gastroenterol. 1995;90:722-726. [PubMed] |
| 10. | Ponchon T, Gagnon P, Berger F, Labadie M, Liaras A, Chavaillon A, Bory R. Value of endobiliary brush cytology and biopsies for the diagnosis of malignant bile duct stenosis: results of a prospective study. Gastrointest Endosc. 1995;42:565-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 264] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 11. | Pugliese V, Conio M, Nicolò G, Saccomanno S, Gatteschi B. Endoscopic retrograde forceps biopsy and brush cytology of biliary strictures: a prospective study. Gastrointest Endosc. 1995;42:520-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 190] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 12. | Howell DA, Parsons WG, Jones MA, Bosco JJ, Hanson BL. Complete tissue sampling of biliary strictures at ERCP using a new device. Gastrointest Endosc. 1996;43:498-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 74] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 13. | Sugiyama M, Atomi Y, Wada N, Kuroda A, Muto T. Endoscopic transpapillary bile duct biopsy without sphincterotomy for diagnosing biliary strictures: a prospective comparative study with bile and brush cytology. Am J Gastroenterol. 1996;91:465-467. [PubMed] |
| 14. | Mansfield JC, Griffin SM, Wadehra V, Matthewson K. A prospective evaluation of cytology from biliary strictures. Gut. 1997;40:671-677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 159] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 15. | Schoefl R, Haefner M, Wrba F, Pfeffel F, Stain C, Poetzi R, Gangl A. Forceps biopsy and brush cytology during endoscopic retrograde cholangiopancreatography for the diagnosis of biliary stenoses. Scand J Gastroenterol. 1997;32:363-368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 129] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 16. | Glasbrenner B, Ardan M, Boeck W, Preclik G, Möller P, Adler G. Prospective evaluation of brush cytology of biliary strictures during endoscopic retrograde cholangiopancreatography. Endoscopy. 1999;31:712-717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 153] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 17. | Jailwala J, Fogel EL, Sherman S, Gottlieb K, Flueckiger J, Bucksot LG, Lehman GA. Triple-tissue sampling at ERCP in malignant biliary obstruction. Gastrointest Endosc. 2000;51:383-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 232] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 18. | Macken E, Drijkoningen M, Van Aken E, Van Steenbergen W. Brush cytology of ductal strictures during ERCP. Acta Gastroenterol Belg. 2000;63:254-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 29] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 19. | de Bellis M, Sherman S, Fogel EL, Cramer H, Chappo J, McHenry L Jr, Watkins JL, Lehman GA. Tissue sampling at ERCP in suspected malignant biliary strictures (Part 2). Gastrointest Endosc. 2002;56:720-730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 53] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 20. | Khan SA, Davidson BR, Goldin R, Pereira SP, Rosenberg WM, Taylor-Robinson SD, Thillainayagam AV, Thomas HC, Thursz MR, Wasan H; British Society of Gastroenterology. Guidelines for the diagnosis and treatment of cholangiocarcinoma: consensus document. Gut. 2002;51 Suppl 6:VI1-VI9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 323] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 21. | Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, Ogino S, Birren BW, Huttenhower C, Garrett WS, Meyerson M. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1199] [Cited by in RCA: 1498] [Article Influence: 107.0] [Reference Citation Analysis (0)] |
| 22. | Yamamura K, Baba Y, Nakagawa S, Mima K, Miyake K, Nakamura K, Sawayama H, Kinoshita K, Ishimoto T, Iwatsuki M, Sakamoto Y, Yamashita Y, Yoshida N, Watanabe M, Baba H. Human Microbiome Fusobacterium Nucleatum in Esophageal Cancer Tissue Is Associated with Prognosis. Clin Cancer Res. 2016;22:5574-5581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 324] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 23. | Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, Goedert JJ, Hayes RB, Yang L. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst. 2013;105:1907-1911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 566] [Cited by in RCA: 713] [Article Influence: 59.4] [Reference Citation Analysis (1)] |
