Published online Aug 18, 2016. doi: 10.4254/wjh.v8.i23.985
Peer-review started: April 8, 2016
First decision: May 23, 2016
Revised: May 25, 2016
Accepted: June 14, 2016
Article in press: June 16, 2016
Published online: August 18, 2016
Processing time: 131 Days and 16.6 Hours
AIM: To interfere with the activation of nuclear factor-κB (NF-κB) with metformin and explore its effect in reversing multidrug resistance (MDR) of hepatocellular carcinoma (HCC) cells.
METHODS: Expression of P-glycoprotein (P-gp) and NF-κB in human HepG2 or HepG2/adriamycin (ADM) cells treated with pCMV-NF-κB-small interference RNA (siRNA) with or without metformin, was analyzed by Western blot or fluorescence quantitative PCR. Cell viability was tested by CCK-8 assay. Cell cycle and apoptosis were measured by flow cytometry and Annexin-V-PE/7-AnnexinV apoptosis detection double staining assay, respectively.
RESULTS: P-gp overexpression in HepG2 and HepG2/ADM cells was closely related to mdr1 mRNA (3.310 ± 0.154) and NF-κB mRNA (2.580 ± 0.040) expression. NF-κB gene transcription was inhibited by specific siRNA with significant down-regulation of P-gp and enhanced HCC cell chemosensitivity to doxorubicin. After pretreatment with metformin, HepG2/ADM cells were sensitized to doxorubicin and P-gp was decreased through the NF-κB signaling pathway. The synergistic effect of metformin and NF-κB siRNA were found in HepG2/ADM cells with regard to proliferation inhibition, cell cycle arrest and inducing cell apoptosis.
CONCLUSION: Metformin via silencing NF-κB signaling could effectively reverse MDR of HCC cells by down-regulating MDR1/P-gp expression.
Core tip: Metformin might target AMP-activated protein kinase mammalian target of rapamycin pathway, suppress hypoxiainducible factor-1α and transcriptionally down-regulate P-glycoprotein (P-gp) and multidrug resistance (MDR)-associated protein 1, suggesting that metformin may reverse MDR by targeting the AMP-activated protein kinase/mammalian target of rapamycin/hypoxia-inducible factor-1α/P-gp and MDR-associated protein 1 pathways. In the present study, HepG2/ADM cells pretreated with metformin were sensitized to doxorubicin and P-gp was decreased through the nuclear factor-κB (NF-κB) signaling pathway. The synergistic effects were found in the cells with regard to proliferation inhibition, cell cycle arrest and inducing apoptosis, and inhibiting P-gp expression via the NF-κB signaling pathway effectively reversed MDR by down-regulating MDR1/P-gp expression.
- Citation: Wu W, Yang JL, Wang YL, Wang H, Yao M, Wang L, Gu JJ, Cai Y, Shi Y, Yao DF. Reversal of multidrug resistance of hepatocellular carcinoma cells by metformin through inhibiting NF-κB gene transcription. World J Hepatol 2016; 8(23): 985-993
- URL: https://www.wjgnet.com/1948-5182/full/v8/i23/985.htm
- DOI: https://dx.doi.org/10.4254/wjh.v8.i23.985
Hepatocellular carcinoma (HCC) is one of the most common cancers and causes of cancer-related mortality worldwide[1-3]. Due to the lack of specific symptoms, the vast majority of HCCs are diagnosed at late and/or advanced stages[4,5]. Although recent advances in surgical techniques and interventional therapy have improved survival, the emergence of multidrug resistance (MDR) to a series of clinical chemotherapeutics with different structures or different target sites severely blocks the successful management of HCC[6,7]. The well recognized mechanism of classical MDR is the significant overexpression of human MDR1 gene encoding MDR1/P-glycoprotein (P-gp) that acts as an efflux pump on cell surface[8,9]. Intracellular anti-cancer drugs increasingly flow from cells through the efflux pump, thus drug concentrations become lower and cancer cells become resistant to chemotherapeutic drugs such as doxorubicin[10,11].
Recently, some studies have found diverse anticancer effects of metformin in the cells of lung, gastric, endometrial, breast, and other types of cancer[12,13]. Metformin exhibits anti-proliferative effects in tumor cells in vitro and in vivo[14,15]. Metformin might target the AMP-activated protein kinase (AMPK)/mammalian target of rapamycin (mTOR) pathway[16,17], suppress the hypoxia-inducible factor-1α (HIF-1α)[18,19] and transcriptionally down-regulate P-gp and MDR-associated protein 1 (MRP1), suggesting that metformin may reverse MDR by targeting the AMPK/mTOR/HIF-1α/P-gp and MRP1 pathways[20,21]. In addition, the activation of nuclear factor-kappa B (NF-κB) pathway plays an important role in the development of HCC[22-24], but whether it is related to MDR and the underlying molecular mechanisms remain to be explored[25,26]. In this study, we silenced NF-κB gene transcription with specific small interference RNA (siRNA) in human resistant HepG2/adriamycin (HepG2/ADM) cells, and explored the impact of metformin and NF-κB silencing, alone or in combination, on MDR1 regulation and MDR in HCC cells.
