Published online Nov 15, 2020. doi: 10.4251/wjgo.v12.i11.1255
Peer-review started: June 12, 2020
First decision: July 21, 2020
Revised: August 6, 2020
Accepted: September 25, 2020
Article in press: September 25, 2020
Published online: November 15, 2020
Processing time: 153 Days and 1.9 Hours
The exact regulation network of programmed death 1 (PD-1), programmed death ligand 1 (PD-L1), and programmed death ligand 2 (PD-L2) signaling in immune escape is largely unknown. We aimed to describe the gene expression profiles related to PD-1 as well as its ligands PD-L1 and PD-L2, thus deciphering their possible biological processes in hepatocellular carcinoma (HCC).
To find the possible mechanism of function of PD-1, PD-L1, and PD-L2 in HCC.
Based on the expression data of HCC from The Cancer Genome Atlas, the PD-1/PD-L1/PD-L2 related genes were screened by weighted correlation network analysis method and the biological processes of certain genes were enriched. Relation of PD1/PD-L1/PD-L2 with immune infiltration and checkpoints was investigated by co-expression analysis. The roles of PD-1/PD-L1/PD-L2 in determination of clinical outcome were also analyzed.
Mutations of calcium voltage-gated channel subunit alpha1 E, catenin beta 1, ryanodine receptor 2, tumor suppressor protein p53, and Titin altered PD-1/PD-L1/PD-L2 expression profiles in HCC. PD-1, PD-L1, and PD-L2 related genes were mainly enriched in biological procedures of T cell activation, cell adhesion, and other important lymphocyte effects. In addition, PD-1/PD-L1/PD-L2 was related with immune infiltration of CD8 T cells, cytotoxic lymphocytes, fibroblasts, and myeloid dendritic cells. Immune checkpoints of CTLA4, CD27, CD80, CD86, and CD28 were significantly related to the PD-1/PD-L1/PD-L2 axis. Clinically, PD-1 and PD-L2 expression was correlated with recurrence (P = 0.005 for both), but there was no significant correlation between their expression and HCC patient survival.
Mutations of key genes influence PD-1, PD-L1, and PD-L2 expression. PD-1, PD-L1, and PD-L2 related genes participate in T cell activation, cell adhesion, and other important lymphocyte effects. The finding that PD-1/PD-L1/PD-L2 is related to immune infiltration and other immune checkpoints would expand our understanding of promising anti-PD-1 immunotherapy.
Core Tip: This study mainly investigated the relationship between programmed death 1 (PD-1)/programmed death ligand 1 (PD-L1)/programmed death ligand 2 (PD-L2) and hepatocellular carcinoma, and explored the possible mechanism of PD-1/PD-L1/PD-L2 in this malignancy.
- Citation: Sheng QJ, Tian WY, Dou XG, Zhang C, Li YW, Han C, Fan YX, Lai PP, Ding Y. Programmed death 1, ligand 1 and 2 correlated genes and their association with mutation, immune infiltration and clinical outcomes of hepatocellular carcinoma. World J Gastrointest Oncol 2020; 12(11): 1255-1271
- URL: https://www.wjgnet.com/1948-5204/full/v12/i11/1255.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v12.i11.1255
In the past decade, the tumor immunotherapy targeting critical immune checkpoints has tremendously changed the anti-cancer therapy[1]. One of the most effective immune checkpoints is programmed death 1 (PD-1) and its ligand programmed death ligand 1 (PD-L1)[2]. The interaction of PD-1 expressed on T cells with PD-L1 on the membrane of cancer cells leads to T cell exhaustion and inhibits subsequent immune reaction, thus bypassing immune surveillance[3,4]. Besides, it has been found that programmed death ligand 2 (PD-L2) is another ligand that interacts with PD1, which suppresses T cell proliferation and cytokine release[5]. Until now, the specific mechanism of PD-L1 and PD-L2 regulation in tumor immune escape remains largely elusive[6].
Hepatocellular carcinoma (HCC), one of the most frequent malignant tumor of the digestive tract, remains a leading cause of cancer-related death worldwide with limited therapeutic regimens[7,8]. It has been proved that HCC is caused mainly by hepatitis virus infection, alcohol drinking, drug abuse, and unhealthy habits[9]. In addition to surgery, chemical therapy, and liver transplantation, a number of molecular therapy regimens have been approved for treating advanced HCC, such as sorafenib and lenvatinib[10]. Recently, immune checkpoint blockade therapy targeting essential immune markers including PD-1, PD-L1 and CTLA-4 has been approved for HCC in which chemotherapeutics are ineffective[11-13].
Previously, the effect and mechanisms of antibodies to PD-1 and its ligands have been investigated in HCC therapy by a number of studies. It has been reported that CD8 positive T cells promoted PD-L1 expression on HCC cells in a IFN-γ-dependent manner, which in turn leads to apoptosis of CD8(+) T cells[14,15]. PD-1 expression was significantly elevated in CD8+ and CD4+ T cells obtained from HCC tissue compared with control tissue as well as blood. Antibodies to PD-L1, TIM3, or LAG3 elicit reactions of HCC-derived T cells against tumor antigens, which might become essential treatment strategies for HCC[16]. In addition, PD-1 expression in HCC has also been suggested to increase tumor proliferation independently of adaptive immunity via interacting with downstream target of rapamycin effectors eukaryotic initiation factor 4E and ribosomal protein S6. Moreover, PD-1 checkpoint blockade in combination with rapamycin inhibitor resulted in more durable and synergistic tumor regression[17,18]. For HCC prognosis, it was suggested that increased expression of PD-L1 instead of PD-L2 predicted an unfavorable survival in HCC patients[19].
Although satisfactory effect of anti-PD-1/PD-L1 therapy has been observed in several types of cancers, the potential complicated interaction network of PD-1/PD-L1/PD-L2 related genes in immune escape and immune surveillance remains unclear. In the present study, we analyzed the gene expression profiles associated with PD-1 and its ligands PD-L1 and PD-L2, deciphered the possible biological processes of the identified genes based on transcriptional data of HCC from The Cancer Genome Atlas (TCGA), explored the influence of PD-1 and its ligands on immune cell infiltration and other immune checkpoints, and investigated the prognostic potential of PD-1, PD-L1, and PD-L2 in HCC to explore their prognostic potential as predictors of survival.
The RNA expression, copy-number variants, and clinical information of HCC individuals of TCGA datasets were obtained via UCSC XENA (https://xena.ucsc.edu/). We marked gene expression data as transcripts per million reads (TPM). Clinical information included age, gender, tumor stage, recurrence, and survival time. We explored the association of PD-1, PD-L1, and PD-L2 expression with these clinical parameters.
Weighted correlation network analysis (WGCNA)[20] represents a method to identify gene-gene interactions considering the weighted aspect. Co-expression genes identified by the WGCNA method can generate more specific and robust results. Through WGCNA exploration, we analyzed the genes co-expressed with PD-1, PD-L1, and PD-L2. Gene expression variation was evaluated via median absolute deviation (MAD). After identification of the interaction genes, protein-protein interaction investigation was performed to identify the interaction of genes with STRING
Multiple studies have reported that immune cell infiltration was related to tumor in many aspects. MCP-counter is a R package to evaluate immune infiltration of individuals[23]. Considering the matrix of gene expression, it generates CD3 + T cells, B lymphocytes, cytotoxic lymphocytes, NK cells, CD8 + T cells, cells derived from monocytes, myeloid dendritic cells, neutrophils, endothelial cells, and fibroblasts for each sample. Thus, the relation of PD-1, PD-L1, and PD-L2 with immune infiltration was investigated. Since immune checkpoints were the key indicators of immune status, we then explored the relation of PD-1/PD-L1/PD-L2 with immune checkpoints.
