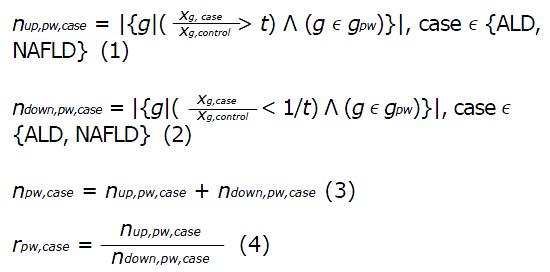Published online Mar 18, 2017. doi: 10.4254/wjh.v9.i8.443
Peer-review started: October 17, 2016
First decision: November 14, 2016
Revised: November 29, 2016
Accepted: December 13, 2016
Article in press: December 14, 2016
Published online: March 18, 2017
Processing time: 152 Days and 11.3 Hours
To compare transcriptomes of non-alcoholic fatty liver disease (NAFLD) and alcoholic liver disease (ALD) in a meta-analysis of liver biopsies.
Employing transcriptome data from patient liver biopsies retrieved from several public repositories we performed a meta-analysis comparing ALD and NAFLD.
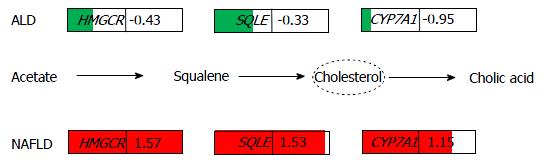
We observed predominating commonalities at the transcriptome level between ALD and NAFLD, most prominently numerous down-regulated metabolic pathways and cytochrome-related pathways and a few up-regulated pathways which include ECM-receptor interaction, phagosome and lysosome. However some pathways were regulated in opposite directions in ALD and NAFLD, for example, glycolysis was down-regulated in ALD and up-regulated in NAFLD. Interestingly, we found rate-limiting genes such as HMGCR, SQLE and CYP7A1 which are associated with cholesterol processes adversely regulated between ALD (down-regulated) and NAFLD (up-regulated). We propose that similar phenotypes in both diseases may be due to a lower level of the enzyme CYP7A1 compared to the cholesterol synthesis enzymes HMGCR and SQLE. Additionally, we provide a compendium of comparative KEGG pathways regulation in ALD and NAFLD.
Our finding of adversely regulated cholesterol processes in ALD and NAFLD draws the focus to regulation of cholesterol secretion into bile. Thus, it will be interesting to further investigate CYP7A1-mediated cholesterol secretion into bile - also as possible drug targets. The list of potential novel biomarkers may assist differential diagnosis of ALD and NAFLD.
Core tip: With a meta-analysis of newly published liver biopsy-derived transcriptome datasets we identified multiple key genes and pathways in common and mutually exclusive in alcoholic liver disease (ALD) and non-alcoholic fatty liver disease (NAFLD). We provide a compendium of comparative regulation for all KEGG pathways in both diseases and propose a list of biomarkers distinguishing both diseases. One surprising finding was that cholesterol metabolism was up-regulated in NAFLD and down-regulated in ALD although leading to the same steatosis phenotype which might be explained by an insufficient conversion rate to bile acids under both conditions.
- Citation: Wruck W, Adjaye J. Meta-analysis reveals up-regulation of cholesterol processes in non-alcoholic and down-regulation in alcoholic fatty liver disease. World J Hepatol 2017; 9(8): 443-454
- URL: https://www.wjgnet.com/1948-5182/full/v9/i8/443.htm
- DOI: https://dx.doi.org/10.4254/wjh.v9.i8.443
Non-alcoholic fatty liver disease (NAFLD) and alcoholic liver disease (ALD) have nearly identical symptoms and in the first report non-alcoholic steatohepatitis (NASH) was described as histologically mimicking alcoholic hepatitis[1]. While the cause of ALD is excessive alcohol, the cause of NAFLD is excessive fat resulting from an imbalance between diet and physical activity often associated with insulin resistance and obesity.
We are working on the hypothesis that alcohol is metabolized to fat and beyond this pathway both diseases share a common phenotype. Therefore we place special emphasis on alcohol metabolism which naturally plays a crucial role in ALD. Associations of variants in alcohol and aldehyde dehydrogenases with alcoholism have already been proposed[2]. Most variants protective against alcoholism result in a higher acetaldehyde level either by accelerating alcohol dehydrogenase (most common variants in ADH1B) metabolizing alcohol to acetaldehyde or by reducing aldehyde dehydrogenase (most common variants in ALDH2) metabolizing acetaldehyde to acetic acid. Acetaldehyde is a carcinogen and causes severe reactions such as flushing, accelerated heart rate and nausea. These severe reactions will impose on most carriers of these variants to abstain from alcohol and thus reduce their risk of becoming alcohol addicts. Furthermore, it has been reported that aldehyde dehydrogenases are down-regulated in alcoholics[3] or animals continually exposed to alcohol had lower ethanol elimination rates[4]. However, this is a matter of debate as no significant down-regulation of aldehyde dehydrogenases was reported by Vidal et al[5] but instead a down-regulation in cirrhotic livers independent of alcoholism. Acetic acid - the product of ethanol metabolism, can be further metabolized by acyl-CoA synthetases (ACSS1 and ACSS2) to acetyl-CoA, the substrate for fatty acid synthesis[6]. The expression and activity of Acyl-CoA synthetases in turn are controlled by the sterol regulatory element-binding protein which has been reported to be activated by ethanol[7].