Human hepatoma cell line HepG2, HepG2/ADM cell line and hepatocyte cell line LO2 were purchased from Aibio Biotech Company (Shanghai, China). LO2 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, KeyGen Biotech Co., Ltd, Nanjing, China) containing 10% fetal bovine serum (FBS, Invitrogen, United States), penicillin (100 U/mL)/streptomycin (100 U/mL), at 37 °C with 5% CO2. HepG2 and HepG2/ADM cells were cultured in RPMI 1640 (KeyGen Biotech Co., Ltd, Nanjing, China) complete medium supplemented with 10% FBS, penicillin (100 U/mL)/streptomycin (100 U/mL) at 37 °C in a humidified incubator containing 5% CO2.
The cultured cells were washed with phosphate buffered saline (PBS) twice and lysed in phenylmethane sulfonyl fluoride (PMSF, Beyotime, Nantong, China) cell lysis buffer (1:1000), and the protein concentrations were determined with the bicinchoninic acid (BCA, Beyotime, Nantong, China) protein assay kit. The protein samples were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride (PVDF, Millipore, United States) membranes. After blocking with 5% skim milk in Tris-buffered saline with tween (TBST) at room temperature for 3 h, the membranes were incubated with the primary antibody overnight at 4 °C. The primary antibodies were diluted as follows: p65 and P-p65 (rabbit anti-human, 1:1000, Cell Signaling, United States), MDR1 (rabbit anti-human, 1:500, Abcam, United States) and β-actin (mouse anti-human, 1:2000, internal reference, Proteintech, United States). Then the membranes were washed three times with TBST and incubated with a horseradish peroxidase-conjugated secondary antibody (mouse or rabbit anti-human, 1:1000, Univ-bio, Nanjing, China) for 2.5 h at room temperature. Finally, the samples were detected with Quantity One software using the electrochemiluminescence kit (Millipore, United States). All Western blot experiments were repeated three times.
The cultured cells were digested with trypsin. Total RNA was extracted with TRIzol (Invitrogen, United States) reagent according to the protocol of the manufacturer. The quantity of total RNA was determined based on absorbance at 260 nm, and the purity of total RNA was analyzed based on the absorbance ratio at 260 and 280 nm (A260/280). Reverse transcription of total RNA to complementary DNA (cDNA) was performed with RevertAidTM First Strand cDNA Synthesis Kit (MBI Fermentas, CA, United States). PCR was carried with an SYBR Premix Ex TaqTMII kit (TaKaRa, Dalian, China), and GAPDH was used as an internal reference. The sequences of the primers used[27] were: NF-κB/p65 (forward: 5’-CTATCAGTCAGCGCATCCAG-3 and reverse: 5’-GCCAGAGTTTCGGTTCACTC-3’); mdr1 (forward: 5’-CCGGTTTGGAGCCTACTTG-3’ and reverse: 5’-TCCAATGTGTTCGGCATTAG-3’); and GAPDH (forward: 5’-CAAGGTCATCCATGACAACTTTG-3’ and reverse: 5’-GTCCACCACCCTGTTGCTGTAG-3’). Real-time PCR cycling parameters consisted of initial denaturation at 94 °C for 2 min and 40 cycles of 95 °C for 10 s, 55 °C for 30 s, and 70 °C for 45 s. The amplification specificity was confirmed by the melting curves. Ct values were calculated based on duplicates and normalized to GAPDH. The relative expression was calculated using the 2−ΔΔCt method. All PCR experiments were repeated three times.
Cell viability was evaluated with CCK-8 kit (Dojindo, Japan). Cells were divided into blank, negative control and experimental groups. Briefly, cells in logarithmic growth phase were digested with trypsin, and the cell suspension liquid (100 μL) was seeded in 96-well plates. Toxicity tests were performed with different concentrations of ADM added to 96-well plates in the experimental group. The micro-plates were pre-cultured at 37 °C in a humidified incubator containing 5% CO2, and liquid was changed at a fixed time interval. Then 10 μL/well CCK-8 solution was added and incubated at 37 °C for 4 h. The absorbance (A) was measured with a microplate reader at a wavelength of 450 nm. Cell survival rate was calculated as Aexp/Acon× 100%. Values of IC50 were evaluated with the Graphpad Prism5 software. Each individual experiment was performed at least three times.
HepG2/ADM cells were divided into three groups: Blank, control and experiment. The experimental group was treated with 1 μmol/L metformin for 24 h, and then continued to be cultured for 48 h with 1.5 μmol/L doxorubicin. The control group was only treated with doxorubicin, and the blank group did not undergo any treatment.
HepG2/ADM cells were treated with drugs for 48 h, and then continued to be cultured for 24 h with another culture solution. Cells were harvested using trypsin without EDTA and washed with cold PBS twice. Cell cycle and apoptosis (n = 3) were measured by flow cytometry and Annexin-V-PE/7-AnnexinV apoptosis detection double staining assay (BD, United States), respectively.