In the present study, we performed statistical analyses via R language
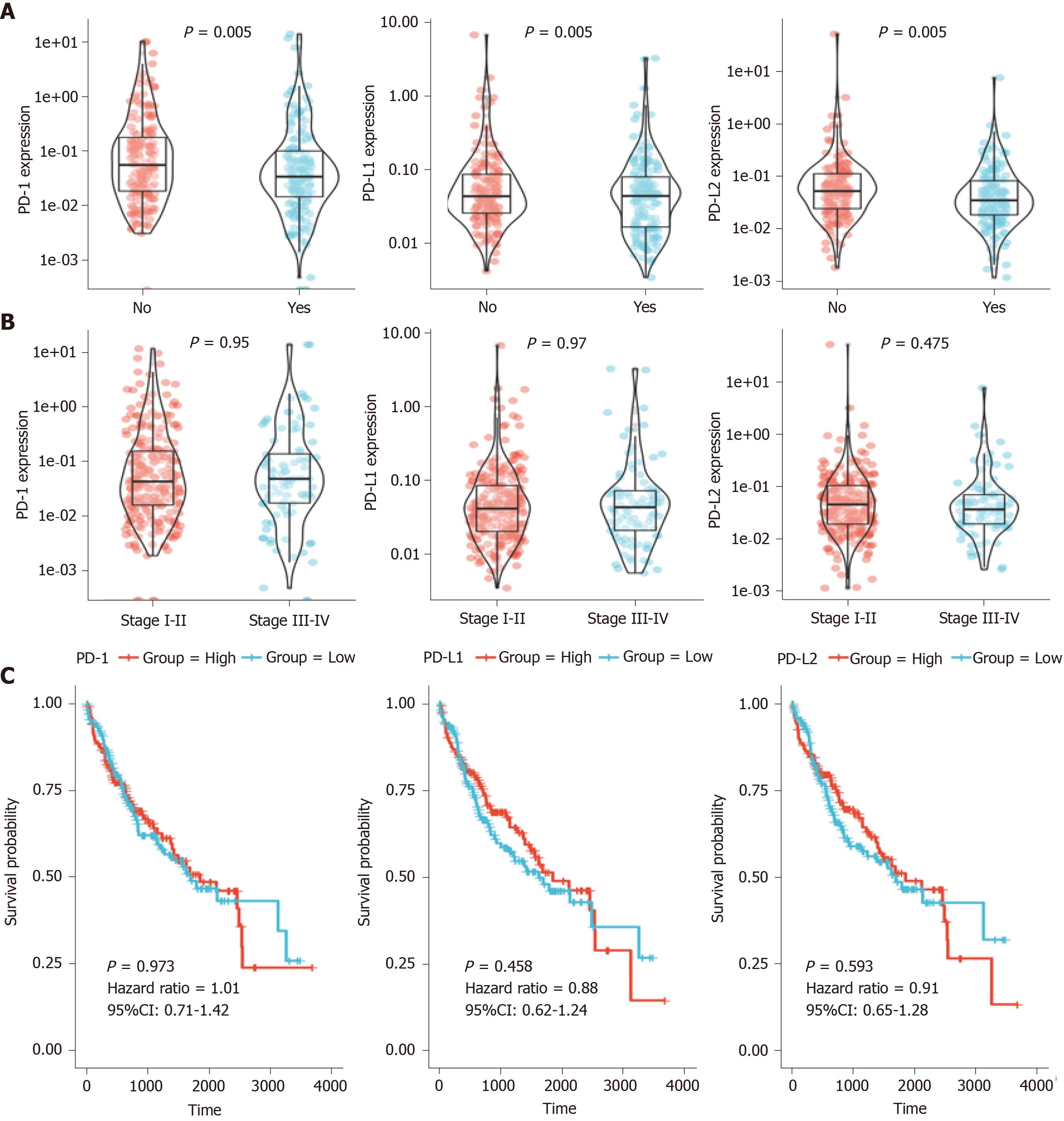
After the preliminary screening, we finally included 374 HCC patients from the TCGA database for the following analysis. Using TCGA datasets, we analyzed the PD-1/PD-L1/PD-L2 expression in different groups according to the clinical data. As shown in Figure 1A, PD-1 and PD-L2 expression was correlated with the recurrence events of HCC patients (P = 0.005), while the expression of PD-L1 showed no significant association with recurrence events (P = 0.155). Moreover, all of the three genes showed no correlation with clinical stage (P = 0.95, 0.97, and 0.475, respectively) (Figure 1B).
Survival of the HCC patients showed no significant difference in the overall survival (OS) analysis of patients with high PD-1/PD-L1/PD-L2 expression (defined as over median expression) or low PD-1/PD-L1/PD-L2 expression (defined as below median expression) with a hazard ratio (HR) of 1.01, 0.88, and 0.91, respectively (P > 0.05) (Figure 1C).
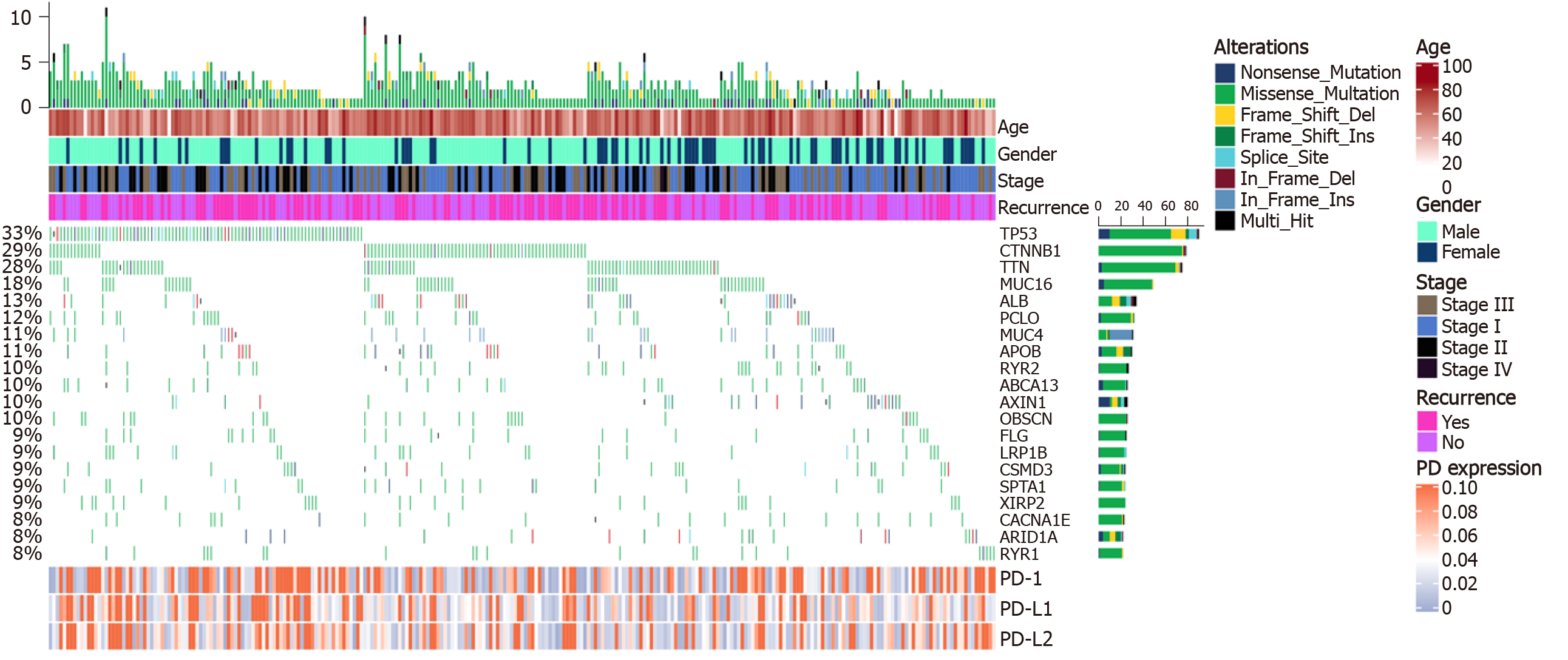
Copy number variants of 270 patients derived from the TCGA database were analyzed. A total of 20 mutations with high occurrence were selected (Figure 2). Expression of PD-1/PD-L1/PD-L2 was not directly correlated to the total mutation load of each patient (r = 0.02/0.07/0.04, respectively). However, after differential expression analysis, mutations of genes including calcium voltage-gated channel subunit alpha1 E (CACNA1E, P = 0.046), catenin beta 1 (CTNNB1, P = 0.020), ryanodine receptor 2 (RYR2, P = 0.030), tumor suppressor protein p53 (TP53, P = 0.016), and Titin (TTN, P = 0.014) could alter PD-1 expression; TP53 mutations (P = 0.003) correlated with high PD-L1 expression; TP53 (P = 0.041) and TTN mutations (P = 0.028) were associated with PD-L2 expression (Table 1).