The progression of NAFLD from mild steatosis up to severe NASH or from ALD to alcoholic hepatitis varies widely between individual patients. Oxidative stress and dysregulation of cytokines as a basis for inflammation appear to foster progression to NASH[8] as well as alcoholic hepatitis (AH)[9]. A two-hit progression from simple steatosis to steatohepatitis and fibrosis has been proposed[10], and suggests that after fat accumulation in the liver, lipids are peroxidized by oxidative stress induced by factors such as CYP2E1. The microsomal enzyme CYP2E1 metabolizes ethanol to acetaldehyde under conditions of alcohol dehydrogenase overload and generates oxidative stress as a by-product, however fatty acids also can be a substrate of CYP2E1[9].
Recently the role of the gut has attracted attention. Under alcoholic or high-fat conditions lipopolysaccharides can pass the border of the intestine to the portal vein and circulate to the liver where they trigger inflammation in ALD[11] and in NAFLD[12].
Some studies have already compared ALD and NAFLD[13], e.g., Wilfred de Alwis and Day[14] compared the genetics of both diseases addressing the question why only a small percentage of heavy drinkers and obese people progress from steatosis to severe liver disease. Here, we provide an analytical comparison of transcriptomic and metabolic processes involved in the progression of ALD and NAFLD. Employing transcriptome data derived from patient liver biopsies retrieved from several public repositories we performed a meta-analysis and report a signature of biomarkers distinguishing AH from NASH samples. Furthermore, we found predominating commonalities between both diseases at the level of biological pathways thus implying a large mechanistic similarity between both diseases.
Datasets of microarray gene expression data from liver biopsies were downloaded from the public repositories at NCBI GEO and EBI Array-Express. The compendium consisted of the ALD datasets GSE28619[15] and E-MTAB-2664[16] and the NAFLD datasets GSE61260[17], GSE59045[18], GSE48452[19] and GSE46300[12]. Illumina data was processed via R/Bioconductor[20] and packages lumi[21], limma[22] and qvalue[23]. Background-corrected log2-transformed data was normalized via quantile normalization from the lumi package. Affymetrix data was processed via R/Bioconductor and packages affy[24], limma, qvalue employing the rma normalization method.
Measurements from the multiple platforms were brought together in terms of mean ratios between ALD cases and controls and between NAFLD cases and controls. As controls, healthy liver biopsies or liver biopsies with a low grade of fat accumulation were used. For details we refer to the methods sections of the publications associated with the employed datasets[12,15-19]. Heterogeneity of the datasets was assessed via the meta-analysis R package metafor[25] generating forest and funnel plots (supplementary Figure 1A and B). The ratios were transformed to a log2 scale and normalized via quantile normalization. The results were again assessed with forest and funnel plots (supplementary Figure 1C and D).
In order to disentangle commonalities and differences between ALD and NAFLD, KEGG pathways[26] were analysed with respect to common pathways, up- and down-regulation and discordant up- and down-regulation. The ratios between ALD and control and NAFLD and control were employed to count the numbers of up- and down-regulated genes for each pathway. A pathway was considered up-regulated when it contained more up- than down-regulated genes. Genes with a ratio > t were termed up-regulated and genes with a ratio < 1/t were termed down-regulated. The threshold t was determined at the 95-quantile of the mean ratios between ALD and NAFLD vs control and was set accordingly to t = 4/5. Up- and down-regulation of a pathway was determined via the ratio of numbers of up-and down-regulated genes and via a binomial test assuming an equal probability of P = 0.5 for a gene to be up- or down-regulated.
Math 1
Here, nup,pw,case and ndown,pw,case are the numbers of up- and down-regulated genes in a pathway pw, gpw are the gene symbols associated with a pathway, xg,case is the gene expression value in a case which can be ALD or NAFLD, xg,control is the gene expression value in the control case, rpw,case is the ratio indicating up-regulation (rpw,case > 1) or down-regulation (rpw,case < 1) of pathway pw. Significance of up- or down-regulation of a pathway is assessed via the Binomial test with the Null hypothesis H0:p ≤ p0 and the test statistic B(p0, npw,case). Because of assumed equal distribution of up- and down-regulation the probability for the binomial distribution is set to p0 = 0.5.
Pathway charts of KEGG pathways indicating up- and down-regulation of genes in ALD and NAFLD were generated via the R/Bioconductor package pathview[27].
