Copyright
©The Author(s) 2017.
World J Hepatol. Mar 18, 2017; 9(8): 443-454
Published online Mar 18, 2017. doi: 10.4254/wjh.v9.i8.443
Published online Mar 18, 2017. doi: 10.4254/wjh.v9.i8.443
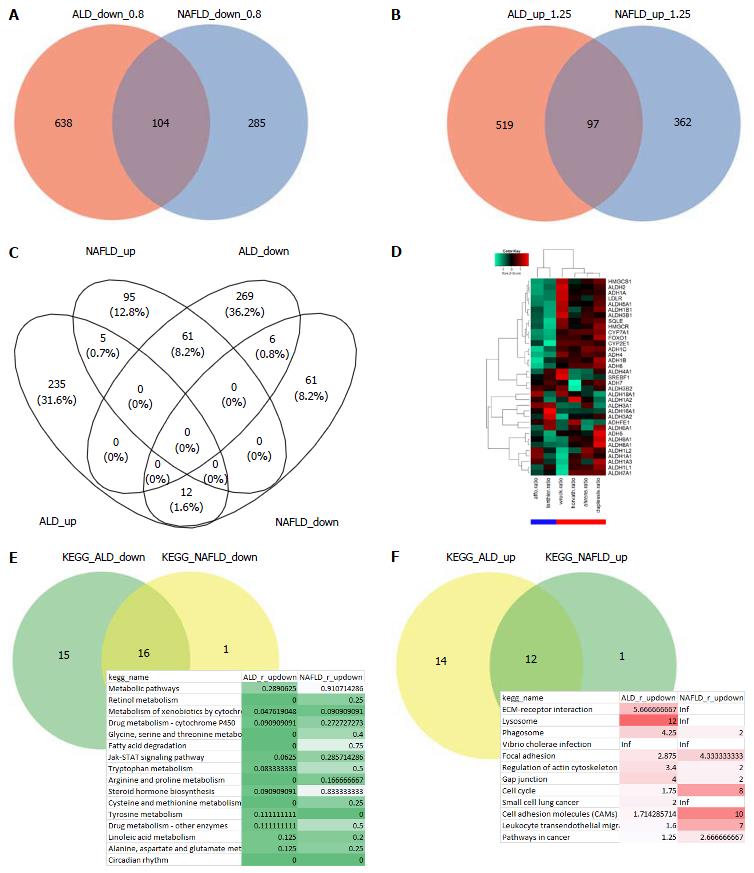
Figure 2 Most biological pathways are regulated in the same direction in alcoholic liver disease and non-alcoholic fatty liver disease but a subset of metabolism-associated genes are oppositely regulated.
A: Compares ALD and NAFLD in terms of down-regulated genes (ratio < 0.8); B: In terms of up-regulated genes (ratio > 1.25). There are more distinct than overlapping genes - in contrary to the KEGG pathways where most pathways overlap (E and F); C: Interestingly, when regulation is further restricted with a P-value < 0.05 more genes are oppositely than commonly regulated - but most are exclusively regulated. Many of the oppositely regulated genes are associated with cholesterol processes, e.g., HMGCR, SQLE and CYP7A1, and are co-expressed with alcohol (ADH) and aldehyd dehydrogeneases (ALDH) as seen in the heatmap (ALD: Blue bar, NAFLD: Red bar) (D). A pathway is considered down-regulated (E) when it contains more down- than up-regulated genes as tested by the binomial test and the ratio, analogously up-regulated pathways are determined (F). The table of common down-regulated pathways includes metabolic, retinol, cytochrome and fatty acid degradation pathways, the up-regulated include ECM-receptor, lysosome and phagosome. ALD: Alcoholic liver disease; NAFLD: Non-alcoholic fatty liver disease.
- Citation: Wruck W, Adjaye J. Meta-analysis reveals up-regulation of cholesterol processes in non-alcoholic and down-regulation in alcoholic fatty liver disease. World J Hepatol 2017; 9(8): 443-454
- URL: https://www.wjgnet.com/1948-5182/full/v9/i8/443.htm
- DOI: https://dx.doi.org/10.4254/wjh.v9.i8.443