| Gene | Mutation | PD-1 (mean ± SD) | PD-1 P value | PD-L1 (mean ± SD) | PD-L1 P value | PD-L2 (mean ± SD) | PD-L2 P value |
| ABCA13 | NO | 0.38 ± 1.47 | 0.14 ± 0.54 | 0.34 ± 3.31 | |||
| Yes | 0.3 ± 0.69 | 0.675 | 0.1 ± 0.21 | 0.431 | 0.08 ± 0.1 | 0.705 | |
| ALB | NO | 0.38 ± 1.46 | 0.14 ± 0.53 | 0.33 ± 3.26 | |||
| Yes | 0.11 ± 0.23 | 0.242 | 0.12 ± 0.27 | 0.545 | 0.11 ± 0.27 | 0.121 | |
| APOB | NO | 0.39 ± 1.46 | 0.14 ± 0.53 | 0.34 ± 3.27 | |||
| Yes | 0.04 ± 0.05 | 0.102 | 0.05 ± 0.03 | 0.858 | 0.05 ± 0.04 | 0.835 | |
| ARID1A | NO | 0.37 ± 1.43 | 0.14 ± 0.52 | 0.33 ± 3.22 | |||
| Yes | 0.59 ± 1.03 | 0.316 | 0.1 ± 0.14 | 0.699 | 0.13 ± 0.17 | 0.495 | |
| AXIN1 | NO | 0.37 ± 1.43 | 0.14 ± 0.52 | 0.32 ± 3.2 | |||
| Yes | 0.16 ± 0.08 | 0.206 | 0.06 ± 0.02 | 0.418 | 0.03 ± 0 | 0.741 | |
| CACNA1E | NO | 0.39 ± 1.48 | 0.14 ± 0.53 | 0.34 ± 3.32 | |||
| Yes | 0.13 ± 0.44 | 0.046 | 0.09 ± 0.26 | 0.083 | 0.07 ± 0.17 | 0.076 | |
| CSMD3 | NO | 0.35 ± 1.42 | 0.14 ± 0.53 | 0.34 ± 3.29 | |||
| Yes | 0.67 ± 1.49 | 0.286 | 0.07 ± 0.09 | 0.981 | 0.12 ± 0.14 | 0.633 | |
| CTNNB1 | NO | 0.43 ± 1.61 | 0.17 ± 0.6 | 0.42 ± 3.75 | |||
| Yes | 0.22 ± 0.75 | 0.02 | 0.06 ± 0.1 | 0.428 | 0.07 ± 0.09 | 0.554 | |
| FLG | NO | 0.39 ± 1.48 | 0.1 ± 0.27 | 0.13 ± 0.55 | |||
| Yes | 0.22 ± 0.52 | 0.78 | 0.49 ± 1.53 | 0.893 | 2.34 ± 10.8 | 0.204 | |
| LRP1B | NO | 0.39 ± 1.48 | 0.14 ± 0.54 | 0.34 ± 3.32 | |||
| Yes | 0.2 ± 0.41 | 0.519 | 0.1 ± 0.17 | 0.554 | 0.12 ± 0.3 | 0.886 | |
| MUC16 | NO | 0.32 ± 1.31 | 0.14 ± 0.55 | 0.35 ± 3.47 | |||
| Yes | 0.64 ± 1.93 | 0.697 | 0.13 ± 0.3 | 0.521 | 0.18 ± 0.53 | 0.997 | |
| MUC4 | NO | 0.38 ± 1.44 | 0.14 ± 0.52 | 0.33 ± 3.23 | |||
| Yes | 0.13 ± 0.3 | 0.196 | 0.1 ± 0.11 | 0.641 | 0.08 ± 0.1 | 0.753 | |
| OBSCN | NO | 0.4 ± 1.49 | 0.1 ± 0.27 | 0.14 ± 0.55 | |||
| Yes | 0.15 ± 0.42 | 0.267 | 0.46 ± 1.46 | 0.087 | 2.13 ± 10.37 | 0.518 | |
| PCLO | NO | 0.33 ± 1.3 | 0.14 ± 0.54 | 0.35 ± 3.36 | |||
| Yes | 0.75 ± 2.29 | 0.712 | 0.1 ± 0.22 | 0.422 | 0.1 ± 0.17 | 0.974 | |
| RYR1 | NO | 0.34 ± 1.29 | 0.14 ± 0.54 | 0.34 ± 3.31 | |||
| Yes | 0.74 ± 2.59 | 0.101 | 0.05 ± 0.04 | 0.872 | 0.06 ± 0.05 | 0.705 | |
| RYR2 | NO | 0.36 ± 1.41 | 0.13 ± 0.5 | 0.34 ± 3.34 | |||
| Yes | 0.47 ± 1.58 | 0.03 | 0.24 ± 0.67 | 0.113 | 0.12 ± 0.2 | 0.321 | |
| SPTA1 | NO | 0.39 ± 1.48 | 0.14 ± 0.54 | 0.33 ± 3.31 | |||
| Yes | 0.18 ± 0.4 | 0.737 | 0.1 ± 0.21 | 0.201 | 0.15 ± 0.36 | 0.384 | |
| TP53 | NO | 0.3 ± 1.22 | 0.08 ± 0.18 | 0.1 ± 0.28 | |||
| Yes | 0.65 ± 2.04 | 0.016 | 0.35 ± 1.08 | 0.003 | 1.19 ± 7.04 | 0.041 | |
| TTN | NO | 0.45 ± 1.62 | 0.15 ± 0.59 | 0.4 ± 3.66 | |||
| Yes | 0.14 ± 0.38 | 0.014 | 0.08 ± 0.18 | 0.291 | 0.08 ± 0.2 | 0.028 | |
| XIRP2 | NO | 0.31 ± 1.12 | 0.13 ± 0.51 | 0.31 ± 3.31 | |||
| Yes | 1.02 ± 3.15 | 0.26 | 0.19 ± 0.63 | 0.235 | 0.39 ± 1.54 | 0.329 |
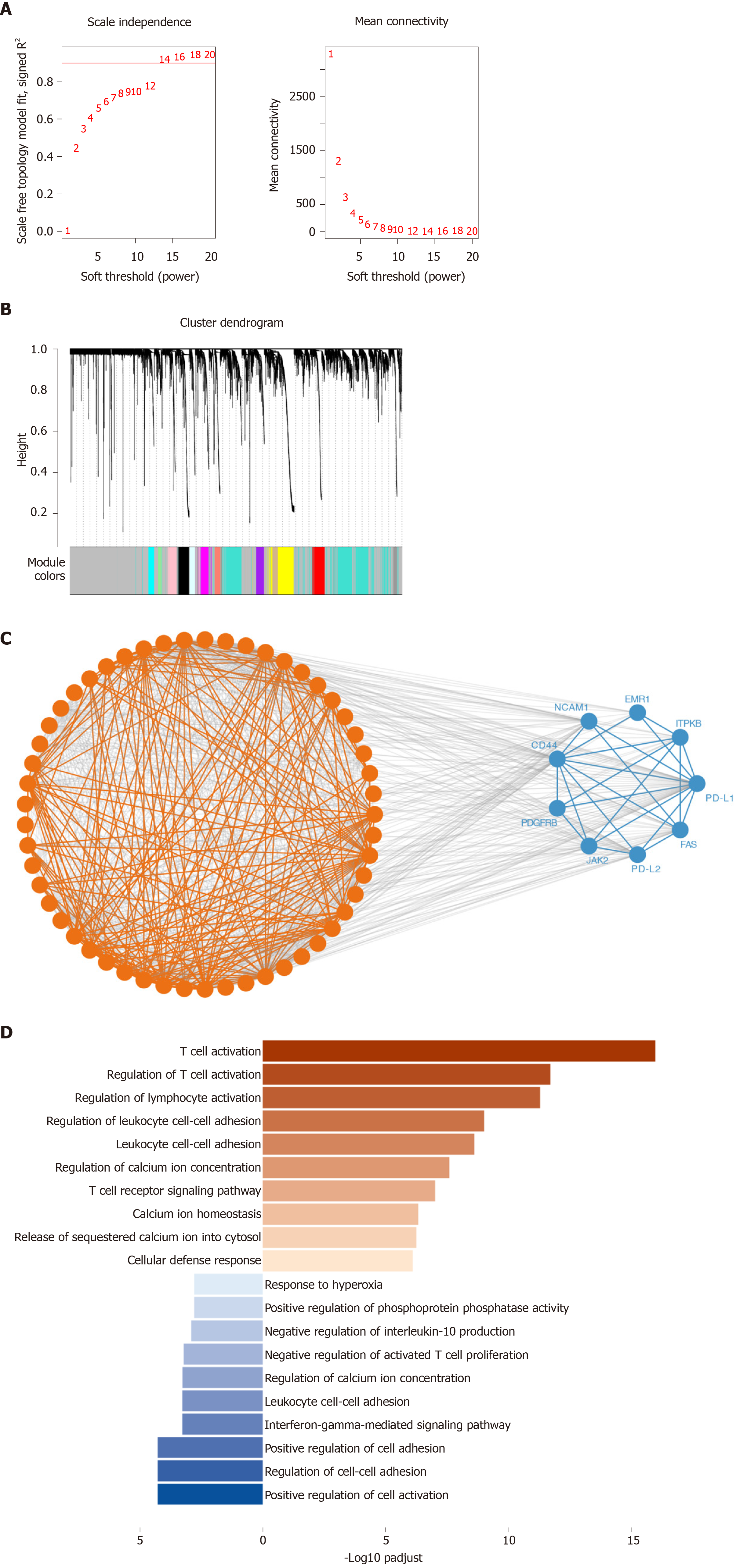
Using WGCNA, we analyzed the co-expressed genes with PD-1, PD-L1, and PD-L2. The connectivity among genes had a scale-free network distribution when the value of soft thresholding power β equals to 14 (Figure 3A). Altogether 22 modules were obtained according to WGCNA analysis (Figure 3B). Among these modules, PD-1 belonged to the brown module while PD-L1 and PD-L2 belonged to the blue module. We finally obtained 371 genes that interacted with PD-1 and 747 PD-L1/PD-L2 related genes. Then, we verified the two module interaction in STRING datasets (Figure 3C). After verification, PD-L1/PD-L2 interacted with 7 genes while PD-1 showed co-expression with 39 genes (Supplementary Table 1). Finally, we selected the interacted genes for further enrichment analysis. PD-1 related genes were mainly enriched in biological processes of T cell activation, regulation of T cell activation, regulation of lymphocyte activation, leukocyte cell-cell adhesion, cytosolic calcium ion concentration, T cell receptor signaling pathway, calcium ion homeostasis, release of sequestered calcium ion into cytosol, and cellular defense response; PD-L1/PD-L2 related genes were enriched in cellular defense response, positive regulation of cell activation, regulation of cell-cell adhesion, interferon-gamma-mediated signaling pathway, negative regulation of activated T cell proliferation, interleukin-10 production, and response to hyperoxia (Figure 3D and Table 2).