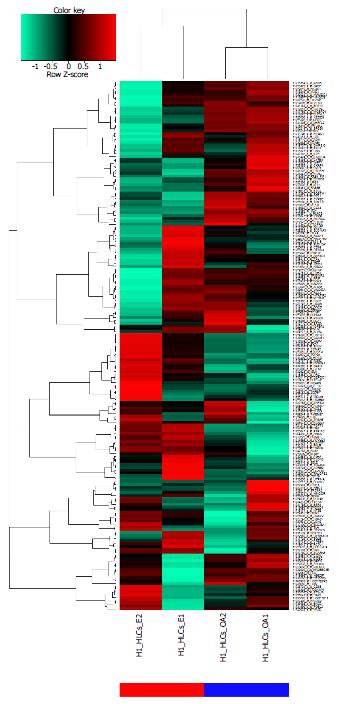
The differences between ALD and NAFLD at the transcriptome level could be condensed to a signature of 187 genes which are differentially expressed between both conditions with a P-value < 0.01 from the limma test and a ratio > 3/2 or a ratio < 2/3. The heatmap in Figure 1A shows a cluster analysis of this signature of gene expression data from ALD liver biopsies (blue bar) and NAFLD liver biopsies (red bar). The table in Figure 1B shows the 20 most up-regulated and 20 most-down-regulated genes from the signature indicating their log2-ratios and their P- and Q-values for the comparison ALD vs NAFLD. The most up-regulated gene between ALD and NAFLD was SPINK1. SPINK1 is secreted in the pancreatic juice to reversibly inhibit activated trypsin thus preventing pancreatic auto-digestion[28] and variants in this gene have been associated with pancreatitis[29]. Obesity and more prominent alcohol abuse are other causative factors for pancreatitis[28] which by its effects on insulin may contribute to liver disease. Lanthier et al[16] revealed the association of SPINK1 with inflammation and proliferation via correlation with the inflammatory macrophage marker CD68 and the cell cycle markers Cdk1 and CyclinB1. At the lower part of the table in Figure 1B two RGS (regulator of G-protein signalling) encoding genes, RGS1 and RGS2 are down-regulated in ALD but up-regulated in NAFLD. Nunn et al[30] reported reduced fat deposits, decreased serum lipids, and low Leptin levels in RGS2 deficient mice.
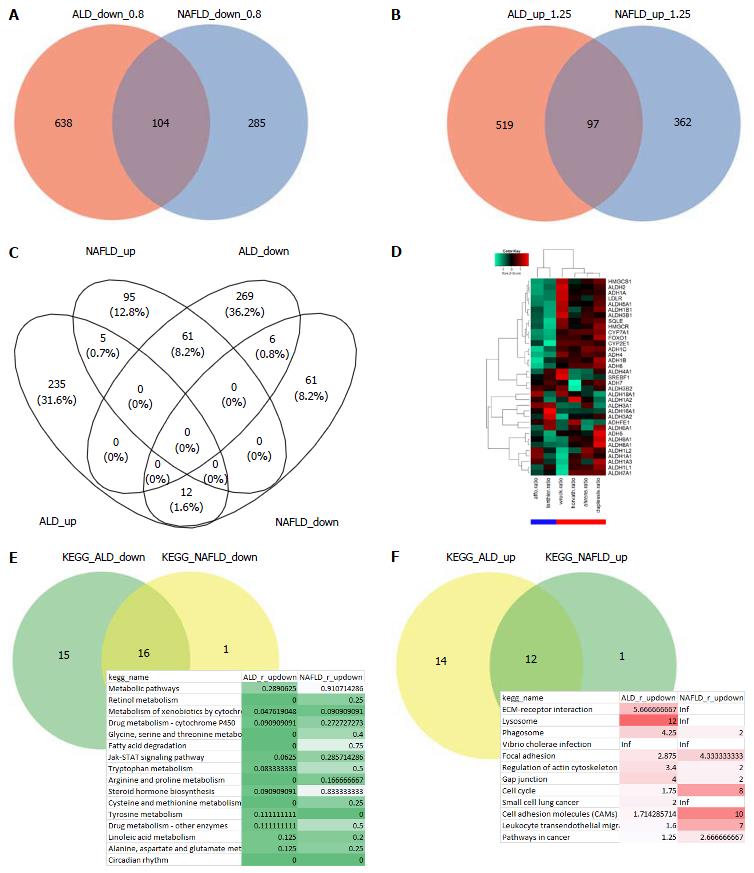
Analysis of the common genes between ALD and NAFLD was subdivided into analysis of down- and up-regulated genes. Figure 2A shows that 104 genes are down-regulated in ALD and NAFLD (ratio < 0.8) while 638 genes are exclusively down-regulated in ALD and 285 in NAFLD. Figure 2B shows that 97 genes are up-regulated in ALD and NAFLD (ratio > 1.25) while 519 genes are exclusively up-regulated in ALD and 362 in NAFLD. There are more distinctly expressed than overlapping genes - in contrary to the KEGG pathways where most pathways overlap (Figure 2E and F). Gene regulation was further restricted with a threshold for the limma test for differential expression of P < 0.05. Figure 2C shows a venn diagram of the four resulting sets of up/down-regulated genes in ALD and NAFLD. Here most genes are exclusively regulated but interestingly from the genes regulated in both diseases more genes are oppositely than commonly regulated: 61 genes are up-regulated in NAFLD but down-regulated in ALD and 12 are up-regulated in ALD and down-regulated in NAFLD while only 5 were commonly up and 6 commonly down-regulated. Supplementary Table 1 shows the corresponding gene sets. The genes up-regulated in NAFLD but down in ALD refer to major players in cholesterol processes such as HMGCS1, HMGCR, SQLE, CYP7A1 and LDLR. This would confirm the involvement of cholesterol biological processes in the etiology of NAFLD as we previously reported[31] and which distinguish it from the etiology of ALD. The opposite regulation of cholesterol processes as down in ALD and up in NAFLD can also be observed in the corresponding KEGG pathways Steroid biosynthesis, Primary bile acid biosynthesis and Terpenoid backbone biosynthesis (Supplementary file 1, p22, 34 and 84). These findings are in line with reports of a 29% decrease in HMGCR and a 56% decrease in cholesterol 7α-hydroxylase alias CYP7A1 by Lakshmanan et al[32], they suggested that increased ethanol leads to a reduced rate of cholesterol degradation to bile acids and accumulation of cholesterol in the liver. We also found (Supplementary Table 2) a stronger down-regulation of CYP7A1 (log2-ratio = -0.95) than of the upstream cholesterol genes HMGCR (log2-ratio = -0.429) and SQLE (log2-ratio = -0.33) in ALD while in NAFLD, CYP7A1 (log2-ratio = 1.15) was weaker up-regulated than HMGCR (log2-ratio = 1.57) and SQLE (log2-ratio = 1.53). Thus although oppositely regulated in ALD and NAFLD in both diseases more cholesterol is produced than can be secreted by the bile via CYP7A1.