| 24. | Mai V, Morris JG Jr. Need for prospective cohort studies to establish human gut microbiome contributions to disease risk. J Natl Cancer Inst. 2013;105:1850-1851. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 25. | Krishnan S, Eslick GD. Streptococcus bovis infection and colorectal neoplasia: a meta-analysis. Colorectal Dis. 2014;16:672-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 26. | Komiya Y, Shimomura Y, Higurashi T, Sugi Y, Arimoto J, Umezawa S, Uchiyama S, Matsumoto M, Nakajima A. Patients with colorectal cancer have identical strains of Fusobacterium nucleatum in their colorectal cancer and oral cavity. Gut. 2019;68:1335-1337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 206] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 27. | Fan X, Alekseyenko AV, Wu J, Peters BA, Jacobs EJ, Gapstur SM, Purdue MP, Abnet CC, Stolzenberg-Solomon R, Miller G, Ravel J, Hayes RB, Ahn J. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut. 2018;67:120-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 386] [Cited by in RCA: 552] [Article Influence: 78.9] [Reference Citation Analysis (0)] |
| 28. | Farrell JJ, Zhang L, Zhou H, Chia D, Elashoff D, Akin D, Paster BJ, Joshipura K, Wong DT. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut. 2012;61:582-588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 403] [Cited by in RCA: 487] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 29. | Torres PJ, Fletcher EM, Gibbons SM, Bouvet M, Doran KS, Kelley ST. Characterization of the salivary microbiome in patients with pancreatic cancer. PeerJ. 2015;3:e1373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 158] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 30. | Michaud DS, Joshipura K, Giovannucci E, Fuchs CS. A prospective study of periodontal disease and pancreatic cancer in US male health professionals. J Natl Cancer Inst. 2007;99:171-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 210] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 31. | Michaud DS, Izard J, Wilhelm-Benartzi CS, You DH, Grote VA, Tjønneland A, Dahm CC, Overvad K, Jenab M, Fedirko V, Boutron-Ruault MC, Clavel-Chapelon F, Racine A, Kaaks R, Boeing H, Foerster J, Trichopoulou A, Lagiou P, Trichopoulos D, Sacerdote C, Sieri S, Palli D, Tumino R, Panico S, Siersema PD, Peeters PH, Lund E, Barricarte A, Huerta JM, Molina-Montes E, Dorronsoro M, Quirós JR, Duell EJ, Ye W, Sund M, Lindkvist B, Johansen D, Khaw KT, Wareham N, Travis RC, Vineis P, Bueno-de-Mesquita HB, Riboli E. Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut. 2013;62:1764-1770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 295] [Cited by in RCA: 310] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 32. | Tanaka M, Fernández-Del Castillo C, Kamisawa T, Jang JY, Levy P, Ohtsuka T, Salvia R, Shimizu Y, Tada M, Wolfgang CL. Revisions of international consensus Fukuoka guidelines for the management of IPMN of the pancreas. Pancreatology. 2017;17:738-753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 868] [Cited by in RCA: 1156] [Article Influence: 144.5] [Reference Citation Analysis (1)] |
| 33. | Takahashi S, Tomita J, Nishioka K, Hisada T, Nishijima M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS One. 2014;9:e105592. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 736] [Cited by in RCA: 894] [Article Influence: 81.3] [Reference Citation Analysis (0)] |
| 34. | Nagashima K, Hisada T, Sato M, Mochizuki J. Application of new primer-enzyme combinations to terminal restriction fragment length polymorphism profiling of bacterial populations in human feces. Appl Environ Microbiol. 2003;69:1251-1262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 175] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 35. | Nagashima K, Mochizuki J, Hisada T, Suzuki S, Shimomura K. Phylogenetic analysis of 16S rRNA gene sequences from human fecal microbiota and improved utility of T-RFLP profiling. Biosci Microflora. 2006;25:99-107. [RCA] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 82] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 36. | Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022-1023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7222] [Cited by in RCA: 6407] [Article Influence: 337.2] [Reference Citation Analysis (0)] |
| 37. | Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. Human gut microbiome viewed across age and geography. Nature. 2012;486:222-227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4762] [Cited by in RCA: 5465] [Article Influence: 420.4] [Reference Citation Analysis (0)] |