| ID | Description | Gene ratio | P value | P adjust | Gene ID | Count |
| GO: 0042110 | T cell activation | 16/27 | 1.23 × 10-19 | 1.10 × 10-16 | PTPN6/CD3E/LCK/PDCD1/CD3G/IDO1/TNFRSF4/TIGIT/PTPRC/CD40LG/ | 16 |
| GO: 0050863 | Regulation of T cell activation | 12/27 | 4.55 × 10-15 | 2.05 × 10-12 | PTPN6/CD3E/LCK/PDCD1/IDO1/TIGIT/PTPRC/CD40LG/ | 12 |
| GO: 0051249 | Regulation of lymphocyte activation | 13/27 | 2.40 × 10-14 | 5.40 × 10-12 | PTPN6/CD3E/LCK/PDCD1/IDO1/TNFRSF4/TIGIT/PTPRC/ | 13 |
| GO: 1903037 | Regulation of leukocyte cell-cell adhesion | 10/27 | 5.69 × 10-12 | 1.02 × 10-9 | PTPN6/CD3E/LCK/PDCD1/IDO1/TIGIT/PTPRC/CD40LG/CD5/ITGA4 | 10 |
| GO: 0007159 | Leukocyte cell-cell adhesion | 10/27 | 1.68 × 10-11 | 2.53 × 10-9 | PTPN6/CD3E/LCK/PDCD1/IDO1/TIGIT/PTPRC/CD40LG/CD5/ITGA4 | 10 |
| GO: 0007204 | Positive regulation of cytosolic calcium ion concentration | 9/27 | 3.25 × 10-10 | 2.66 × 10-8 | PTPN6/LCK/CD19/PTPRC/CXCR3/FASLG/CXCR4/CD52/CXCL9 | 9 |
| GO: 0050852 | T cell receptor signaling pathway | 9/27 | 1.56 × 10-9 | 1.00 × 10-7 | PTPN6/CD3E/LCK/HLA-DQB1/CD3G/PTPRC/CD247 | 7 |
| GO: 0055074 | Calcium ion homeostasis | 9/27 | 1.08 × 10-8 | 4.87 × 10-7 | PTPN6/LCK/CD19/PTPRC/CXCR3/FASLG/CXCR4/CD52/CXCL9 | 9 |
| GO: 0051209 | Release of sequestered calcium ion into cytosol | 6/27 | 1.42 × 10-8 | 5.81 × 10-7 | PTPN6/LCK/CD19/PTPRC/FASLG/CXCL9 | 6 |
| GO: 0006968 | Cellular defense response | 5/27 | 2.52 × 10-8 | 8.11 × 10-7 | TNFRSF4/CD19/PRF1/CXCL9/SH2D1A | 5 |
| GO: 0050867 | Positive regulation of cell activation | 5/8 | 2.00 × 10-7 | 5.23 × 10-5 | PDCD1LG2/JAK2/CD274/ITPKB/PDGFRB | 5 |
| GO: 0022407 | Regulation of cell-cell adhesion | 5/8 | 2.11 × 10-7 | 5.23 × 10-5 | PDCD1LG2/JAK2/CD44/CD274/ITPKB | 5 |
| GO: 0045785 | Positive regulation of cell adhesion | 5/8 | 2.36 × 10-7 | 5.23 × 10-5 | PDCD1LG2/JAK2/CD44/CD274/ITPKB | 5 |
| GO: 0060333 | Interferon-gamma-mediated signaling pathway | 3/8 | 6.13 × 10-6 | 0.000528087 | JAK2/CD44/NCAM1 | 3 |