Amongst the genes up-regulated in ALD but down in NAFLD are TNFSF14 in line with the major role of TNF-alpha in ALD[11] and SPINK1 which was described above in “a gene signature distinguishes ALD from NAFLD”.
To further investigate the mechanisms by which ethanol induces these changes in cholesterol processes we analysed expression clusters of genes involved in ethanol and cholesterol related processes. The analysis revealed a cluster of genes down-regulated in ALD and up-regulated in NAFLD including among others the genes encoding for ALDH2, ADH1A, LDLR, SQLE, HMGCR, CYP7A1, CYP2E1 and FOXO1 (Figure 2D). FOXO Transcription factors such as FOXO1, whose expression has been reported to be altered by ethanol[33] and may play a role in the regulation of several genes from this cluster. Interestingly, the heatmap (Figure 2D) shows a much higher degree of co-regulation of FOXO1 with the rate-limiting cholesterol synthesis enzymes HMGCR and SQLE than of SREBF1 which is known as the main regulator of cholesterol[34].
The five genes up-regulated in common between ALD and NAFLD include two collagen encoding genes - COL1A1 and COL3A1, thus demonstrating overlapping disease pathology in the development of fibrotic tissue. The six down-regulated genes in ALD and NAFLD include HPRT1 which has been reported to be down-regulated in severe liver disease[35].
Most biological pathways are regulated in the same direction in ALD and NAFLD. A pathway is considered down-regulated (Figure 2E) when it contains more down- than up-regulated genes as tested by the binomial test and the ratio is less than 1. Up-regulated pathways are determined accordingly (Figure 2F). The table of common down-regulated pathways includes metabolic, retinol, cytochrome and fatty acid degradation pathways, the up-regulated pathways include ECM-receptor, lysosome and phagosome.
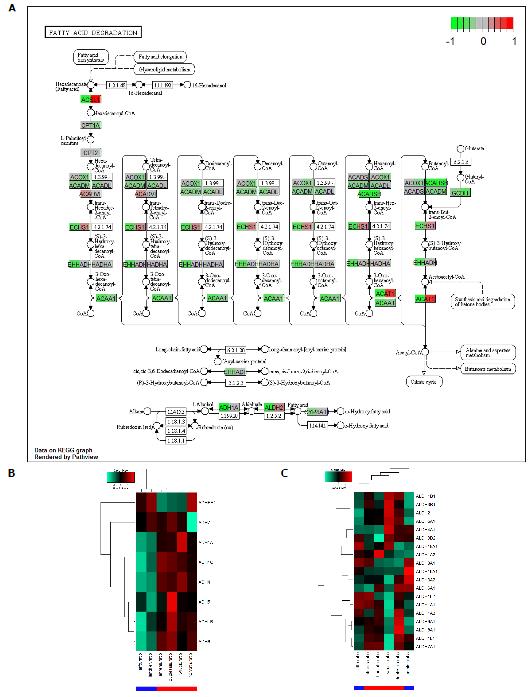
Sixteen common pathways are down-regulated in ALD and NAFLD. A pathway with high relevance to both diseases is Fatty acid degradation which is down-regulated in ALD and NAFLD but more so in ALD. The KEGG graph in Figure 3A shows down-regulation (green) in nearly all genes for ALD (left part of the gene boxes) while for NAFLD (right part of the gene boxes) there are up-regulated genes such as ACSL1 and ACAT1 but more are down-regulated. Interestingly, in the alcohol metabolism at the bottom of the chart, genes are down-regulated in ALD. At the bottom of Figure 3A, alcohol metabolism is shown in a schematic view. In a more detailed view we examined the behaviour of the alcohol dehydrogenase (ADH) encoding genes in the heatmap in Figure 3B and in the aldehyde dehydrogenase genes in Figure 3C. This resulted in a clear image for the ADHs which were down-regulated in ALD. The heatmap for the ALDHs (Figure 3C) looked more complex showing consistently ALD-down-regulated ALDHs only in a cluster at the top including ALDH2 while most genes were heterogeneously regulated between ALD and NAFLD.
Few pathways (12) were up-regulated in ALD and NAFLD. One of these is ECM-receptor interaction (Supplementary file 1, p. 142). Up-regulation of this pathway might indicate the onset of fibrosis which is accompanied by excessive accumulation of extracellular matrix proteins including collagen[36]. Here, the involvement of the collagen COL1A1 is shown.