NF-κB-siRNAs were designed according to the previously reported sequences[28] and synthesized by the Biomics Company (Nantong, China) according to Rel A sequence obtained from Gene ID 5970. The sequences of siRNAs were: NF-κB/p65 siRNA (forward, 5′-TGCTGTTCATCTCCTGAAAGGAGGCCGTTTTGGCCACTGACTGACGGCCTCCTCAGGAGATGAA-3′ and reverse, 5′-CCTGTTCATCTCCTGAGGAGGCCGTCAGTCAGTGGCCAAAACGGCCTCCTTTCAGGAGATGAAC-3′; and negative-siRNA (forward, 5′-TGCTGAAATGTACTGCGCGTGGAGACGTTTTGGCCACTGACTGACGTCTCCACGCAGTACATTT-3′ and reverse, 5′-CCTGAAATGTACTGCGTGGAGACGTCAGTCAGTGGCCAAAACGTCTCCACGCGCAGTACATTTC-3′. Each siRNA was inserted to a pcDNA™ 6.2-GW/EmGFPmiR vector (Invitrogen, United States). HepG2/ADM cells were divided into blank control, negative siRNA control and NF-κB/p65 siRNA transfection groups. After cells were planted into microwell plates at a density of 70%, the plasmids were transfected into cells for incubation for 24 h according to the manufacturer’s instructions. The medium was removed on another day and replaced with the fresh one, and the transfection efficiency was observed with a fluorescence microscope. These experiments were performed in triplicate.
Data are expressed as the mean ± SD. Statistical analyses were done using the SPSS21.0 software package. Differences between groups were assessed using analysis of variance or t-test. P≤ 0.05 was regarded as statistically significant.
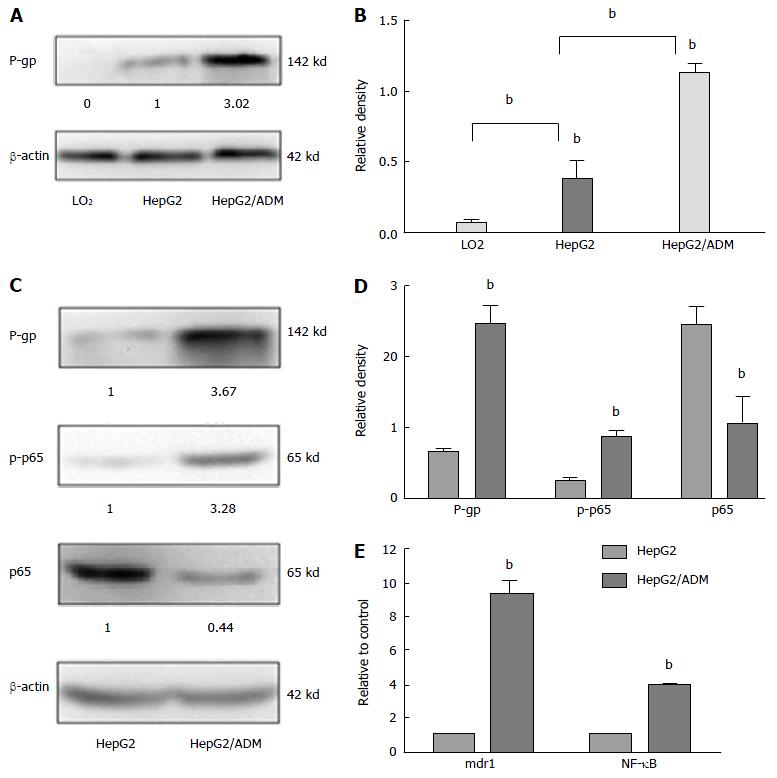
The levels of P-gp, mdr1, and NF-κB expression in different liver cell lines are shown in Figure 1. The proliferation of HepG2 and HepG2/ADM cells was decreased along with the increase of the concentration of doxorubicin, and the ability of proliferation was higher in HepG2/ADM cells than in HepG2 cells. At 24, 48 and 72 h, the IC50 values of doxorubicin against HepG2 cells were 0.489, 0.221 and 0.224 μmol/L, respectively, and the IC50 values of doxorubicin against HepG2/ADM cells were 4.166, 1.522 and 1.380 μmol/L, respectively. The resistance index (RI, μmol/L) of HepG2/ADM cells was 8.519 at 24 h, 6.874 at 48 h and 6.166 at 72 h. There was almost no P-gp expression in LO2 cells. Different degrees of expression of P-gp protein were observed in HepG2 and HepG2/ADM cells, but the P-gp expression in HepG2/ADM cells was significantly higher than that in HepG2 cells (Figure 1A and B). The p-p65 expression was significantly increased, while the expression of p65 was significantly decreased in HepG2/ADM cells (Figure 1C and D). The levels of mdr1 mRNA and NF-κB mRNA were 3.310 ± 0.154 and 2.580 ± 0.040, respectively, in HepG2/ADM cells, and 0.084 ± 0.038 and 0.607 ± 0.032, respectively, in HepG2 cells; the former was significantly higher than the latter (P < 0.01). Relative transcript levels (2-∆∆ct) of mdr1 mRNA and NF-κB mRNA were 9.381 ± 0.750 and 3.927 ± 0.069, respectively (Figure 1E).