| 38. | Kanda Y. Investigation of the freely available easy-to-use software 'EZR' for medical statistics. Bone Marrow Transplant. 2013;48:452-458. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9275] [Cited by in RCA: 13281] [Article Influence: 1106.8] [Reference Citation Analysis (0)] |
| 39. | Olson SH, Satagopan J, Xu Y, Ling L, Leong S, Orlow I, Saldia A, Li P, Nunes P, Madonia V, Allen PJ, O'Reilly E, Pamer E, Kurtz RC. The oral microbiota in patients with pancreatic cancer, patients with IPMNs, and controls: a pilot study. Cancer Causes Control. 2017;28:959-969. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 40. | Lu H, Ren Z, Li A, Li J, Xu S, Zhang H, Jiang J, Yang J, Luo Q, Zhou K, Zheng S, Li L. Tongue coating microbiome data distinguish patients with pancreatic head cancer from healthy controls. J Oral Microbiol. 2019;11:1563409. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 96] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 41. | Lockhart PB, Brennan MT, Sasser HC, Fox PC, Paster BJ, Bahrani-Mougeot FK. Bacteremia associated with toothbrushing and dental extraction. Circulation. 2008;117:3118-3125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 648] [Cited by in RCA: 549] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 42. | Crasta K, Daly CG, Mitchell D, Curtis B, Stewart D, Heitz-Mayfield LJ. Bacteraemia due to dental flossing. J Clin Periodontol. 2009;36:323-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 77] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 43. | Mitsuhashi K, Nosho K, Sukawa Y, Matsunaga Y, Ito M, Kurihara H, Kanno S, Igarashi H, Naito T, Adachi Y, Tachibana M, Tanuma T, Maguchi H, Shinohara T, Hasegawa T, Imamura M, Kimura Y, Hirata K, Maruyama R, Suzuki H, Imai K, Yamamoto H, Shinomura Y. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget. 2015;6:7209-7220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 301] [Cited by in RCA: 288] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 44. | Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, Quesada P, Sahin I, Chandra V, San Lucas A, Scheet P, Xu H, Hanash SM, Feng L, Burks JK, Do KA, Peterson CB, Nejman D, Tzeng CD, Kim MP, Sears CL, Ajami N, Petrosino J, Wood LD, Maitra A, Straussman R, Katz M, White JR, Jenq R, Wargo J, McAllister F. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell. 2019;178:795-806.e12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 469] [Cited by in RCA: 1038] [Article Influence: 207.6] [Reference Citation Analysis (0)] |
| 45. | Di Carlo P, Serra N, D'Arpa F, Agrusa A, Gulotta G, Fasciana T, Rodolico V, Giammanco A, Sergi C. The microbiota of the bilio-pancreatic system: a cohort, STROBE-compliant study. Infect Drug Resist. 2019;12:1513-1527. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 46. | Mei QX, Huang CL, Luo SZ, Zhang XM, Zeng Y, Lu YY. Characterization of the duodenal bacterial microbiota in patients with pancreatic head cancer vs. healthy controls. Pancreatology. 2018;18:438-445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 75] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 47. | Gaiser RA, Halimi A, Alkharaan H, Lu L, Davanian H, Healy K, Hugerth LW, Ateeb Z, Valente R, Fernández Moro C, Del Chiaro M, Sällberg Chen M. Enrichment of oral microbiota in early cystic precursors to invasive pancreatic cancer. Gut. 2019;68:2186-2194. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 173] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 48. | Duncan L, Yoshioka M, Chandad F, Grenier D. Loss of lipopolysaccharide receptor CD14 from the surface of human macrophage-like cells mediated by Porphyromonas gingivalis outer membrane vesicles. Microb Pathog. 2004;36:319-325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 92] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 49. | Stathopoulou PG, Benakanakere MR, Galicia JC, Kinane DF. The host cytokine response to Porphyromonas gingivalis is modified by gingipains. Oral Microbiol Immunol. 2009;24:11-17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 93] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 50. | Singh A, Wyant T, Anaya-Bergman C, Aduse-Opoku J, Brunner J, Laine ML, Curtis MA, Lewis JP. The capsule of Porphyromonas gingivalis leads to a reduction in the host inflammatory response, evasion of phagocytosis, and increase in virulence. Infect Immun. 