Of the oppositely regulated pathways, sixteen were down-regulated in ALD and up-regulated in NAFLD while only one was up-regulated in ALD and down in NAFLD (Supplementary Table 3). The Glycolysis pathway was down-regulated in ALD and up-regulated in NAFLD. The KEGG graph (Supplementary file 1, p. 11) shows more down- (green, e.g., PGM1, ENO1) than up-regulated (red, e.g., PFKL) genes for ALD (left part of gene boxes) while for NAFLD (right part of gene boxes) up-regulated genes predominate. Reduction of glycolysis by ethanol has been brought into context with consumption of oxygen for the alcohol metabolism and has been reported by several authors[37,38]. Berry et al[38] reported that ethanol oxidation inhibits glycolysis in rat hepatocytes via competition of the reducing equivalents generated during ethanol oxidation with those arising in glycolysis for transfer to the mitochondria.
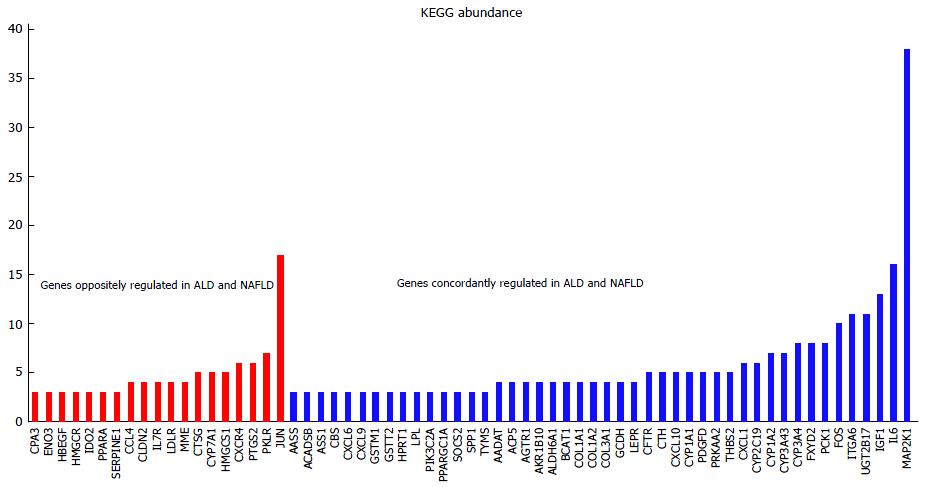
In “genes regulated in common between ALD and NAFLD” we described that after filtering genes with a P-value < 0.05 for differential expression more genes were oppositely than concordantly regulated in ALD and NAFLD. This filtering revealed the interesting genes described above but was very restrictive due to the low number of replicates in the condensed ratios - the P-values were relatively high. However, the condensed ratios were themselves based on numerous replicates so we consider them as reliable. In a second approach, we filtered genes only by fold change 1.25 and checked on the pathway-level if there were significantly more up- or down-regulated genes based on the binomial test. With this method more genes were concordantly than oppositely regulated in ALD and NAFLD. Figure 4 shows the abundance of concordantly and oppositely regulated genes in KEGG pathways (for abundances > 3). The most abundant MAP2K1 (MEK1) refers to the MAPK/RAS-signalling module acting in many KEGG-pathways. JUN which appears in 17 KEGG pathways and is down-regulated in ALD and up-regulated in NAFLD shows that there are mechanistic differences in molecular basis of these diseases. JUN which is directly connected to c-Jun N-terminal kinase (JNK) was down-regulated in ALD and up-regulated in NAFLD. The up-regulation of JUN in NAFLD is in line with reports from Samuel et al[39] showing that activated PKC-ε and JNK can induce insulin resistance via impaired IRS1 and IRS2 tyrosine phosphorylation in rats fed with high fat diet.
We recently described a disease-in-a-dish model of steatosis[40]. Pluripotent stem cells, both human embryonic stem cells and induced pluripotent stem cells were differentiated into hepatocyte-like cells and afterwards challenged with ethanol (E) and oleic acid. In order to test how close these models are to the modeled disease we applied our gene signature distinguishing ALD from NAFLD to gene expression data described in Graffmann et al[40]. Figure 5 demonstrates that our gene signature can clearly separate two clusters of the ALD and the NAFLD model in a heatmap generated from this gene expression dataset. Furthermore, relevant regulating or rate-limiting genes described above such as CYP7A1, CYP2E1, HMGCS1, FOXO1 are down-regulated in the ALD-model and up-regulated in the NAFLD-model similar to the liver biopsy-derived dataset.
In this comparative analysis of gene expression in ALD and NAFLD liver biopsies we unveiled many commonalities in pathways regulated in the same direction in both diseases. However, there were also pathways regulated in the opposite direction and maybe even more important, essential rate-limiting or regulating genes were adversely regulated. This adverse effect was unexpected as in our working hypothesis, we stated that alcohol is metabolized to fat and beyond this pathway both diseases share a common phenotype. It could hardly be brought together with the common phenotype that of the genes significantly dysregulated between ALD and NAFLD there were more genes regulated in the opposite than in the same direction. One major complex within the adversely regulated genes were cholesterol-related processes including the rate-limiting genes HMGCR, SQLE, CYP7A1 and LDLR. These were down-regulated in ALD and up-regulated in NAFLD (each compared vs healthy control). However, we found in both cases that the gene encoding CYP7A1 - the enzyme responsible for cholesterol removal by catalysing the conversion of cholesterol to bile acids was regulated at a lower level than the genes encoding for the cholesterol synthesis determining enzymes HMGCR and SQLE. This would explain cholesterol accumulation in the liver because more cholesterol is produced than secreted into bile - regardless if the cholesterol processes are down-regulated in total (in ALD) or up-regulated (in NAFLD). Moreover, the strong down-regulation of CYP7A1 in ALD might be a clue for the higher risk of cholestasis in ALD than in NAFLD[41]. Briefly, these findings emphasize the importance of cholesterol efflux from the liver via CYP7A1 and may suggest that the cause of the disease might be that the rate of cholesterol efflux is too low. Negative feedback loops down-regulating CYP7A1 by bile acids have already been described[42]: Bile acids can down-regulate CYP7A1 via (1) FXR and SHP; or (2) by interaction with liver macrophages (Kupffer cells) whose role in fibrosis has been established as they produce cytokines such as transforming growth factor beta leading to the transformation of stellate cells into myofibroblasts[43]. Furthermore, Kupffer cells secrete cytokines, e.g., tumor necrosis factor (TNFα) and interleukin (IL-1β) which in turn induce protein kinase, c-Jun N-terminal kinase and thus inhibit hepatocyte nuclear factor and consequently CYP7A1[44,45]. This gives rise to the question if the lower CYP7A1 levels are a cause of steatosis or are a consequence of the profibrotic stage. Here, systems biology modelling of cholesterol fluxes in the liver including bile acids and regulatory mechanisms of CYP7A1 could be useful in determining under which condition efflux rates are too low.