| GO: 0007159 | Leukocyte cell-cell adhesion | 4/8 | 6.35 × 10-6 | 0.000528087 | PDCD1LG2/CD44/CD274/ITPKB | 4 |
| GO: 2001269 | Positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 2/8 | 7.36 × 10-6 | 0.000543547 | JAK2/FAS | 2 |
| GO: 0046007 | Negative regulation of activated T cell proliferation | 2/8 | 8.99 × 10-6 | 0.000597772 | PDCD1LG2/CD274 | 2 |
| GO: 0032693 | Negative regulation of interleukin-10 production | 2/8 | 2.22 × 10-5 | 0.001230174 | PDCD1LG2/CD274 | 2 |
| GO: 0032516 | Positive regulation of phosphoprotein phosphatase activity | 2/8 | 3.42 × 10-5 | 0.001626763 | JAK2/PDGFRB | 2 |
| GO: 0055093 | Response to hyperoxia | 2/8 | 3.42 × 10-5 | 0.001626763 | FAS/PDGFRB | 2 |
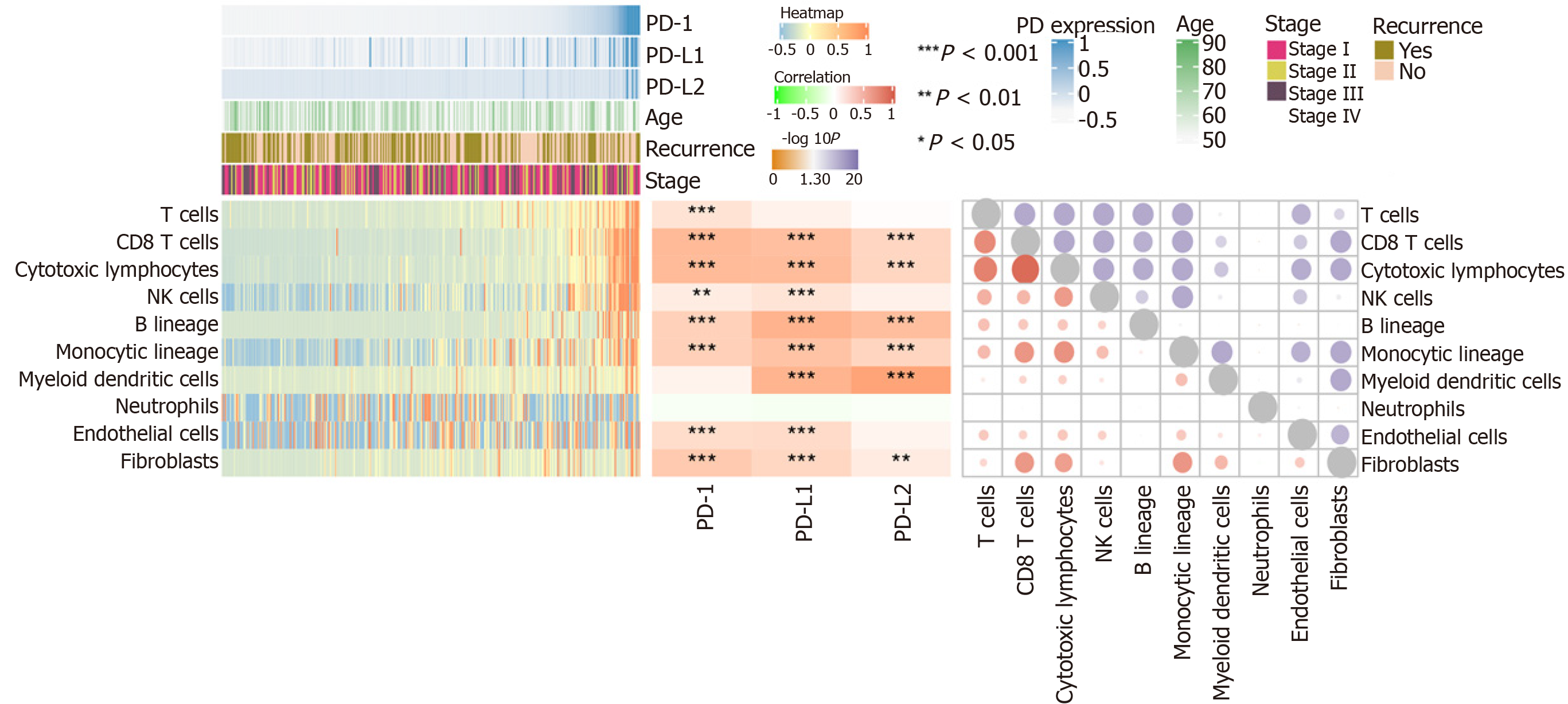
Using MCP-counter, we evaluated the profiles of immune infiltration among various subtypes and stages of HCC (Figure 4). The heatmap in Figure 4 (middle) shows the associations of PD-1, PD-L1, and PD-L2 with immune cell populations according to analysis of the transcriptomic data. The results indicated that PD-1 was mainly related with CD8 T cells (r = 0.608) and cytotoxic lymphocytes (r = 0.60); PD-L1 was mainly related with fibroblasts (r = 0.671); PD-L2 showed a significant correlation with myeloid dendritic cells (r = 0.805). Moreover, we also presented the correlation among different infiltrated immune cell types. As shown in the right correlation heatmap in Figure 4, CD8 T cells were associated with cytotoxic lymphocytes (r = 0.93).
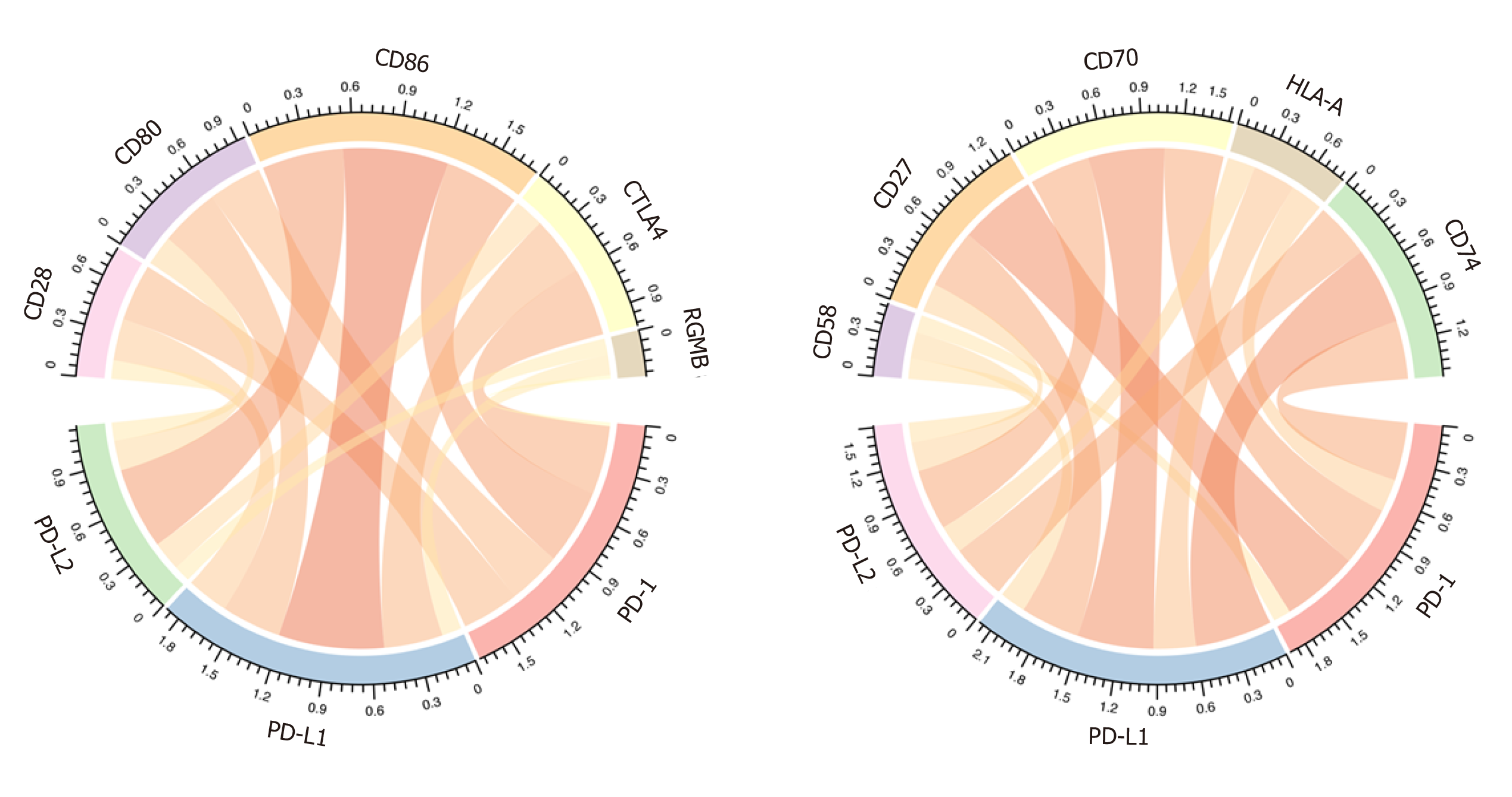
As previous reported, the immune checkpoints of the PD1/PD-L1/PDL2 regulatory axis mainly included CD28, CD80, CD86, CTLA4, RGMB, CD58, CD27, CD70, HLA-A, and CD74. We then analyzed the correlation between PD-1/PD-L1/PD-L2 expression and these important immune checkpoints. As shown in Figure 5 and Table 3, PD-1 was mainly associated with CTLA4 (r = 0.828) and CD27 (r = 0.855), PD-L1 correlated with CD80 (r = 0.675) and CD86 (r = 0.695), and PD-L2 correlated with CD86 (r = 0.797) and CD28 (r = 0.714).