The effect of metformin on the proliferation of HepG2/ADM cells was concentration- and time-dependent (Table 1). Metformin showed no significant effect on HepG2/ADM cells when its concentration was less than 3 mmol/L, but had different degrees of inhibition on the proliferation of HepG2/ADM cells when its concentration was between 3-10 mmol/L (P < 0.05). The HepG2/ADM cells were divided into experimental and control groups. After pretreatment with metformin, the experimental group cells were treated with different concentrations of doxorubicin. The effect of adriamycin combined with metformin on the proliferation of HepG2/ADM cells is shown in Table 2. After treatment with metformin, HepG2/ADM cells were more sensitive to adriamycin.
| Adriamycin (μmol/L) | 24 h | 48 h | 72 h | |||
| Control | Metformin | Control | Metformin | Control | Metformin | |
| 0 | 1.434 ± 0.03 | 1.327 ± 0.04a | 1.477 ± 0.08 | 1.357 ± 0.01 | 1.695 ± 0.08 | 1.507 ± 0.05a |
| 0.01 | 1.280 ± 0.06 | 1.160 ± 0.01a | 1.489 ± 0.03 | 1.314 ± 0.03a | 1.505 ± 0.01 | 1.378 ± 0.07a |
| 0.1 | 1.194 ± 0.10 | 1.111 ± 0.09 | 1.418 ± 0.01 | 1.213 ± 0.02a | 1.453 ± 0.02 | 1.249 ± 0.04a |
| 1 | 0.847 ± 0.02 | 0.662 ± 0.02a | 0.661 ± 0.01 | 0.661 ± 0.06 | 0.753 ± 0.04 | 0.508 ± 0.04a |
| 5 | 0.628 ± 0.08 | 0.458 ± 0.02a | 0.358 ± 0.02 | 0.208 ± 0.03a | 0.347 ± 0.03 | 0.194 ± 0.03a |
| 10 | 0.531 ± 0.00 | 0.399 ± 0.01a | 0.162 ± 0.01 | 0.062 ± 0.01a | 0.122 ± 0.01 | 0.049 ± 0.01a |
| 20 | 0.284 ± 0.01 | 0.162 ± 0.01a | 0.143 ± 0.01 | 0.051 ± 0.00a | 0.084 ± 0.01 | 0.027 ± 0.00a |
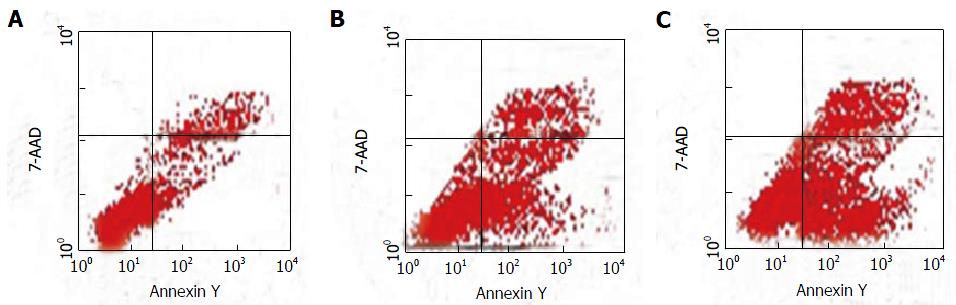
The levels of HepG2/ADM cell apoptosis in the experimental (treated with metformin plus adriamycin), control (only treated with adriamycin) and blank (without adriamycin or metformin) groups are shown in Figure 2. After the cells were pretreated with 1 mmol/L metformin for 24 h, adriamycin was added. MDR1 in HepG2/ADM cells was down-regulated, the cell cycle was blocked at G0/G1 phase, and apoptosis was enhanced. Significant differences in the apoptosis rates were found among different groups (F = 3726.97, P < 0.001), and the apoptosis rate was significantly higher in the experimental group (22.17% ± 0.37%) than in the control group (14.86% ± 0.21%) or the blank group (4.17% ± 0.13%).
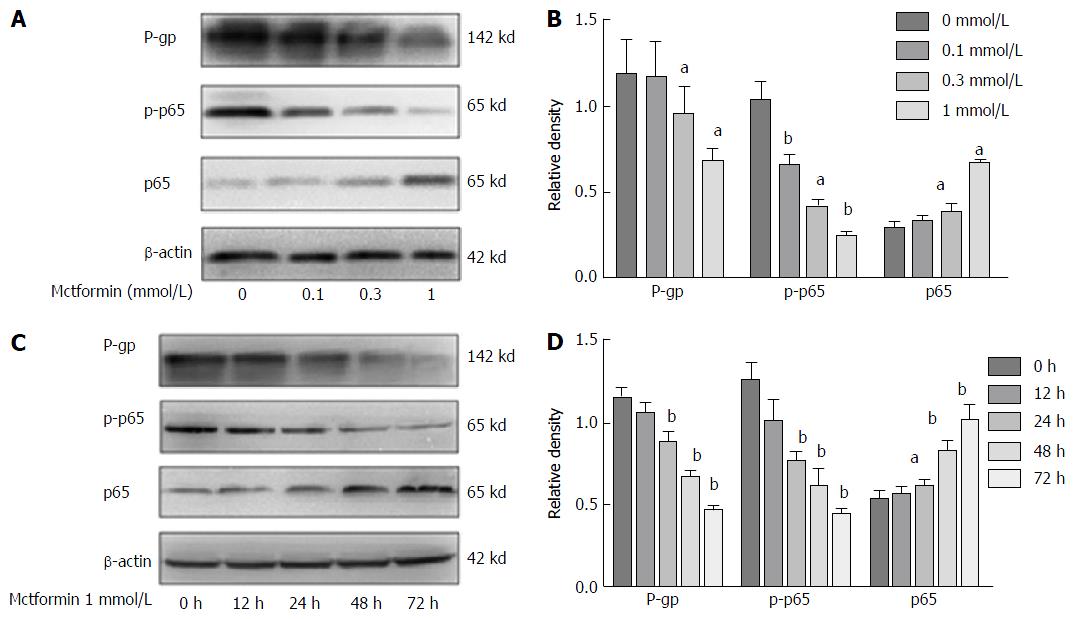
Metformin reversed the MDR of HCC cells via the NF-κB signaling pathway (Figure 3). The levels of P-gp expression in the HepG2/ADM cells were decreased with the increasing dose of metformin, and the phosphorylated p65 expression in the nucleus was also decreased. Metformin could down-regulate P-gp expression by inhibiting NF-κB activation in a dose- and time-dependent manner.