2011;79:4533-4542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 95] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 51. | Taxman DJ, Swanson KV, Broglie PM, Wen H, Holley-Guthrie E, Huang MT, Callaway JB, Eitas TK, Duncan JA, Ting JP. Porphyromonas gingivalis mediates inflammasome repression in polymicrobial cultures through a novel mechanism involving reduced endocytosis. J Biol Chem. 2012;287:32791-32799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 69] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 52. | Palm E, Khalaf H, Bengtsson T. Porphyromonas gingivalis downregulates the immune response of fibroblasts. BMC Microbiol. 2013;13:155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 53. | Hayashi C, Papadopoulos G, Gudino CV, Weinberg EO, Barth KR, Madrigal AG, Chen Y, Ning H, LaValley M, Gibson FC 3rd, Hamilton JA, Genco CA. Protective role for TLR4 signaling in atherosclerosis progression as revealed by infection with a common oral pathogen. J Immunol. 2012;189:3681-3688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 54. | Zambirinis CP, Levie E, Nguy S, Avanzi A, Barilla R, Xu Y, Seifert L, Daley D, Greco SH, Deutsch M, Jonnadula S, Torres-Hernandez A, Tippens D, Pushalkar S, Eisenthal A, Saxena D, Ahn J, Hajdu C, Engle DD, Tuveson D, Miller G. TLR9 Ligation in pancreatic stellate cells promotes tumorigenesis. J Exp Med. 2015;212:2077-2094. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 143] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 55. | Zhang JJ, Wu HS, Wang L, Tian Y, Zhang JH, Wu HL. Expression and significance of TLR4 and HIF-1alpha in pancreatic ductal adenocarcinoma. World J Gastroenterol. 2010;16:2881-2888. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 64] [Cited by in RCA: 81] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 56. | Ochi A, Nguyen AH, Bedrosian AS, Mushlin HM, Zarbakhsh S, Barilla R, Zambirinis CP, Fallon NC, Rehman A, Pylayeva-Gupta Y, Badar S, Hajdu CH, Frey AB, Bar-Sagi D, Miller G. MyD88 inhibition amplifies dendritic cell capacity to promote pancreatic carcinogenesis via Th2 cells. J Exp Med. 2012;209:1671-1687. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 205] [Cited by in RCA: 242] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 57. | Bracci PM. Oral Health and the Oral Microbiome in Pancreatic Cancer: An Overview of Epidemiological Studies. Cancer J. 2017;23:310-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 58. | Sethi V, Vitiello GA, Saxena D, Miller G, Dudeja V. The Role of the Microbiome in Immunologic Development and its Implication For Pancreatic Cancer Immunotherapy. Gastroenterology. 2019;156:2097-2115.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 79] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 59. | Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M, Honda K. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1875] [Cited by in RCA: 2138] [Article Influence: 178.2] [Reference Citation Analysis (2)] |
| 60. | Bazewicz CG, Dinavahi SS, Schell TD, Robertson GP. Aldehyde dehydrogenase in regulatory T-cell development, immunity and cancer. Immunology. 2019;156:47-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 61. | Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3108] [Cited by in RCA: 3614] [Article Influence: 278.0] [Reference Citation Analysis (0)] |
| 62. | Protti MP, De Monte L, Di Lullo G. Tumor antigen-specific CD4+ T cells in cancer immunity: from antigen identification to tumor prognosis and development of therapeutic strategies. Tissue Antigens. 2014;83:237-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 47] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 63. | Saito T, Nishikawa H, Wada H, Nagano Y, Sugiyama D, Atarashi K, Maeda Y, Hamaguchi M, Ohkura N, Sato E, Nagase H, Nishimura J, Yamamoto H, Takiguchi S, Tanoue T, Suda W, Morita H, Hattori M, Honda K, Mori M, Doki Y, Sakaguchi S. Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat Med. 2016;22:679-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 471] [Cited by in RCA: 657] [Article Influence: 73.0] [Reference Citation Analysis (0)] |
| 64. | Tanaka A, Sakaguchi S. Regulatory T cells in cancer immunotherapy. Cell Res. 2017;27:109-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 773] [Cited by in RCA: 1357] [Article Influence: 150.8] [Reference Citation Analysis (0)] |
| 65. | Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Jian M, Zhou Y, Li Y, Zhang X, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J; MetaHIT Consortium, Bork P, Ehrlich SD, Wang J. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59-65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9101] [Cited by in RCA: 7837] [Article Influence: 522.5] [Reference Citation Analysis (4)] |