| PD-1 r | PD-1 padj | PD-L1 r | PD-L1 padj | PD-L2 r | PD-L2 padj | |
| CD28 | 0.57465488 | 4.28 × 10-34 | 0.6660232 | 9.08 × 10-49 | 0.71414661 | 6.86 × 10-59 |
| CD80 | 0.63623359 | 1.59 × 10-43 | 0.67510422 | 2.19 × 10-50 | 0.70070417 | 4.16 × 10-56 |
| CD86 | 0.72311298 | 2.38 × 10-61 | 0.69521618 | 2.70 × 10-54 | 0.79715204 | 1.59 × 10-82 |
| CTLA4 | 0.82827652 | 6.53 × 10-95 | 0.53400839 | 9.77 × 10-29 | 0.62325084 | 2.09 × 10-41 |
| RGMB | 0.08079832 | 0.11878686 | 0.31356282 | 5.62 × 10-10 | 0.15655839 | 0.00239497 |
| CD58 | 0.4020193 | 6.49 × 10-16 | 0.44699953 | 1.00 × 10-19 | 0.34510312 | 7.46 × 10-12 |
| CD27 | 0.8554091 | 2.51 × 10-107 | 0.564041 | 1.70 × 10-32 | 0.70361681 | 1.23 × 10-56 |
| CD70 | 0.73757481 | 6.84 × 10-65 | 0.49791513 | 1.14 × 10-24 | 0.5810653 | 5.35 × 10-35 |
| HLA-A | 0.49005962 | 6.82 × 10-24 | 0.45628329 | 1.56 × 10-20 | 0.47535158 | 2.20 × 10-22 |
| CD74 | 0.62377393 | 1.71 × 10-41 | 0.57693716 | 3.59 × 10-34 | 0.68544635 | 6.61 × 10-53 |
Previous clinical trials indicated that immune checkpoint blockade therapies demonstrated satisfactory curative effect for multiple types of cancer, with persistent responses and acceptable toxicity[3]. However, only part of individuals who received antibody immunotherapy greatly benefit from the treatment[26]. It is therefore urgent to unravel the underlying signaling pathway of important immune checkpoints such as PD-1 and its ligands PD-L1 and PD-L2 to improve the therapeutic sensitivity. Here, we presented an integrative analysis of PD-1, PD-L1, and PD-L2 based on the HCC data from TCGA. We identified potential molecular and genetic alterations correlating with the PD-1/PD-L1/PD-L2 and revealed their biological functions. The relation of PD-1/PD-L1/PD-L2 with immune infiltration and HCC survival were also explored to elucidate their role as prognostic biomarkers.
We first described the PD-1/PD-L1/PD-L2 expression profiles in different clinical subtypes. The results indicated that PD-1 and PD-L2 expression was associated with the recurrence events of HCC patients. However, none of the three genes showed a significant correlation with clinical stage. In a previous research of 217 HCC patients, PD-L1 expression demonstrated a significant relation with multiple markers of cancer aggressiveness including satellite nodules, macrovascular invasion, microvascular invasion, and poor differentiation[27]. Besides, PD-1 and PD-L1 expression was upregulated in HCC tissues compared with adjacent normal tissues, which was positively related with the clinical stage and lymph node metastasis, but negatively related with the survival of HCC patients[28]. These results suggested that the PD-1/PD-L1/PD-L2 axis might correlate with multiple clinical parameters of HCC.
Prognostic analysis of the HCC patients showed no significant association between PD-1/PD-L1/PD-L2 expression and the OS of HCC patients. In a study of 85 HCC patients, PD-L1 or PD-L2 expression was associated with a poor survival according to immunohistochemical investigation[29-31]. Elevated PD-L1 expression has been reported to be an independent adverse prognosis predictor of disease-free survival in addition to previously reported factors[32]. One study performing immunohistochemical staining of 136 HCC tissues demonstrated that PD-L1 high expression exhibited a significant correlation with clinical and pathological parameters indicating worse HCC progression and prognosis[33]. In another study of PD-L1 in HCC, however, PD-L1 expression was inversely correlated with P53 and associated with a longer survival of patients with HCC[34]. There was also a study reporting that PD-L1 expression failed to have a markedly significant prognostic association with survival in 143 patients with HCC[35]. According to the analysis of TCGA data and previous studies on PD-1/PD-L1/PD-L2, the relation of PD-1/PD-L1/PD-L2 expression with HCC prognosis still requires to be confirmed in future studies with large samples.
Based on the copy number variations and mutation analysis, a total of 20 mutations with high occurrence were selected. Expression of PD-1/PD-L1/PD-L2 was not directly correlated to the total mutation load of each patient. After differential expression analysis, however, mutations of genes including CACNA1E, CTNNB1, RYR2, TP53, and TTN could alter PD-1 expression; TP53 mutations correlated with high PD-L1 expression, and TP53 and TTN mutations were associated with PD-L2 expression. Interestingly, TP53 mutation significantly increased the expression of PD-1, PD-L1, and PD-L2, suggesting a probable molecular link between TP53 and PD-1 axis regulation. Admittedly, change of PD-1-PD-L1 immune regulator and p53 alternation promote cancer development in activated B-cell diffuse large B-cell lymphomas[36]. In addition, the prognosis of Kras-p53-associated lung cancer is regulated by MEK and PD-1/PD-L1 immune checkpoint[37-40]. The underlying mechanisms of critical mutations such as TP53, TTN, and CTNNB1 mutations in modulating PD-1 signaling might provide novel strategies for immunotherapies. In the future, we may be able to perform different immunotherapies by stratifying patients based on the mutation types of these genes.
Using WGCNA, we next analyzed the co-expressed genes with PD-1, PD-L1, and PD-L2. After verification, PD-L1/PD-L2 interacted with 7 genes while PD-1 showed co-expression with 39 genes. PD-1 related genes were mainly enriched in T cell activation, lymphocyte activation, leukocyte cell-cell adhesion, cytosolic calcium ion concentration, T cell receptor signaling pathway, calcium ion homeostasis, release of sequestered calcium ion into cytosol, and cellular defense response; PD-L1/PD-L2 related genes were enriched in cellular defense response, cell activation, cell-cell adhesion, interferon-gamma-mediated signaling pathway, negative regulation of activated T cell proliferation, interleukin-10 production, and response to hyperoxia. As indicated by the enrichment analysis, PD-1/PD-L1/PD-L2 signaling is a key regulator of T cell activation and other important lymphocyte functions, which was consistent with the results of multiple previous investigations[41-44]. It is worthy elucidating the immune modulating mechanisms of PD-1 axis in the future to unravel its specific role in cancer immunity.
Immune cell infiltration reflects the immune microenvironment around the tumor tissues and is reportedly correlated with outcome of cancer progression. We subsequently evaluated PD-1/PD-L1/PD-L2 expression and immune infiltration in HCC, the results of which suggested that PD-1 was correlated with CD8 T cells and cytotoxic lymphocytes, PD-L1 was related with fibroblasts, and PD-L2 was significantly correlated with myeloid dendritic cells. It has been reported that CD8+ cytotoxic T lymphocytes greatly increase PD-L1 expression on cancer cell lines, and PD-L1 expression and CD8+ T-cell density showed a significant positive correlation in HCC patients[45]. Besides, PD-1 and PD-L1 expression was suggested to be significantly related to high levels of CD8+ tumor-infiltrating lymphocytes (TILs)[32,44]. In addition to T lymphocytes, PD-1 expression on dendritic cells has been found to restrict CD8+ T cell function and anti-cancer immunity[46]. Understanding the correlation of PD-1/PD-L1/PD-L2 immune checkpoints with immune cell infiltration might greatly benefit the tumor immunotherapy targeting PD-1 signaling. After analyzing the immune checkpoints of PD1/PD-L1/PDL2 regulatory axis including CD28, CD74, CD86, CD58, CTLA4, RGMB, CD70, CD27, CD80, and HLA-A, we suggested that PD-1 was mainly associated with CD27 and CTLA4, PD-L1 related with CD86 as well as CD80, and PD-L2 related with CD28 and CD86. The combined inhibition of different immune checkpoints might generate more satisfactory clinical outcome. Thus, future studies focusing on PD-1/PD-L1/PD-L2 related immune checkpoints including CTLA4, CD27, CD80, CD86, and CD28 are required to obtain an optimal immunotherapy effect.
Mutations of CACNA1E, CTNNB1, RYR2, TP53, and TTN alter PD-1/PD-L1/PD-L2 expression profiles in HCC. The limitation on the effect of mutations on gene expression is that only statistical differences have been observed so far. We will conduct follow-up research on its detailed mechanism. PD-1/PD-L1/PD-L2 related genes are enriched in the biological processes of T cell activation, cell-cell adhesion, and other important lymphocyte effects. In addition, PD-1/PD-L1/PD-L2 is related to immune infiltration of CD8 T cells, cytotoxic lymphocytes, fibroblasts, and myeloid dendritic cells. Immune checkpoints CTLA4, CD27, CD80, CD86, and CD28 are significantly associated with PD-1/PD-L1/PD-L2 axis. Clinically, PD-1 and PD-L2 expression is correlated with recurrence, but there is no significant correlation between PD-1/PD-L1/PD-L2 expression and survival of HCC patients.
The potential regulating network of programmed death 1 (PD-1)/programmed death ligand 1 (PD-L1)/programmed death ligand 2 (PD-L2) signaling in the immune escape is unclear. We aimed to describe the gene expression profiles related with PD-1 and its ligands PD-L1 and PD-L2 to decipher their possible biological processes in hepatocellular carcinoma (HCC).