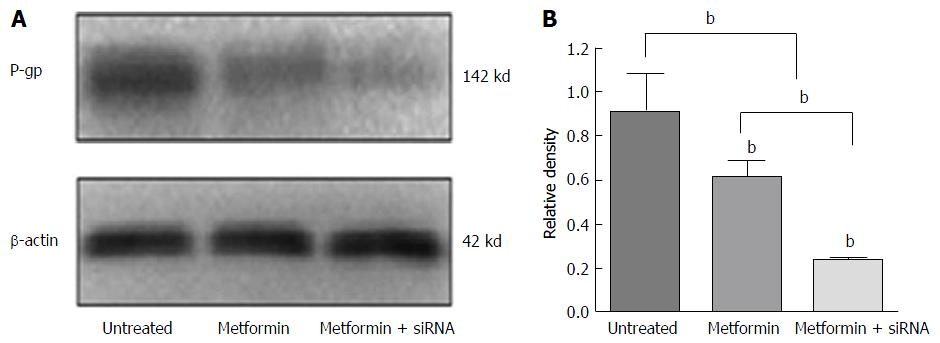
The synergistic effects of metformin combined with NF-κB siRNA in reversing MDR are shown in Figure 4. HepG2/ADM cells were divided into three groups: Untreated cells, cells treated with metformin alone and those treated with metformin combined with NF-κB-siRNA. In the NF-κB-siRNA group, NF-κB-siRNA was transfected into HepG2/ADM cells for 24 h, and then cells were treated with 1 mmol/L metformin for 48 h. The levels of P-gp expression were 0.91 ± 0.24, 0.63 ± 0.13 and 0.22 ± 0.02 (F = 14.47, P = 0.005) in untreated, metformin and the metformin combined with NF-κB-siRNA groups, respectively. The expression of P-gp was significantly reduced in cells treated with metformin plus NF-κB-siRNA compared with that in cells only treated with metformin (t = 5.39, P = 0.006).
Recent advances in surgical techniques and interventional therapy have improved survival of HCC patients[6,7,29]. However, the emergence of MDR to a series of clinical chemotherapeutics with different structures or target sites severely blocks the successful management of HCC and still is a difficult problem to be solved in clinical practice[30,31]. MDR in HCC could result from several biochemical mechanisms including decreased drug influx, increased drug efflux, altered cell cycle checkpoints, altered drug targets, increased drug metabolism and/or resistance to drug-induced apoptosis. Therefore, it is very important to find safe and effective MDR reversal agents for HCC[32]. In the present study, metformin with silencing NF-κB gene transcription was used to reverse MDR of HepG2/ADM cells with high NF-κB expression.
Anti-cancer drug efflux is one of the most common mechanisms of MDR of HCC cells, and it is mediated by ATP-binding cassette transporters[33,34], such as P-gp encoded by MDR1 gene, which is located downstream of the NF-κB signaling pathway. P-gp expression regulated by MDR1 is the most important and common cause of MDR, and weakened the apoptosis of cancer cells induced by chemotherapeutic drugs. Both P-gp expression and NF-κB activation are linked closely with HCC progression[35]. Usually NF-κB takes part in gene transcription by means of homodimers or heterodimers, such as p50/p65, p65/p65, and p65/Rel. In quiescent cells, they are predominantly cytoplasmic, associating with members of inhibitory IκB family and forming NF-κB/IκB complexes without activity. Both P-gp and NF-κB at the protein or transcriptional level were significantly higher (Figure 1), with p65 expression decreasing in HepG2/ADM cells, indicating that abnormal P-gp and NF-κB expression could associate with the MDR formation of HCC cells[20].
Metformin is a safe, low-cost drug, and therefore remains one of the most commonly prescribed drugs worldwide[16,36]. The anticancer effects of metformin indicate the possibility that certain diabetes-associated types of cancer[37,38] may be circumvented, and metformin has anti-proliferative potential against cancer cells or reversing MDR in vitro and in vivo[39,40]. However, the precise molecular mechanisms whereby metformin works in cancer prevention remain multi-factorial and ill-defined. Metformin affected HepG2/ADM cell proliferation in a dose- and time-dependent manner (Table 1). Metformin at < 3 mmol/L had no significant impact on HepG2/ADM cells, but the cells treated with metformin between 3-10 mmol/L were more sensitive to adriamycin with regard to promoting cell apoptosis (Figure 2) and inhibiting cell proliferation (Table 2), suggesting that metformin could increase the sensitivity of HepG2/ADM cells to anti-cancer drugs.