Beside the differences in cholesterol processes we could also confirm effects which had been much disputed before such as the ethanol-mediated down-regulation of glycolysis and of alcohol and aldehyde dehydrogenases.
The common up-regulated pathways might provide synergies for research into ALD and NAFLD. We found similar mechanisms underlying the progression of both diseases and could identify the common up-regulated ECM-receptor interactions and also associated collagen encoding genes COL1A1 and COL3A1 which indicate development of fibrotic tissue.
Finally, we provide a comprehensive compendium displaying comparative regulation of all KEGG pathways in ALD vs NAFLD which may serve as an encyclopaedic tool to lookup regulation of dedicated pathways associated with ALD and NAFLD.
In the current study we performed a meta-analysis of gene expression data of liver-derived biopsies from ALD and NAFLD patients, and report a gene signature which clearly separates the transcriptomes of ALD and NAFLD derived liver biopsies. Furthermore, we uncovered predominating commonalities between both diseases at the level of biological pathways, e.g., common down-regulation of the Fatty acid degradation pathway and common up-regulation of the ECM-receptor interaction pathway which may explain common progression of both diseases by cytokines being exchanged between hepatocytes, Kupffer cells and stellate cells at the fibrosis stage. This is confirmed by the common expression of COL1A1 and COL3A1 which are associated with fibrotic tissue.
Interestingly, we found rate-limiting genes of cholesterol processes such as HMGCR, SQLE and CYP7A1 adversely regulated (Figure 6) between ALD (down-regulated) and NAFLD (up-regulated). The fact that both diseases have the same phenotype may be due to a lower level of the enzyme CYP7A1 compared to the cholesterol synthesis enzymes HMGCR and SQLE. Thus, it will be interesting to further investigate CYP7A1-mediated cholesterol secretion into bile - possibly by systems biology modeling of cholesterol fluxes in the liver. For future therapy, drugs able to adjust CYP7A1 to levels amenable with cholesterol synthesized in or transported to the liver will be useful.
Non-alcoholic fatty liver disease (NAFLD) and alcoholic liver disease (ALD) are highly prevalent liver diseases and in an increasing number of developed countries NAFLD is becoming the most common cause of liver disease. Although NAFLD and ALD have distinct etiologies the manifestation and the potential progression of both diseases to hepatitis, cirrhosis and cancer is similar.
A two-hit hypothesis is the established explanation for disease progression to alcoholic hepatitis (AH) and non-alcoholic steatohepatitis (NASH). After steatotic fat accumulation due to metabolic disorders such as insulin resistance (NAFLD) or due to alcohol (ALD) oxidative stress and dysregulation of cytokines initiate inflammation and hence the progression to NASH as well as AH.
The authors found that rate-limiting enzymes of cholesterol metabolism such as HMGCR, SQLE and CYP7A1 are down-regulated in ALD and up-regulated in NAFLD compared to a healthy control. However, in ALD and NAFLD CYP7A1 - associated with conversion of cholesterol into bile acids - is regulated at a lower level than HMGCR and SQLE. That might explain the accumulation of cholesterol by the reduced efflux into bile acids.
CYP7A1 is a potential drug target and the proposed gene signature distinguishing ALD from NAFLD consists of biomarkers which may be exploited for diagnostic tests. The compendium of KEGG pathway regulation in ALD and NAFLD and the finding of the adverse regulation of cholesterol metabolism in ALD and NAFLD are promising start points for future research.
NAFLD is the disease related to fat accumulation (steatosis) in the liver in the absence of alcohol abuse (usually the threshold is set at 30 g/d of alcohol for men and 20 g/d for women). It ranges from the relatively benign steatosis to NASH, cirrhosis and hepatocellular carcinoma.