Although satisfactory effect of anti-PD-1/PD-L1 therapy has been observed in several types of cancers, the potential complicated interaction network of PD-1/PD-L1/PD-L2 related genes in immune escape and immune surveillance still remains unclear.
The aim of the study was to explore the possible mechanism of function of PD-1, PD-L1, and PD-L2 in HCC.
Based on transcriptional data of HCC from TCGA, PD-1/PD-L1/PD-L2 related genes were screened by weighted correlation network analysis and the biological processes of certain genes were enriched. The relation of PD1/PD-L1/PD-L2 expression with immune infiltration and checkpoints was investigated by co-expression analysis. The role of PD-1/PD-L1/PD-L2 in the determination of clinical outcome was also analyzed.
Mutations of calcium voltage-gated channel subunit alpha1 E (CACNA1E), catenin beta 1 (CTNNB1), ryanodine receptor 2 (RYR2), tumor suppressor protein p53 (TP53), and Titin (TTN) altered PD-1/PD-L1/PD-L2 expression profiles in HCC. PD-1/PD-L1/PD-L2 related genes were mainly enriched in biological processes of T cell activation, cell-cell adhesion, and other important lymphocyte effects. In addition, PD-1/PD-L1/PD-L2 was related with immune infiltration of CD8 T cells, cytotoxic lymphocytes, fibroblasts, and myeloid dendritic cells. Immune checkpoints CTLA4, CD27, CD80, CD86, and CD28 were significantly correlated with PD-1/PD-L1/PD-L2 axis. Clinically, PD-1 and PD-L2 expression was correlated with recurrence (P = 0.005 for both), but there was no significant correlation between PD-1/PD-L1/PD-L2 expression and HCC patient survival.
Mutations of key genes influence PD-1/PD-L1/PD-L2 expression. PD-1/PD-L1/PD-L2 related genes participate in T cell activation, cell-cell adhesion, and other important lymphocyte effects. Correlation of PD-1/PD-L1/PD-L2 with immune infiltration and other immune checkpoints would expand our understanding of promising anti-PD-1 immunotherapy.
Mutations of CACNA1E, CTNNB1, RYR2, TP53, and TTN altered PD-1/PD-L1/PD-L2 expression profiles in HCC. The limitation on the effect of mutations on gene expression is that only statistical differences have been observed so far. We will conduct follow-up research on its detailed mechanism. PD-1/PD-L1/PD-L2 related genes were enriched in biological processes of T cell activation, cell-cell adhesion, and other important lymphocyte effects. In addition, PD-1/PD-L1/PD-L2 was related with immune infiltration of CD8 T cells, cytotoxic lymphocytes, fibroblasts, and myeloid dendritic cells. Immune checkpoints CTLA4, CD27, CD80, CD86, and CD28 were significantly correlated with PD-1/PD-L1/PD-L2 axis. Clinically, PD-1 and PD-L2 expression was correlated with recurrence, but there was no significant correlation between PD-1/PD-L1/PD-L2 and survival of HCC patients.
Manuscript source: Unsolicited manuscript
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Emi M, Lucchesi A S-Editor: Gao CC L-Editor: Wang TQ P-Editor: Li JH
| 1. | Yang Y. Cancer immunotherapy: harnessing the immune system to battle cancer. J Clin Invest. 2015;125:3335-3337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 653] [Cited by in RCA: 994] [Article Influence: 99.4] [Reference Citation Analysis (0)] |
| 2. | Zak KM, Grudnik P, Magiera K, Dömling A, Dubin G, Holak TA. Structural Biology of the Immune Checkpoint Receptor PD-1 and Its Ligands PD-L1/PD-L2. Structure. 2017;25:1163-1174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 265] [Article Influence: 37.9] [Reference Citation Analysis (0)] |
| 3. | Alsaab HO, Sau S, Alzhrani R, Tatiparti K, Bhise K, Kashaw SK, Iyer AK. PD-1 and PD-L1 Checkpoint Signaling Inhibition for Cancer Immunotherapy: Mechanism, Combinations, and Clinical Outcome. Front Pharmacol. 2017;8:561. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1367] [Cited by in RCA: 1243] [Article Influence: 155.4] [Reference Citation Analysis (0)] |
| 4. | Iwai Y, Hamanishi J, Chamoto K, Honjo T. Cancer immunotherapies targeting the PD-1 signaling pathway. J Biomed Sci. 2017;24:26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 346] [Cited by in RCA: 484] [Article Influence: 60.5] [Reference Citation Analysis (0)] |
| 5. | Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol. 2008;26:677-704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3785] [Cited by in RCA: 4140] [Article Influence: 243.5] [Reference Citation Analysis (0)] |
| 6. | Dyck L, Mills KHG. Immune checkpoints and their inhibition in cancer and infectious diseases. Eur J Immunol. 2017;47:765-779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 295] [Cited by in RCA: 404] [Article Influence: 50.5] [Reference Citation Analysis (0)] |
| 7. | Sia D, Villanueva A, Friedman SL, Llovet JM. Liver Cancer Cell of Origin, Molecular Class, and Effects on Patient Prognosis. Gastroenterology. 2017;152:745-761. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 619] [Cited by in RCA: 839] [Article Influence: 104.9] [Reference Citation Analysis (2)] |
| 8. | Yu LX, Schwabe RF. The gut microbiome and liver cancer: mechanisms and clinical translation. Nat Rev Gastroenterol Hepatol. 2017;14:527-539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 414] [Article Influence: 51.8] [Reference Citation Analysis (0)] |
| 9. | Allaire M, Nault JC. Advances in management of hepatocellular carcinoma. Curr Opin Oncol. 2017;29:288-295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 10. | Hartke J, Johnson M, Ghabril M. The diagnosis and treatment of hepatocellular carcinoma. Semin Diagn Pathol. 2017;34:153-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 467] [Article Influence: 58.4] [Reference Citation Analysis (0)] |
| 11. | Iñarrairaegui M, Melero I, Sangro B. Immunotherapy of Hepatocellular Carcinoma: Facts and Hopes. Clin Cancer Res. 2018;24:1518-1524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 182] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 12. | Kudo M. Immune Checkpoint Inhibition in Hepatocellular Carcinoma: Basics and Ongoing Clinical Trials. Oncology. 2017;92 Suppl 1:50-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 140] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 13. | Abad-Belando R, Varas-Lorenzo MJ, Pons-Vilardell C, Puig-Torrus X, Pla-Alcaraz M, Monleón-Getino A, Sánchez-Vizcaíno-Mengual E. Canalization technique to obtain deep tissue biopsy of gastrointestinal subepithelial tumors as an alternative to conventional known techniques. Endosc Ultrasound. 2018;7:184-190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 14. | Shi F, Shi M, Zeng Z, Qi RZ, Liu ZW, Zhang JY, Yang YP, Tien P, Wang FS. PD-1 and PD-L1 upregulation promotes CD8(+) T-cell apoptosis and postoperative recurrence in hepatocellular carcinoma patients. Int J Cancer. 2011;128:887-896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 371] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 15. | Adler DG. Single-operator experience with a 20-mm diameter lumen apposing metal stent to treat patients with large pancreatic fluid collections from pancreatic necrosis. Endosc Ultrasound. 2018;7:422-423. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 16. | Zhou G, Sprengers D, Boor PPC, Doukas M, Schutz H, Mancham S, Pedroza-Gonzalez A, Polak WG, de Jonge J, Gaspersz M, Dong H, Thielemans K, Pan Q, IJzermans JNM, Bruno MJ, Kwekkeboom J. Antibodies Against Immune Checkpoint Molecules Restore Functions of Tumor-Infiltrating T Cells in Hepatocellular Carcinomas. Gastroenterology 2017; 153: 1107-1119. e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 322] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 17. | Li H, Li X, Liu S, Guo L, Zhang B, Zhang J, Ye Q. Programmed cell death-1 (PD-1) checkpoint blockade in combination with a mammalian target of rapamycin inhibitor restrains hepatocellular carcinoma growth induced by hepatoma cell-intrinsic PD-1. Hepatology. 2017;66:1920-1933. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 152] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 18. | Adler DG. EUS-guided gallbladder drainage: Current status and future prospects. Endosc Ultrasound. 2018;7:1-3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 19. | Ma LJ, Feng FL, Dong LQ, Zhang Z, Duan M, Liu LZ, Shi JY, Yang LX, Wang ZC, Zhang S, Ding ZB, Ke AW, Cao Y, Zhang XM, Zhou J, Fan J, Wang XY, Gao Q. Clinical significance of PD-1/PD-Ls gene amplification and overexpression in patients with hepatocellular carcinoma. Theranostics. 2018;8:5690-5702. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 20. | Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10254] [Cited by in RCA: 16471] [Article Influence: 968.9] [Reference Citation Analysis (0)] |