There are few studies on the effect of metformin on MDR of HCC cells. siRNA strategy is a powerful technique to inhibit specific gene expression, which has highlighted the potential use of siRNA molecules to study gene function or explore new HCC therapeutic agents[41,42]. The expression of NF-κB gene transcription was inhibited by specific siRNA, which significantly down-regulated P-gp and enhanced the chemosensitivity of HCC cells to doxorubicin, confirming the mechanism of decreasing P-gp via the NF-κB signaling pathway. The synergistic effects of metformin and NF-κB siRNA were found in HepG2/ADM cells with regard to cell proliferation inhibition, cell cycle arrest, and inducing cell apoptosis. These data confirm that the metformin could enhance the HepG2/ADM cells sensitivity to adriamycin and reverse MDR via the NF-κB signaling pathway (Figure 4).
In conclusion, the development of MDR still is one of major causes of HCC chemotherapy failure[43,44]. Although specific NF-κB siRNA is a powerful small molecule reagent designed to silence expression of NF-κB and MDR1/P-gp related to MDR to increase tumor cell sensitivity to anti-cancer drugs, how to apply metformin plus interfering NF-κB activation for effective reversal of MDR of HCC cells still needs to be further explored.
The authors thank Dr. FitzGibbon T for comments on earlier drafts of the manuscript.
Hepatocellular carcinoma (HCC) multidrug resistance (MDR) to a series of clinical chemotherapeutics with different structures or different target sites severely blocks the successful management of HCC. The mechanism of classical MDR is the significant overexpression of MDR1/P-glycoprotein (P-gp) that acts as an efflux pump on cell surface. Intracellular anti-cancer drugs increasingly flow from cells through the efflux pump, thus drug concentrations become lower and cancer cells become resistant to chemotherapeutic drugs such as doxorubicin.
Metformin could target AMP-activated protein kinase mammalian target of rapamycin pathway, suppress hypoxia-inducible factor-1α (HIF-1α) and transcriptionally down-regulate P-gp and MDR-associated protein 1, suggesting that metformin may reverse MDR by targeting the AMP-activated protein kinase/mammalian target of rapamycin/HIF-1α/P-gp and MDR-associated protein 1 pathways. However, whether metformin plus nuclear factor-κB (NF-κB) inhibition might effectively reverse MDR of HCC cells remains to be explored.
Recently, there are few studies on the effects of metformin on MDR of HCC cells. In this study, the data suggested that the abnormal expression of MDR1/P-gp and NF-κB activation during HCC development were related to MDR formation, which might be down-regulated through inhibiting activation of the NF-κB signaling pathway with specific small interference RNA (siRNA). The combination of metformin with interfering NF-κB gene transcription could effectively reverse the MDR of HCC cells.
The abnormal expression of MDR1/P-gp in HCC was related to MDR formation, which could be down-regulated through inhibiting activation of the NF-κB signaling pathway with specific siRNA and increasing sensitivity of HCC cells to chemotherapy drugs. Interfering NF-κB activation with metformin is effective to reverse MDR of HCC cells. However, how to apply metformin plus interfering NF-κB activation for effective reversal of MDR of HCC cells still needs to be explored.
Metformin is a safe, low-cost drug. The anticancer effects of metformin indicate the possibility that certain diabetes-associated types of cancer may be circumvented. Indeed, many retrospective meta-analyses have shown that metformin possesses anti-cancer activities and decreases the incidence of primary cancer development in those taking metformin routinely, and a multitude of clinical cancer trials are actively assessing its benefits in non-diabetic population who have already developed cancer. However, the precise molecular mechanisms whereby metformin works in cancer prevention remain multi-factorial and ill-defined.
Authors have done excellent work in this present study. They have explored the effect of metformin and interfering NF-κB gene transcription with specific siRNA, alone or in combination, on MDR1 gene regulation. The application of interfering NF-κB activation with metformin was more effective to reverse MDR of HCC cells.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P- Reviewer: Chiang TA, Jamall IS S- Editor: Qi Y L- Editor: Wang TQ E- Editor: Li D
| 1. | Singal AG, El-Serag HB. Hepatocellular Carcinoma From Epidemiology to Prevention: Translating Knowledge into Practice. Clin Gastroenterol Hepatol. 2015;13:2140-2151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 406] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 2. | de Martel C, Maucort-Boulch D, Plummer M, Franceschi S. World-wide relative contribution of hepatitis B and C viruses in hepatocellular carcinoma. Hepatology. 2015;62:1190-1200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 348] [Cited by in RCA: 381] [Article Influence: 38.1] [Reference Citation Analysis (0)] |
| 3. | Ashtari S, Pourhoseingholi MA, Sharifian A, Zali MR. Hepatocellular carcinoma in Asia: Prevention strategy and planning. World J Hepatol. 2015;7:1708-1717. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 92] [Article Influence: 9.2] [Reference Citation Analysis (2)] |