This manuscript was informative. The authors found commonalities between both ALD and NAFLD at the level of biological pathways implying some mechanistic similarity between both diseases.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Germany
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: de Oliveira CPMS, Qu BG, Sanal MG, Tarantino G S- Editor: Ji FF L- Editor: A E- Editor: Li D
| 1. | Ludwig J, Viggiano TR, McGill DB, Oh BJ. Nonalcoholic steatohepatitis: Mayo Clinic experiences with a hitherto unnamed disease. Mayo Clin Proc. 1980;55:434-438. [PubMed] |
| 2. | Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health. 2007;30:5-13. [PubMed] |
| 3. | Jenkins WJ, Cakebread K, Palmer KR. Effect of alcohol consumption on hepatic aldehyde dehydrogenase activity in alcoholic patients. Lancet. 1984;1:1048-1049. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 45] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 4. | Samson HH, Morgan DC, Price CM, Tang M, Falk JL. Ethanol elimination rates in normal and ethanol dependent animals. Pharmacol Biochem Behav. 1976;5:335-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 5. | Vidal F, Toda R, Gutiérrez C, Broch M, Fernández-Muixí F, Lorenzo A, Richart C. Influence of chronic alcohol abuse and liver disease on hepatic aldehyde dehydrogenase activity. Alcohol. 1998;15:3-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 21] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 6. | Luong A, Hannah VC, Brown MS, Goldstein JL. Molecular characterization of human acetyl-CoA synthetase, an enzyme regulated by sterol regulatory element-binding proteins. J Biol Chem. 2000;275:26458-26466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 221] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 7. | You M, Fischer M, Deeg MA, Crabb DW. Ethanol induces fatty acid synthesis pathways by activation of sterol regulatory element-binding protein (SREBP). J Biol Chem. 2002;277:29342-29347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 382] [Cited by in RCA: 415] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 8. | Marra F, Gastaldelli A, Svegliati Baroni G, Tell G, Tiribelli C. Molecular basis and mechanisms of progression of non-alcoholic steatohepatitis. Trends Mol Med. 2008;14:72-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 331] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 9. | Lieber CS. Alcoholic fatty liver: its pathogenesis and mechanism of progression to inflammation and fibrosis. Alcohol. 2004;34:9-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 424] [Cited by in RCA: 450] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 10. | Day CP, James OF. Steatohepatitis: a tale of two “hits”? Gastroenterology. 1998;114:842-845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2953] [Cited by in RCA: 3130] [Article Influence: 115.9] [Reference Citation Analysis (36)] |
| 11. | Lucey MR, Mathurin P, Morgan TR. Alcoholic hepatitis. N Engl J Med. 2009;360:2758-2769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 659] [Cited by in RCA: 693] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 12. | Wruck W, Kashofer K, Rehman S, Daskalaki A, Berg D, Gralka E, Jozefczuk J, Drews K, Pandey V, Regenbrecht C. Multi-omic profiles of human non-alcoholic fatty liver disease tissue highlight heterogenic phenotypes. Sci Data. 2015;2:150068. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 13. | Sookoian S, Pirola CJ. Systems biology elucidates common pathogenic mechanisms between nonalcoholic and alcoholic-fatty liver disease. PLoS One. 2013;8:e58895. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 14. | Wilfred de Alwis NM, Day CP. Genetics of alcoholic liver disease and nonalcoholic fatty liver disease. Semin Liver Dis. 2007;27:44-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 123] [Article Influence: 6.8] [Reference Citation Analysis (2)] |
| 15. | Affò S, Dominguez M, Lozano JJ, Sancho-Bru P, Rodrigo-Torres D, Morales-Ibanez O, Moreno M, Millán C, Loaeza-del-Castillo A, Altamirano J. Transcriptome analysis identifies TNF superfamily receptors as potential therapeutic targets in alcoholic hepatitis. Gut. 2013;62:452-460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 170] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 16. | Lanthier N, Rubbia-Brandt L, Lin-Marq N, Clément S, Frossard JL, Goossens N, Hadengue A, Spahr L. Hepatic cell proliferation plays a pivotal role in the prognosis of alcoholic hepatitis. J Hepatol. 2015;63:609-621. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 55] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 17. | Horvath S, Erhart W, Brosch M, Ammerpohl O, von Schönfels W, Ahrens M, Heits N, Bell JT, Tsai PC, Spector TD. Obesity accelerates epigenetic aging of human liver. Proc Natl Acad Sci USA. 2014;111:15538-15543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 480] [Cited by in RCA: 535] [Article Influence: 48.6] [Reference Citation Analysis (0)] |
| 18. | du Plessis J, van Pelt J, Korf H, Mathieu C, van der Schueren B, Lannoo M, Oyen T, Topal B, Fetter G, Nayler S. Association of Adipose Tissue Inflammation With Histologic Severity of Nonalcoholic Fatty Liver Disease. Gastroenterology. 2015;149:635-648.e14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 242] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 19. | Ahrens M, Ammerpohl O, von Schönfels W, Kolarova J, Bens S, Itzel T, Teufel A, Herrmann A, Brosch M, Hinrichsen H. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013;18:296-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 335] [Cited by in RCA: 417] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 20. | Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9318] [Cited by in RCA: 9601] [Article Influence: 457.2] [Reference Citation Analysis (0)] |
| 21. | Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547-1548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1676] [Cited by in RCA: 1713] [Article Influence: 100.8] [Reference Citation Analysis (0)] |