| 21. | Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362-D368. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4345] [Cited by in RCA: 5081] [Article Influence: 564.6] [Reference Citation Analysis (0)] |
| 22. | The Gene Ontology Consortium. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017;45:D331-D338. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1342] [Cited by in RCA: 1382] [Article Influence: 153.6] [Reference Citation Analysis (0)] |
| 23. | Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, Selves J, Laurent-Puig P, Sautès-Fridman C, Fridman WH, de Reyniès A. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17:218. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 987] [Cited by in RCA: 2242] [Article Influence: 249.1] [Reference Citation Analysis (0)] |
| 24. | Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847-2849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3200] [Cited by in RCA: 5883] [Article Influence: 653.7] [Reference Citation Analysis (0)] |
| 25. | Gu Z, Gu L, Eils R, Schlesner M, Brors B. circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811-2812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1625] [Cited by in RCA: 2428] [Article Influence: 220.7] [Reference Citation Analysis (0)] |
| 26. | Curry WT, Lim M. Immunomodulation: checkpoint blockade etc. Neuro Oncol. 2015;17 Suppl 7:vii26-vii31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 27. | Calderaro J, Rousseau B, Amaddeo G, Mercey M, Charpy C, Costentin C, Luciani A, Zafrani ES, Laurent A, Azoulay D, Lafdil F, Pawlotsky JM. Programmed death ligand 1 expression in hepatocellular carcinoma: Relationship With clinical and pathological features. Hepatology. 2016;64:2038-2046. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 260] [Cited by in RCA: 331] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 28. | Long J, Qu T, Pan XF, Tang X, Wan HH, Qiu P, Xu YH. Expression of programmed death ligand-1 and programmed death 1 in hepatocellular carcinoma and its clinical significance. J Cancer Res Ther. 2018;14:S1188-S1192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 29. | Jung HI, Jeong D, Ji S, Ahn TS, Bae SH, Chin S, Chung JC, Kim HC, Lee MS, Baek MJ. Overexpression of PD-L1 and PD-L2 Is Associated with Poor Prognosis in Patients with Hepatocellular Carcinoma. Cancer Res Treat. 2017;49:246-254. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 170] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 30. | Adler DG, Mallery S, Amateau S, Nieto J, Taylor LJ, Siddiqui A. A pilot study of a 20-mm lumen-apposing metal stent to treat pancreatic fluid collections: First reported multicenter use of a new device. Endosc Ultrasound. 2019;8:136-138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 31. | Adler DG, Muthusamy VR, Ehrlich DS, Parasher G, Thosani NC, Chen A, Buscaglia JM, Appannagari A, Quintero E, Aslanian H, Taylor LJ, Siddiqui A. A multicenter evaluation of a new EUS core biopsy needle: Experience in 200 patients. Endosc Ultrasound. 2019;8:99-104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (1)] |
| 32. | Chang H, Jung W, Kim A, Kim HK, Kim WB, Kim JH, Kim BH. Expression and prognostic significance of programmed death protein 1 and programmed death ligand-1, and cytotoxic T lymphocyte-associated molecule-4 in hepatocellular carcinoma. APMIS. 2017;125:690-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 55] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 33. | Hu K, Wang ZM, Li JN, Zhang S, Xiao ZF, Tao YM. CLEC1B Expression and PD-L1 Expression Predict Clinical Outcome in Hepatocellular Carcinoma with Tumor Hemorrhage. Transl Oncol. 2018;11:552-558. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 34. | Kan G, Dong W. The expression of PD-L1 APE1 and P53 in hepatocellular carcinoma and its relationship to clinical pathology. Eur Rev Med Pharmacol Sci. 2015;19:3063-3071. [PubMed] |
| 35. | Pei R, Zhang W, Wang S, Huang X, Zou Y. Prognostic Value of PD-L1 in Patients with Hepatocellular Carcinoma. Clin Lab. 2019;65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 36. | Pascual M, Mena-Varas M, Robles EF, Garcia-Barchino MJ, Panizo C, Hervas-Stubbs S, Alignani D, Sagardoy A, Martinez-Ferrandis JI, Bunting KL, Meier S, Sagaert X, Bagnara D, Guruceaga E, Blanco O, Celay J, Martínez-Baztan A, Casares N, Lasarte JJ, MacCarthy T, Melnick A, Martinez-Climent JA, Roa S. PD-1/PD-L1 immune checkpoint and p53 Loss facilitate tumor progression in activated B-cell diffuse large B-cell lymphomas. Blood. 2019;133:2401-2412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 37. | Lee JW, Zhang Y, Eoh KJ, Sharma R, Sanmamed MF, Wu J, Choi J, Park HS, Iwasaki A, Kaftan E, Chen L, Papadimitrakopoulou V, Herbst RS, Koo JS. The Combination of MEK Inhibitor With Immunomodulatory Antibodies Targeting Programmed Death 1 and Programmed Death Ligand 1 Results in Prolonged Survival in Kras/p53-Driven Lung Cancer. J Thorac Oncol. 2019;14:1046-1060. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 65] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 38. | Ang TL, Li JW, Kwek ABE, Thurairajah PH, Wang LM. The difference in histological yield between 19G EUS-FNA and EUS-fine-needle biopsy needles. Endosc Ultrasound. 2019;8:255-260. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 39. | Antonini F, Delconte G, Fuccio L, De Nucci G, Fabbri C, Armellini E, Frazzoni L, Fornelli A, Magarotto A, Mandelli E, Occhipinti P, Masci E, Manes G, Macarri G. EUS-guided tissue sampling with a 20-gauge core biopsy needle for the characterization of gastrointestinal subepithelial lesions: A multicenter study. Endosc Ultrasound. 2019;8:105-110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 40. | Bhatia V, Dhir V. Radial EUS imaging of the liver: A pictorial guide. Endosc Ultrasound. 2019;8:76-81. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 41. | Cubillos-Zapata C, Avendaño-Ortiz J, Hernandez-Jimenez E, Toledano V, Casas-Martin J, Varela-Serrano A, Torres M, Almendros I, Casitas R, Fernández-Navarro I, Garcia-Sanchez A, Aguirre LA, Farre R, López-Collazo E, García-Rio F. Hypoxia-induced PD-L1/PD-1 crosstalk impairs T-cell function in sleep apnoea. Eur Respir J. 2017;50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 91] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 42. | Hui E, Cheung J, Zhu J, Su X, Taylor MJ, Wallweber HA, Sasmal DK, Huang J, Kim JM, Mellman I, Vale RD. T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science. 2017;355:1428-1433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 860] [Cited by in RCA: 1221] [Article Influence: 152.6] [Reference Citation Analysis (0)] |
| 43. | Seifert AM, Zeng S, Zhang JQ, Kim TS, Cohen NA, Beckman MJ, Medina BD, Maltbaek JH, Loo JK, Crawley MH, Rossi F, Besmer P, Antonescu CR, DeMatteo RP. PD-1/PD-L1 Blockade Enhances T-cell Activity and Antitumor Efficacy of Imatinib in Gastrointestinal Stromal Tumors. Clin Cancer Res. 2017;23:454-465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 116] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 44. | Adler DG, Gabr M, Taylor LJ, Witt B, Pleskow D. Initial report of transesophageal EUS-guided intraparenchymal lung mass core biopsy: Findings and outcomes in two cases. Endosc Ultrasound. 2018;7:413-417. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 45. | Huang CY, Wang Y, Luo GY, Han F, Li YQ, Zhou ZG, Xu GL. Relationship Between PD-L1 Expression and CD8+ T-cell Immune Responses in Hepatocellular Carcinoma. J Immunother. 2017;40:323-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 67] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 46. | Lim TS, Chew V, Sieow JL, Goh S, Yeong JP, Soon AL, Ricciardi-Castagnoli P. PD-1 expression on dendritic cells suppresses CD8+ T cell function and antitumor immunity. Oncoimmunology. 2016;5:e1085146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 141] [Article Influence: 14.1] [Reference Citation Analysis (0)] |