| 4. | Yao M, Wang L, Yao Y, Gu HB, Yao DF. Biomarker-based MicroRNA Therapeutic Strategies for Hepatocellular Carcinoma. J Clin Transl Hepatol. 2014;2:253-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 5. | Zhang H, Yao M, Wu W, Qiu L, Sai W, Yang J, Zheng W, Huang J, Yao D. Up-regulation of annexin A2 expression predicates advanced clinicopathological features and poor prognosis in hepatocellular carcinoma. Tumour Biol. 2015;36:9373-9383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 6. | Bruix J, Gores GJ, Mazzaferro V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 2014;63:844-855. [PubMed] |
| 7. | Llovet JM, Bruix J. Molecular targeted therapies in hepatocellular carcinoma. Hepatology. 2008;48:1312-1327. [PubMed] |
| 8. | Fang P, Zhang X, Gao Y, Ding CR, Cui F, Jiao SC. Reversal effect of melanoma differentiation associated gene-7/interleukin-24 on multidrug resistance in human hepatocellular carcinoma cells. Anat Rec (Hoboken). 2012;295:1639-1646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 9. | Liu Y, Lou G, Wu W, Zheng M, Shi Y, Zhao D, Chen Z. Involvement of the NF-κB pathway in multidrug resistance induced by HBx in a hepatoma cell line. J Viral Hepat. 2011;18:e439-e446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 10. | Zheng W, Sai W, Yao M, Gu H, Yao Y, Qian Q, Yao D. Silencing clusterin gene transcription on effects of multidrug resistance reversing of human hepatoma HepG2/ADM cells. Tumour Biol. 2015;36:3995-4003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 11. | Zhao X, Chen Q, Li Y, Tang H, Liu W, Yang X. Doxorubicin and curcumin co-delivery by lipid nanoparticles for enhanced treatment of diethylnitrosamine-induced hepatocellular carcinoma in mice. Eur J Pharm Biopharm. 2015;93:27-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 122] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 12. | CImai A, Ichigo S, Matsunami K, Takagi H, Yasuda K. linical benefits of metformin in gynecologic oncology. Oncol Lett. 2015;10:577-582. [PubMed] |
| 13. | Cazzaniga M, Bonanni B. Breast Cancer Metabolism and Mitochondrial Activity: The Possibility of Chemoprevention with Metformin. Biomed Res Int. 2015;2015:972193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 14. | Hall C, Stone RL, Gehlot A, Zorn KK, Burnett AF. Use of Metformin in Obese Women With Type I Endometrial Cancer Is Associated With a Reduced Incidence of Cancer Recurrence. Int J Gynecol Cancer. 2016;26:313-317. [PubMed] |
| 15. | Cazzaniga M, Bonanni B. Relationship Between Metabolic Reprogramming and Mitochondrial Activity in Cancer Cells. Understanding The Anticancer Effect of Metformin and Its Clinical Implications. Anticancer Res. 2015;35:5789-5796. [PubMed] |
| 16. | Kim HG, Hien TT, Han EH, Hwang YP, Choi JH, Kang KW, Kwon KI, Kim BH, Kim SK, Song GY. Metformin inhibits P-glycoprotein expression via the NF-κB pathway and CRE transcriptional activity through AMPK activation. Br J Pharmacol. 2011;162:1096-1108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 151] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 17. | Chen S, Wang Y, Ruan W, Wang X, Pan C. Reversing multidrug resistance in hepatocellular carcinoma cells by inhibiting extracellular signal-regulated kinase/mitogen-activated protein kinase signaling pathway activity. Oncol Lett. 2014;8:2333-2339. [PubMed] |
| 18. | Li S, Yao D, Wang L, Wu W, Qiu L, Yao M, Yao N, Zhang H, Yu D, Ni Q. Expression characteristics of hypoxia-inducible factor-1α and its clinical values in diagnosis and prognosis of hepatocellular carcinoma. Hepat Mon. 2011;11:821-828. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 30] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 19. | Dong ZZ, Yao DF, Li SS, Yao M, Yu DD, Yao NH, Qian YJ, Qiu LW. Inhibitory effect of miRNA silencing hypoxia-inducible factor alpha subunit gene on the proliferation of HepG2 cells. Zhonghua Gan Zang Bing Za Zhi. 2011;19:281-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 20. | Xiang QF, Zhang DM, Wang JN, Zhang HW, Zheng ZY, Yu DC, Li YJ, Xu J, Chen YJ, Shang CZ. Cabozantinib reverses multidrug resistance of human hepatoma HepG2/adr cells by modulating the function of P-glycoprotein. Liver Int. 2015;35:1010-1023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 21. | Fantappiè O, Sassoli C, Tani A, Nosi D, Marchetti S, Formigli L, Mazzanti R. Mitochondria of a human multidrug-resistant hepatocellular carcinoma cell line constitutively express inducible nitric oxide synthase in the inner membrane. J Cell Mol Med. 2015;19:1410-1417. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 22. | Wu W, Yao DF, Dong ZZ, Bian YZ, Yao NH, Qiu LW, Yang JL, Sai WL. [Abnormality of NF- kappa B expression and the clinical implications in patients with HBV-related hepatocellular carcinoma]. Zhonghua Gan Zang Bing Za Zhi. 2011;19:466-468. [PubMed] |
| 23. | Dong ZZ, Yao DF, Wu W, Yao M, Yu HB, Shen JJ, Qiu LW, Yao NH, Sai WL, Yang JL. Delayed hepatocarcinogenesis through antiangiogenic intervention in the nuclear factor-kappa B activation pathway in rats. Hepatobiliary Pancreat Dis Int. 2010;9:169-174. [PubMed] |