| 22. | Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7784] [Cited by in RCA: 8799] [Article Influence: 419.0] [Reference Citation Analysis (0)] |
| 23. | Storey JD. A direct approach to false discovery rates. J R Stat Soc Ser B Stat Methodol. 2002;64:479-498. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3772] [Cited by in RCA: 3532] [Article Influence: 153.6] [Reference Citation Analysis (0)] |
| 24. | Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3737] [Cited by in RCA: 4049] [Article Influence: 192.8] [Reference Citation Analysis (0)] |
| 25. | Viechtbauer W. Conducting meta-analyses in R with the metafor package. J Stat Softw. 2010;36:1-48. [RCA] [DOI] [Full Text] [Cited by in Crossref: 7317] [Cited by in RCA: 7427] [Article Influence: 495.1] [Reference Citation Analysis (0)] |
| 26. | Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457-D462. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3741] [Cited by in RCA: 4786] [Article Influence: 478.6] [Reference Citation Analysis (0)] |
| 27. | Luo W, Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29:1830-1831. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 955] [Cited by in RCA: 1503] [Article Influence: 125.3] [Reference Citation Analysis (0)] |
| 28. | Mitchell RM, Byrne MF, Baillie J. Pancreatitis. Lancet. 2003;361:1447-1455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 202] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 29. | Whitcomb DC, Gorry MC, Preston RA, Furey W, Sossenheimer MJ, Ulrich CD, Martin SP, Gates LK, Amann ST, Toskes PP. Hereditary pancreatitis is caused by a mutation in the cationic trypsinogen gene. Nat Genet. 1996;14:141-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1123] [Cited by in RCA: 1028] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 30. | Nunn C, Zhao P, Zou MX, Summers K, Guglielmo CG, Chidiac P. Resistance to age-related, normal body weight gain in RGS2 deficient mice. Cell Signal. 2011;23:1375-1386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 31. | Wruck W, Graffmann N, Kawala MA, Adjaye J. Current Status and Future Directions on Research Related to Nonalcoholic Fatty Liver Disease. Stem Cells. 2016;35:89-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 32. | Lakshmanan MR, Veech RL. Short- and long-term effects of ethanol administration in vivo on rat liver HMG-CoA reductase and cholesterol 7alpha-hydroxylase activities. J Lipid Res. 1977;18:325-330. [PubMed] |
| 33. | Lieber CS, Leo MA, Wang X, Decarli LM. Alcohol alters hepatic FoxO1, p53, and mitochondrial SIRT5 deacetylation function. Biochem Biophys Res Commun. 2008;373:246-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 62] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 34. | Ikonen E. Cellular cholesterol trafficking and compartmentalization. Nat Rev Mol Cell Biol. 2008;9:125-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 962] [Cited by in RCA: 1243] [Article Influence: 73.1] [Reference Citation Analysis (1)] |
| 35. | Congiu M, Slavin JL, Desmond PV. Expression of common housekeeping genes is affected by disease in human hepatitis C virus-infected liver. Liver Int. 2011;31:386-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 36. | Bataller R. Liver fibrosis. J Clin Invest. 2005;115:1100-1100. [RCA] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 37. | Young TA, Bailey SM, Van Horn CG, Cunningham CC. Chronic ethanol consumption decreases mitochondrial and glycolytic production of ATP in liver. Alcohol Alcohol. 2006;41:254-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 38. | Berry MN, Gregory RB, Grivell AR, Phillips JW, Schön A. The capacity of reducing-equivalent shuttles limits glycolysis during ethanol oxidation. Eur J Biochem. 1994;225:557-564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 39. | Samuel VT, Liu ZX, Qu X, Elder BD, Bilz S, Befroy D, Romanelli AJ, Shulman GI. Mechanism of hepatic insulin resistance in non-alcoholic fatty liver disease. J Biol Chem. 2004;279:32345-32353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 945] [Cited by in RCA: 985] [Article Influence: 46.9] [Reference Citation Analysis (0)] |
| 40. | Graffmann N, Ring S, Kawala MA, Wruck W, Ncube A, Trompeter HI, Adjaye J. Modeling Nonalcoholic Fatty Liver Disease with Human Pluripotent Stem Cell-Derived Immature Hepatocyte-Like Cells Reveals Activation of PLIN2 and Confirms Regulatory Functions of Peroxisome Proliferator-Activated Receptor Alpha. Stem Cells Dev. 2016;25:1119-1133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 82] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 41. | Tannapfel A, Denk H, Dienes HP, Langner C, Schirmacher P, Trauner M, Flott-Rahmel B. Histopathological diagnosis of non-alcoholic and alcoholic fatty liver disease. Virchows Arch. 2011;458:511-523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 98] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 42. | Davis RA, Miyake JH, Hui TY, Spann NJ. Regulation of cholesterol-7alpha-hydroxylase: BAREly missing a SHP. J Lipid Res. 2002;43:533-543. [PubMed] |
| 43. | Kolios G, Valatas V, Kouroumalis E. Role of Kupffer cells in the pathogenesis of liver disease. World J Gastroenterol. 2006;12:7413-7420. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 315] [Cited by in RCA: 370] [Article Influence: 19.5] [Reference Citation Analysis (1)] |
| 44. | Gupta S, Stravitz RT, Dent P, Hylemon PB. Down-regulation of cholesterol 7alpha-hydroxylase (CYP7A1) gene expression by bile acids in primary rat hepatocytes is mediated by the c-Jun N-terminal kinase pathway. J Biol Chem. 2001;276:15816-15822. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 263] [Cited by in RCA: 262] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 45. | Li T, Jahan A, Chiang JY. Bile acids and cytokines inhibit the human cholesterol 7 alpha-hydroxylase gene via the JNK/c-jun pathway in human liver cells. Hepatology. 2006;43:1202-1210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 129] [Article Influence: 6.8] [Reference Citation Analysis (0)] |