| 24. | Yao DF, Yu HB, Shen JJ, Wang YL, Wu XH, Qiu LW, Wu W. [The effect of thalidomine-induced NF-kappa B activation on malignant transformation of hepatocytes]. Zhonghua Gan Zang Bing Za Zhi. 2009;17:312-314. [PubMed] |
| 25. | Shi Y, Wang SY, Yao M, Sai WL, Wu W, Yang JL, Cai Y, Zheng WJ, Yao DF. Chemosensitization of HepG2 cells by suppression of NF-κB/p65 gene transcription with specific-siRNA. World J Gastroenterol. 2015;21:12814-12821. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 26. | Liu Y, Lou G, Wu W, Shi Y, Zheng M, Chen Z. Interferon-α sensitizes HBx-expressing hepatocarcinoma cells to chemotherapeutic drugs through inhibition of HBx-mediated NF-κB activation. Virol J. 2013;10:168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 27. | Wu W, Yao D, Wang Y, Qiu L, Sai W, Yang J, Yao N, Li S, Bian Y, Wang Z. Suppression of human hepatoma (HepG2) cell growth by nuclear factor-kappaB/p65 specific siRNA. Tumour Biol. 2010;31:605-611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 28. | Wang YL, Yao DF, Wu W, Sai WL, Qiu LW, Yang JL, Zhu JW. [Effect of siRNA-mediated inhibition of nuclear transcription factor-kappa B on apoptosis of hepatocarcinoma cells]. Zhonghua Gan Zang Bing Za Zhi. 2010;18:609-613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 29. | Gillet JP, Andersen JB, Madigan JP, Varma S, Bagni RK, Powell K, Burgan WE, Wu CP, Calcagno AM, Ambudkar SV. A Gene Expression Signature Associated with Overall Survival in Patients with Hepatocellular Carcinoma Suggests a New Treatment Strategy. Mol Pharmacol. 2016;89:263-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 30. | Ho CT, Shang HS, Chang JB, Liu JJ, Liu TZ. Folate deficiency-triggered redox pathways confer drug resistance in hepatocellular carcinoma. Oncotarget. 2015;6:26104-26118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 31. | Xiang Y, Liu Y, Yang Y, Hu H, Hu P, Ren H, Zhang D. A secretomic study on human hepatocellular carcinoma multiple drug-resistant cell lines. Oncol Rep. 2015;34:1249-1260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 32. | Yan J, Zhou Y, Chen D, Li L, Yang X, You Y, Ling X. Effects of mitochondrial translocation of telomerase on drug resistance in hepatocellular carcinoma cells. J Cancer. 2015;6:151-159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 33. | Colombo F, Trombetta E, Cetrangolo P, Maggioni M, Razini P, De Santis F, Torrente Y, Prati D, Torresani E, Porretti L. Giant Lysosomes as a Chemotherapy Resistance Mechanism in Hepatocellular Carcinoma Cells. PLoS One. 2014;9:e114787. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 34. | Shibasaki Y, Sakaguchi T, Hiraide T, Morita Y, Suzuki A, Baba S, Setou M, Konno H. Expression of indocyanine green-related transporters in hepatocellular carcinoma. J Surg Res. 2015;193:567-576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 35. | Wu W, Yao DF, Qiu LW, Sai WL, Shen JJ, Yu HB, Wu XH, Li YM, Wang YL, Gu WJ. Characteristics of hepatic nuclear-transcription factor-kappa B expression and quantitative analysis in rat hepatocarcinogenesis. Hepatobiliary Pancreat Dis Int. 2009;8:504-509. [PubMed] |
| 36. | Pryor R, Cabreiro F. Repurposing metformin: an old drug with new tricks in its binding pockets. Biochem J. 2015;471:307-322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 182] [Cited by in RCA: 199] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 37. | Bhat A, Sebastiani G, Bhat M. Systematic review: Preventive and therapeutic applications of metformin in liver disease. World J Hepatol. 2015;7:1652-1659. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 63] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 38. | Ling S, Tian Y, Zhang H, Jia K, Feng T, Sun D, Gao Z, Xu F, Hou Z, Li Y. Metformin reverses multidrug resistance in human hepatocellular carcinoma Bel7402/5fluorouracil cells. Mol Med Rep. 2014;10:2891-2897. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 39. | Yan F, Bai LP, Gao H, Zhu CM, Lin L, Kang XP. EGF reverses multi-drug resistance via the p-ERK pathway in HepG2/ADM and SMMC7721/ADM hepatocellular carcinoma models. Asian Pac J Cancer Prev. 2014;15:2619-2623. [PubMed] |
| 40. | Qin Y, Bao H, Pan Y, Yin M, Liu Y, Wu S, Li H. SUMOylation alterations are associated with multidrug resistance in hepatocellular carcinoma. Mol Med Rep. 2014;9:877-881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 41. | Shen J, Sun H, Meng Q, Yin Q, Zhang Z, Yu H, Li Y. Simultaneous inhibition of tumor growth and angiogenesis for resistant hepatocellular carcinoma by co-delivery of sorafenib and survivin small hairpin RNA. Mol Pharm. 2014;11:3342-3351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 42. | Yao M, Wang L, Qiu L, Qian Q, Yao D. Encouraging microRNA-based Therapeutic Strategies for Hepatocellular Carcinoma. Anticancer Agents Med Chem. 2015;15:453-460. [PubMed] |
| 43. | Cheng L, Luo S, Jin C, Ma H, Zhou H, Jia L. FUT family mediates the multidrug resistance of human hepatocellular carcinoma via the PI3K/Akt signaling pathway. Cell Death Dis. 2013;4:e923. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 93] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 44. | Gu W, Liu L, Fang FF, Huang F, Cheng BB, Li B. Reversal effect of bufalin on multidrug resistance in human hepatocellular carcinoma BEL-7402/5-FU cells. Oncol Rep. 2014;31:216-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |