Published online Jul 26, 2013. doi: 10.4252/wjsc.v5.i3.73
Revised: April 18, 2013
Accepted: June 5, 2013
Published online: July 26, 2013
Processing time: 173 Days and 13.9 Hours
The study of embryonic stem cells is in the spotlight in many laboratories that study the structure and function of chromatin and epigenetic processes. The key properties of embryonic stem cells are their capacity for self-renewal and their pluripotency. Pluripotent stem cells are able to differentiate into the cells of all three germ layers, and because of this property they represent a promising therapeutic tool in the treatment of diseases such as Parkinson’s disease and diabetes, or in the healing of lesions after heart attack. As the basic nuclear unit, chromatin is responsible for the regulation of the functional status of cells, including pluripotency and differentiation. Therefore, in this review we discuss the functional changes in chromatin during differentiation and the correlation between epigenetics events and the differentiation potential of embryonic stem cells. In particular we focus on post-translational histone modification, DNA methylation and the heterochromatin protein HP1 and its unique function in mouse and human embryonic stem cells.
Core tip: Here, we provided a summary on epigenetics and chromatin structure in pluripotent embryonic stem cells (ESCs) and their differentiated counterpart. We especially aim at histone signature, function of heterochromatin protein 1. Moreover, we summarized published data on nuclear architecture; we especially addressed arrangement of chromosome territories and genes in pluripotent ESCs and after induced differentiation.
- Citation: Přikrylová T, Pacherník J, Kozubek S, Bártová E. Epigenetics and chromatin plasticity in embryonic stem cells. World J Stem Cells 2013; 5(3): 73-85
- URL: https://www.wjgnet.com/1948-0210/full/v5/i3/73.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v5.i3.73
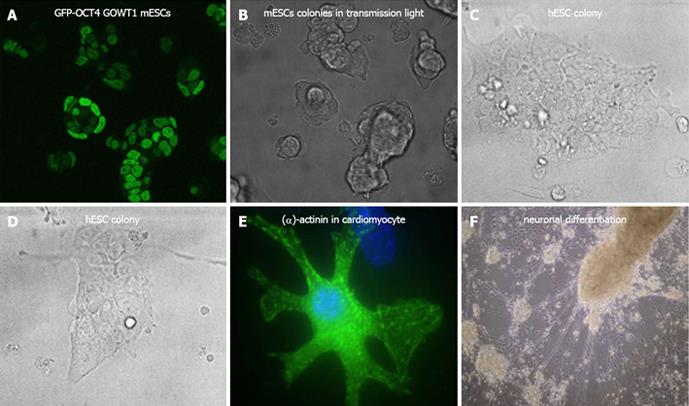
Human embryonic stem cells (hESCs) were first isolated by the American biologist Thomson et al[1]. Revolutionary breakthrough in stem cell biology represents the first isolation of induced pluripotent stem cells in 2006-2007[2-5]. Due to their unique properties, stem cells have become the subject of research for many teams that work on the treatment of several serious diseases. Human and mouse ESCs can be grown in vitro as clearly visible colonies (Figure 1A-D) and effective methods have been developed for the generation of cardiomyocytes (Figure 1E), hepatocytes, melanocytes, osteoblasts, pancreatic β-cells, or neural cells (Figure 1F) from ESCs. Induction of pluripotency in terminally differentiated cells is also considered a key tool for regenerative medicine. Thus, the discovery of induced pluripotent stem cells (iPS) has great potential for the treatment of degenerative diseases without the problems associated with immunogenicity and ethical issues related to the isolation of hESCs from human embryos.
An important characteristic associated with the self-renewal and pluripotency of ESCs is their almost unlimited replication potential, which allows constant proliferation. This is ensured by activation of telomerase, the enzyme that prevents telomere shortening during cell division[1]. The self-renewal of ESCs is linked to their pluripotency and can be manifested as symmetric or asymmetric cell division. Symmetric division generates two identical sister cells whereas during asymmetric division one cell preserves the original phenotype of the mother cell and the second cell acquires a new phenotype. Asymmetric division is the basic mechanism for maintaining cell diversity, but both types of cell division ensure that physiological and morphological properties of the parental cells are transmitted to the next cell generation[6]. In this regard, it is important to understand the extent to which the nuclear pattern, especially that of chromatin is transmitted through mitosis. This issue has been addressed by several authors, although mostly in somatic cells[7-12].
In addition to self-renewal, a major feature of ESCs is their pluripotency. The single cell (zygote) formed after fertilization becomes totipotent and can produce all differentiated cells of an organism, including cells of extra-embryonic tissues. This extremely favorable feature is lost after division into the 8-cell embryonic stage[13]. However, ESCs in the inner cell mass of the blastocyst become pluripotent, and retain the potential to differentiate into cells of all three germ layers: endoderm (cells of the gastrointestinal tract and lungs), mesoderm (muscle cells, bone, blood), and ectoderm (epidermal tissues and nervous system cells) (summarized by Yamanaka and Ralston[14]). In vivo evidence of this differentiation potential is provided by injection of ESCs into immuno-deficient mice, which leads to the generation of tetracarcinomas formed by cells from all three germ layers. Moreover, in vitro evidence of ESC differentiation potential is provided by the formation of highly specific embryonic bodies (EBs)[15].
The greatest progress in understanding the pluripotency of ESCs came with the identification of three key transcription factors: Oct4, Sox2, and Nanog[14,16]. These factors have been shown to be essential for pluripotency in both in vivo and in vitro conditions[17].
Several groups of scientists have used the method of immunoprecipitation (IP) to identify the target genes of these transcription factors in mouse and human ESCs. Their data showed that transcription of many genes was regulated by the combination of these three transcription factors in hESCs[16] and that expression of Oct4 and Nanog is fundamental for the pluripotency of mouse embryonic stem cells (mESCs)[18]. Furthermore, these studies also showed that Oct4 and Nanog function as mutual regulators and self-regulators. The effect of these transcription factors on target genes seems to be significantly different between mouse and human ESCs. Nonetheless, Oct4, Sox2, and Nanog are general factors required for the maintenance of pluripotency in both organisms and other proteins probably cooperate with them to control the expression of individual genes.
The central regulator of pluripotency is Oct4, which belongs to the Pit-Oct-Unc (POU) protein family[17]. Oct4 prevents spontaneous differentiation of ESCs and artificial suppression of its expression results in the differentiation of cells into trophoectoderm[19] due to increased activity of caudal-type homeobox transcription factor 2. Moreover, in some circumstances markedly increased expression of Oct4 can also lead to differentiation, which is the main reason why negative regulators of Oct4 expression are required. For example, liver receptor homolog is believed to be a positive regulator of Oct4, whereas germ cell nuclear factor is a potential negative regulator[20].
The pluripotency regulator Sox2 is expressed in the inner cell mass of blastocysts as well as in early mesoderm[21]. In mESCs, increased expression of Sox2 induces neural differentiation and subsequent cell death. Repression of Sox2 leads to differentiation into trophoectoderm in mESCs, and in hESCs induces differentiation towards an endoderm-like pathway. However, although the above role was established in several experimental models, the main function of Sox2 remains co-operation with Oct4 to activate related target genes[22].
The other gene involved in safeguarding the pluripotent state is Nanog, which belongs to the class of NK-2 transcription factors. In mESCs, increased expression of the Nanog gene helps to maintain pluripotency even in the absence of leukemia inhibitory factor (LIF). However, hESCs can be cultured on a mouse embryonic fibroblast feeder layer, even in the presence of a high level of Nanog. If Nanog is not expressed, ESC morphology is markedly similar to that of cells induced by factor Gata6, which are characterized by the morphology of primitive endoderm[23]. Although stimulation of Nanog expression inhibits cell differentiation into endoderm, there is no direct evidence that Gata6 activity is inhibited by Nanog[24]. Nanog also inhibits neural cell differentiation induced by removal of LIF and BMP factor from the culture[25] and exerts an inhibitory effect on mesodermal differentiation. In conclusion, Nanog is able to inhibit endodermal, neural, and mesodermal differentiation under different cultivation conditions (summarized by Yamanaka and Ralston[14]).
Polycomb group proteins (PcGs) were first identified in Drosophila melanogaster as molecules that selectively inhibit the expression of many regulatory genes during embryonic development. Polycomb proteins form complexes with gene-suppressor activity. The mechanism of PcG function is based on gene silencing mediated by chromatin modifications, including tri-methylation of histone H3 at the position of lysine 27 (H3K27me3). These changes can help to maintain the undifferentiated state of stem cells[26,27]. Despite the high transcriptional activity and high degree of chromatin relaxation in ESCs, non-differentiated hESCs contain the low level of repressive chromatin marker H3K27me3. During hESC differentiation there is pronounced accumulation of H3K27me3, mostly in the inactive X chromosome[28]. PcG proteins form two main, but distinct complexes, polycomb repressive complex (PRC)1 and PRC2. Human cells contain an additional PRC3 complex, consisting of EED, EZH2, SUZ12, and RBBP subunits[29].
The PRC1 complex contains several subunits, including HPH, RING, CBX, MEL18 and BMI1. This complex is responsible for maintaining the stability of chromatin, repressed by PcG proteins. Specific factor from the PRC1 complex is also responsible for monoubiquitination of histone H2AK119[30]. The second complex, PRC2, is composed of SUZ12, EED, EZH2, and RbAP48 and is essential for the initiation of gene silencing. EZH2 functions as a histone methyltransferase that mediates H3K27me3, which acts as a binding landscape for the PRC1 protein complex. PRC2 also plays an important role in the inactivation of the X chromosome, which represents a form of facultative heterochromatin[27]. Chromatin that is modified in this way looks like dense clusters of nucleosomes and this chromatin arrangement is associated with transcriptional repression.
PcG proteins have repressive effects on the expression of many genes, including homeobox (Hox) genes that represent regulatory elements responsible for morphological changes during embryogenesis. PcG proteins maintain repression of these genes as a result of chromatin remodeling and subsequent prevention of transcription factor binding. With regard to nuclear distribution, proteins forming PcG bodies are mainly accumulated in proximity to clusters of pericentric heterochromatin. Moreover, some authors showed on ultrastructural levels PcG bodies located in the nuclear interchromatin regions. Recently, PcG bodies were revealed as as distinct and locally accumulated nuclear domains that are enriched in separated heterochromatin regions[31].
In the first half of the twentieth century, the scientific fields of developmental biology and genetics were still considered to be strictly separate. However, the British professor of genetics Conrad Hal Waddington considered this artificial division needless. In 1942, he introduced the concept of epigenetics, representing a combination of these two disciplines that can be summed up by the CH Waddington equation: epigenesis + genetics = epigenetics[32]. In those days it was still just speculation that each gene affects a number of different processes. It is now well known that genes cooperate to form a specific network and a few decades later epigenetic mechanisms are being widely discussed in the context of DNA and histone modifications.
Nowadays, the term epigenetics represents a science that deals with reversible changes in chromatin structure that are not caused by a change in the nucleotide sequence[33]. Nowadays, an increasing incidence of serious human diseases is considered one of the biggest health problems and it is assumed that epigenetic changes in the human genome are responsible for their pathophysiological states. Thus, practical application of knowledge on the epigenetics and physiology of ESCs has potential implications for further progress in medicine.
Epigenetic modifications of histones can be described as biochemical marks within histones that result in changes to chromatin conformation and thus affect chromatin accessibility to transcription factors.
The N-terminal domains of core histones are susceptible to many post-translational modifications, including acetylation (Ac), methylation (Me), phosphorylation (Ph), ubiquitination (Ub), or SUMOylatation (su)[34,35]. Because histones contain more than 60 residues that can be modified, this translates into countless variations of epigenetic modification. Histones are highly evolutionary conserved, but it is possible to establish general rules for each modification that together form so-called histone code or histone signature[36,37]. In general, histone post-translational modifications fall into two categories, those that support transcription and those that inhibit transcription[38]. Typical examples of modifications, characterized for transcriptionally active chromatin, are DNA hypomethylation, histones H3 and H4 acetylation, and methylation of H3 on Lys 4 (H3K4), H3K36, and H3K79. However, it depends which genomic regions (promoters, coding regions or enhancers) are enriched in these histone marks. Moreover, combination of specific histone marks can dictate gene transcription activity/inactivity. For repressive chromatin state is characterized hypoacetylation of H3 and H4, and methylation of H3 on Lys 9 (H3K9), H4K20, H3K27, or H4K20[39]. However, whether a given modification is gene activating or repressing may also dictate co-interacting factors. Ribosomal genes represent such an example, which transcriptional activity is regulated by ATP-dependent nucleosome remodeling events. In this case repressive H3K9me2, mediated by G9a histone methyltransferase, in a complex with other factors, including Cockayne syndrome protein B and transcription factor TTF-I, promotes transcription elongation[40].
One well-characterized post-translational modification of histones is acetylation of lysine residues, which is catalyzed by histone acetyltransferases (HATs). Histone acetyltransferases always transfer an acetyl group from acetyl-CoA to histones, thereby neutralizing their positive charge[37]. The result is a reduced ability of DNA to bind to histones, which leads to chromatin relaxation and subsequent transcriptional activation[35]. The reverse event is deacetylation, in which the acetyl group is removed in the presence of histone deacetylases (HDACs). As this increases the positive charge of histones, the chromatin becomes more condensed and transcription is repressed[37]. For example, H3K9 deacetylation was observed after induced differentiation of human ESCs[41], and was accompanied by global condensation of chromatin and restriction of global transcription[42]. Interestingly, levels of other histone modifications that characterize the active chromatin state, including H3K14ac, H3K4me3, and H3K36me2/me3, are also increased in pluripotent ESCs compared with differentiated neuronal progenitor cells (summarized by Mattout and Meshorer[43]).
Histone methylation is a covalent modification mediated by highly conserved histone methyltransferases (HMTs). This modification occurs through a biochemical reaction responsible for adding one, two, or three methyl groups to the nitrogen atoms of lysine, arginine, or histidine. The fundamental histone methylation is H3K9me2/me3 which regulates the processes of general heterochromatinization and gene silencing that primarily occur during cell differentiation. Regulation of transcription is based on the formation of specific binding sites in the genome that are attractive to regulatory proteins. In this regard, the interaction of H3K9 methylation and heterochromatin protein 1 (HP1) is particularly important[44-46].
The opposing process of histone methylation is demethylation which is catalyzed by histone demethylases. The first identified histone demethylase was lysine-specific demethylase 1 (LSD1), described by Shi et al[47]. This enzyme demethylates histone H3K4 or H3K9 under specific conditions. Interestingly, when LSD1 attacks repressive histone mark H3K9 methylation by its demethylating activity, it leads to activation of androgen receptor target genes[48]. Conversely, H3K4 demethylation seems to be fundamental for inactivation of enhancer function, which leads to specific gene silencing, according to differentiation demands, especially in ESCs. Whyte et al[49] showed that LSD1 controls the enhancers of transcriptionally active loci, which is essential for maintaining of ESC pluripotency. Interestingly, ESCs that lack LSD1 activity have no potential to differentiate. Moreover, at active enhancers LSD1 associates with the Nucleosome Remodeling and histone Deacetylase (NuRD) complex and this protein interaction appears to be fundamental for ESC differentiation[49]. The importance of the NuRD complex in ESC integrity, and some other functional events in ESC nuclei, was also reported by Reynolds et al[47]. These authors observed that the NuRD complex, which is required for ESC lineage commitment, modulates both transcriptional heterogeneity and the dynamic properties of pluripotency-related genes[50]. Moreover, HDAC1, HDAC2, and HDAC3, are present in four distinct multiprotein complexes Sin3, NuRD, CoREST, NCoR/SMRT (summarized by Hayakawa and Nakayama[51]). Thus, NuRD protein complex, consisting of HDAC1/2, is characterized by both histone deacetylase and nucleosome remodeling activity. These complex plays a role in various cellular processes including cell cycle regulation[52,53], maintenance of stem cell pluripotency[54], self-renewal, and cellular differentiation (summarized by Hayakawa and Nakayawa[51]). These findings partially clarifies our recent data on Oct4 recruitment to locally induced double strand breaks in living ES cells, which occurred simultaneously with HDAC1 recruitment; under specific histone acetylation conditions[55]. Considering that Nanog and Oct4 interact with various core components in the NuRD complex[54], these data collectively suggest that especially Oct4 can associate with specific regulatory complexes to control ESC fate.
The concept of DNA methylation has been recognized since 1948 when the first modified base, 5-methylcytosine, was discovered by Hotchkiss using the method of paper chromatography[56]. DNA methylation can be divided into two categories, maintenance methylation and de novo methylation. De novo methylation takes place on previously modified DNA whereas maintenance methylation occurs on a newly established strand of DNA that is methylated according to the pattern of the old strand[33]. Covalent DNA methylation is a post-replication modification that mainly, but not exclusively, occurs on cytosine residues in CpG dinucleotides. Based on an earlier study, it is thought that 75%-85% of DNA methylation occurs at CpG islands. These CpG dinucleotides are found primarily at the promoters of genes[57]. DNA methylation is mediated by DNA methyltransferases (Dnmts) that catalyze the transfer of a methyl group within a nucleic acid. During DNA methylation the methyl groups are accumulated in the major groove of DNA, preventing the binding of transcription factors to these genomic regions. An accompanying event related to gene silencing is the binding of the methylation-specific protein MeCP2 to methylated CpG sites resulting in chromatin conformational changes[33]. The processes of DNA methylation are associated with the function of particular methyltransferases: Dnmt1 and Dnmt2 are among the enzymes responsible for maintenance methylation whereas the only de novo methyltransferases are Dnmt3a and Dnmt3b. These two enzymes operate in early embryonic development, between the stages of morula and blastocyst, when the methylation profile is determined. In later development methylation is maintained by Dnmt1[58].
It is now well recognized that differentiation of ESCs is accompanied by Oct4 down-regulation as a consequence of de novo DNA methylation of the Oct4 regulatory region. As Athanasiadou et al[59] showed de novo methylation mediated by Dnmt3a and Dnmt3b occurs at two discrete sites: the proximal enhancer and distal promoter of the Oct4 gene. For efficient complexity, the functional inter-connection of Dnmts is essential for Oct4 transcriptional regulation. Interestingly, even H3K9 methyltransferase G9a can recruit Dnmts to the Oct4 locus and other loci upon ESC differentiation[60], but generally, CpG methylation throughout most of the regulatory genomic region accumulates even in the absence of G9a[59]. These data support the notion that histone modifications and DNA methylation are basic transcriptional regulators acting specifically and simultaneously in pluripotency-related genes.
DNA methylation is considered as well explored epigenetic phenomenon in ESCs. However, function of 5-hydroxymethylcytosine (5-hmC) in maintaining ESC pluripotency and differentiation is right now a hot topic of ESC biology. 5-hmC is generated by the TET family of Fe(II) and 2-oxoglutarate-dependent enzymes through oxidation of 5-methylcytosine (5-mC). Recent studies of Tahiliani et al[61] or Kriaucionis and Heintz[62] revealed a high level of 5-hmC in Purkinje neurons (described by Czech physiologist Jan Evangelista Purkyně) and ESCs. Interestingly, the level of 5-hmC can decrease during differentiation of ESCs, thus functional Tet1 enzyme is required for maintenance of ESC pluripotency[63]. Genome-wide analyses showed enrichment of 5-hmC at enhancers abundant on H3K4me1 and H3K27ac. Moreover, binding sites of OCT4 and NANOG were also characterized by cytosine hydroxymethylation[64]. High-throughput sequencing of 5-hmC containing DNA revealed 5-hmC within exons and near transcription start sites enriched in both H3K27me3 and H3K4me3[65]. Interestingly, these authors suggested a model of how 5-hmC contributes to “poised” chromatin signature. They claimed that similarly as 5-mC, 5-hmC at promoters caused gene down-regulation. However, in ESC differentiation-specific genes, “poised” for transcription, 5-hmC could be fundamental for subsequent gene up-regulation responsible for induction of given differentiation pathway[65]. Other report showed a novel phenomenon of how 5-hmC can further modulates epigenetic events responsible for ESC pluripotency[66]. For example, selected enhancers with a high level of 5-hmC were in parallel enriched in H3K4me1, H3K18ac, H4K5ac, H3K27ac in human ESCs. Mentioned experimental approach, and especially genome-wide analyses seems to be a promising tool how to distinguish between epigenetic landscape in pluripotent ESCs and their differentiated counterpart.
HP1 (or chromobox homolog) was first identified in Drosophila melanogaster by the American scientists James and Elgin[67]. HP1 is a highly conserved non-histone protein that plays an important role in epigenetic regulation[37,68]. The main function of HP1 is the regulation of gene expression through the binding of protein complexes to heterochromatin, thus maintaining the integrity of heterochromatin. Complex studies have confirmed this role by demonstrating specific interaction of HP1 with other histone and non-histone proteins[44-46].
HP1 homologues (HP1α, HP1β, HP1γ) have been identified in almost all eukaryotic organisms from yeast to human. HP1 proteins are the basic units of heterochromatin, including centromeres and telomeres[37,68,69]. HP1 protein subtypes are characterized by an N-terminal chromodomain (CD) and C-terminal chromoshadow domain (CSD), which are separated by the so-called “hinge region”[70]. Three-dimensional structures of the CD and CSD were determined by nuclear magnetic resonance spectroscopy[71] and X-ray crystallography[72]. Both of these domains represent globules with a diameter of approximately 30 AA. Each domain is composed of anti-parallel three-fiber β-sheets wrapped around one (α2) or two (α1, α2) α-helices. Conserved non-polar residues represent the backbone of this characteristic fold and form a hydrophobic groove on the β-sheet. This groove is not very accessible in the CSD but is relatively open in the CD, where it provides potential sites for protein-protein interactions[73].
As mentioned above, binding of HP1 protein to methylated H3K9 is important for the formation of heterochromatin[69]. The CSD binds proteins that are responsible for chromatin remodeling. The CD interacts with the N-terminal end of H3, therefore HP1 therefore gradually binds to methylated histone H3 leading to an effective increase in the local concentration of HP1. Direction of HP1 to pericentric heterochromatin requires not only H3 methylation, but also histone deacetylation[73]. Moreover, HP1 protein is not accumulated in facultative heterochromatin, such as the inactivated X chromosome, despite the fact that these chromosome territories contain methylated H3K9, and preferentially H3K27me3 appears in female inactive chromosome X. In addition, mutations in the HP1-associated genomic regions lead to re-location of HP1 from this heterochromatin[74] despite the fact that this area is rich in H3K9 methylation[75]. These observations indicate that, in addition to H3 methylation, the interaction of HP1 with other proteins is also important for directing HP1 to specific chromatin domains.
HP1 was originally characterized as a fundamental component of heterochromatin[76]. This view has been changed by the observation that HP1γ exhibits features highly characteristic of euchromatin[77]. Recent data suggest that the presence of HP1 outside of the constitutive heterochromatin has functional relevance because euchromatin also contains genes whose transcription is repressed by the presence of HP1 sub-types[78]. This is in accordance with the fact that HP1 interacts with Krüppel associated box (KRAB) and KRAB domain-associated protein (KAP-1), co-repressors of the KRAB domain and a zinc finger protein[79]. Considering that KRAB domain proteins represent the largest family of transcription repressors, it is not surprising that HP1 proteins are attracted to areas where gene silencing is expected regardless of whether this region is in heterochromatin or euchromatin[68].
One explanation for the activating and repressive effects of HP1 is that homo- and hetero-oligomers of each of the three HP1 variants play different roles. Although this depends on the chromatin context, HP1γ homodimers seem to have an activating function. HP1α-HP1γ, and HP1β-HP1γ heterodimers act variously and HP1α and HP1β homodimers act exclusively as repressors. This hypothesis is supported by the fact that HP1α and HP1β suppress the activity of several human genes while HP1γ supports their transcription[80]. An example of HP1 acting as a positive regulator of transcription can be seen in the ribosomal genes (rDNA), in which HP1β and HP1γ proteins, in a complex with RNA polymerase I and additional above mentioned proteins, cause activation of transcription rather than silencing[40,81]. Drosophila melanogaster provides another example of the HP1 protein acting as a positive regulator. Analysis of larval stages showed that gene activity of heat shock protein 70 (Hsp70) was increased in the presence of HP1 protein[82].
The nucleus of every eukaryotic cell is characterized by its well-organized structure (summarized by Cremer et al[83]). Structural integrity of the nucleus ensures proper functioning of the whole cell. As a result of co-operation among a number of regulatory factors, the nucleus consists of DNA and histone proteins that are packed into higher order chromatin structures with specific functions. Chromatin is arranged into large compartments called chromosome territories (CTs) that are partially separated by interchromatin space. Intermingling of CTs is a matter of discussion and seems to depend on the method of CT visualization, but certain interactions of CTs must exist because of the presence of chromosome translocations in tumor cells[83-85]. Taken together, current data indicate that the nucleus contains macromolecular complexes that function in basic nuclear processes, including replication, transcription, splicing, and/or DNA repair. Moreover, these processes are influenced by physiological condition of additional structures, such as the nuclear lamina, nucleolus, promyelocytic leukemia (PML) bodies or Cajal bodies that are characterized by well-defined functions[86,87].
A very important compartment of the cell nucleus is the nuclear lamina (NL), which predominantly consists of lamin proteins that form part of the nuclear membrane. The NL is not obvious at all stages of the cell cycle; it is disrupted at the beginning of mitosis by the activity of cyclin-dependent kinases and joins together again in late anaphase[86]. Other clearly distinguished structures are Cajal bodies, described in the early twentieth century by the Spanish neurobiologist Santiago Ramón y Cajal. Cajal bodies are spherical structures with a typical size of 0.1 to 2 µm. They contain high concentrations of the protein p80 coilin, small nuclear ribonucleoprotein particles, and small nucleolar ribonucleoproteins. The majority of these proteins are involved in the proper progression of RNA processing[88]. Moreover, it is generally accepted that formation of Cajal bodies is limited in hESCs, and only low level of coilin is homogeneously dispersed through hESC nucleoplasm (summarized by Morris et al[89]). Other structures with a regulatory function are PML bodies, the number of which varies from 5-30 per cell depending on the cell type and the current phase of the cell cycle[90-92]. PML bodies reach a maximum size of 1 µm and are formed by the accumulation of PML protein isoforms, which are freely distributed in the cell nucleus. PML bodies are discussed in the literature under different names, for example PML oncogenic domains, Kramer (Kr) bodies, or nuclear domains 10. PML bodies are considered protein reservoirs, and their main function is indirect regulation of gene expression, DNA repair, proteolysis, apoptosis, antiviral response, tumor suppressor function, and anti-proliferative processes[92,93]. In ES cells, PML bodies not only differ in their morphology, but also in protein composition.
Chromatin plasticity affects many nuclear processes including transcription, replication, cell cycle kinetics, and dynamics of nuclear proteins. Therefore, chromatin is a basic regulatory unit that controls the developmental and functional status of the cell and in this regard in vitro cultivated ESCs are no exception[94].
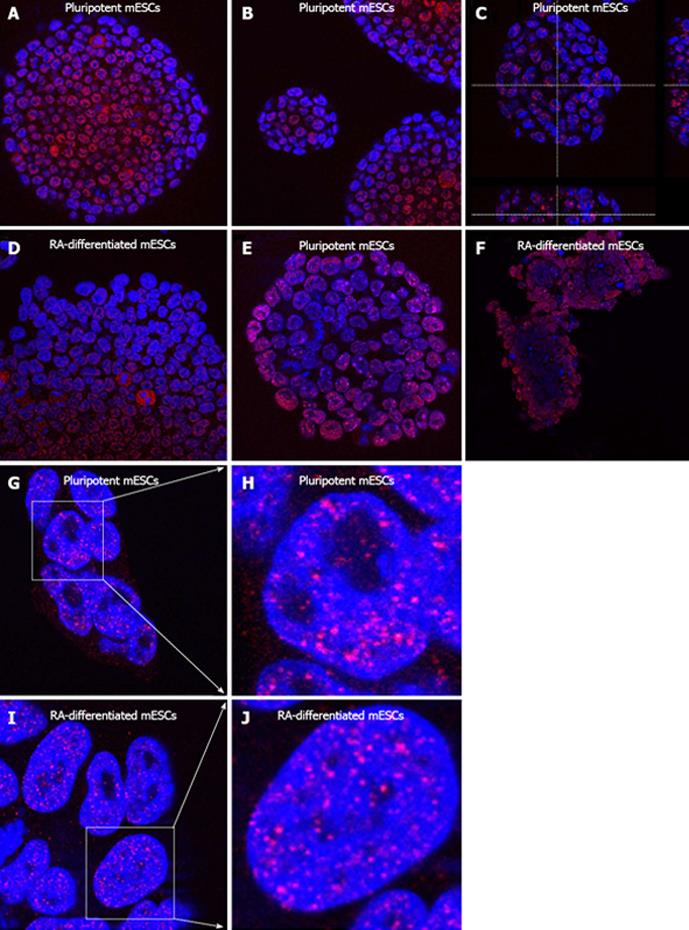
In ESCs an open, more relaxed, chromatin configuration is observed. This chromatin is highly dynamic and is significantly different from the chromatin in terminally differentiated cells[95]. In pluripotent ESCs, highly condensed state of chromatin appears rarely in comparison with differentiated counterparts, characterized by pronounced chromatin compaction. In terminally differentiated cells, especially the periphery of interphase nuclei occupies condensed heterochromatin, while more relaxed euchromatin appears in more central parts of the nucleus[83]. As a consequence, transcriptionally inactive chromatin predominates in differentiated cells whereas in pluripotent ESCs there is a large amount of transcriptionally active chromosomal regions[96]. Concerning morphological differences, chromatin in particular ESCs is spread out in relaxed regions, diffused, and amorphous, compared with smaller condensed chromatin domains in differentiated cells. In addition, nuclear lamina, which is characterized by a reduced level of A-type lamins in ESCs, becomes densely stained by the appropriate lamin antibodies after induced differentiation[97-99]. Specific distribution of euchromatin- and heterochromatin-related factors can be also observed within colonies of ESCs. Recently, we have found that Nanog-positive cells are present in the very interior regions of pluripotent ESC colonies. However, H3K27me3 shows high positivity in the ESC nuclei positioned on the periphery of the mESC colony[100]. Here, we show that nuclei with a high positivity for the splicing factor SC-35 are located in the colony interior (Figure 2A-D), but HP1β-positive cells are mainly at the periphery of the mESC colony, moreover, similar HP1β pattern was found after induced differentiation (Figure 2E, F). PcG-related BMI1 protein showed no preferential positioning within the colonies (Figure 2G-J). These data unambiguously showed that in majority of cases, nuclear pattern of ESC cells must be analyzed in view of the whole colony, because analysis on single cell level could be unsatisfactory in some cases[100].
Chromatin and chromatin-related proteins in pluripotent ESCs are additionally highly dynamic, as proved by living cell studies[98]. For example, hyperdynamic binding of structural chromatin proteins is a functionally important feature of pluripotent ESCs and is probably responsible for the undifferentiated state of ESCs[98]. In ESCs, HP1 protein and histone H1 are only loosely bound to chromatin unlike neural progenitor cells and primary mouse embryonic fibroblasts, in which HP1 and H1 binding is much stronger and results in a more “closed” chromatin configuration[87]. This raises the idea that the dynamics of chromatin proteins is regulated during cellular processes and not just a consequence of the overall biophysical state of chromatin, such as simple diffusion that can be studied by the fluorescence recovery after photobleaching technique (FRAP). Moreover, the formation of an open chromatin structure and the hyperdynamic plasticity of chromatin correlate with biological properties of ESCs[98]. This is again linked to specific transcriptional profiles, associated with fast localized movement of epigenetically important proteins[41,43,94,96].
As outlined above, it is well known that chromatin is present in the nucleus in two forms: basically, heterochromatin, which is compact, highly conserved, and transcriptionally inactive, but euchromatin is relaxed and transcriptionally active. This is also the case in ESCs. In addition, heterochromatin may be facultative, with the ability to change from a transcriptionally active to a silent state during ontogenesis[101]. The inactive chromosome X represents one of the best-known examples of facultative heterochromatin[102]. Moreover, X chromosome inactivation occurs during differentiation of female ESCs and is accompanied by repositioning of the X chromosome closer to the nuclear periphery and increased level of H3K27me3 appears[28].
Territories of autosomes also follow general principles with respect to nuclear architecture[83,103]. The general principle of nuclear organization is based on the polarized distribution of gene-rich and gene-poor chromosomes[104]. Gene-rich chromosome domains, or even whole chromosome 19, are mostly localized inwards the cell nucleus. Gene-poor chromosomes are more frequently oriented towards the nuclear periphery, for example chromosome 18[105]. The radial arrangement of territories of chromosomes 6, 8, 10, 12, 15, 17, and 19 is similar in differentiated and pluripotent hESCs (Bártová et al[28]; radial distribution is the average distance from the nuclear fluorescence center and is normalized to the average nuclear radius). For example, in hESCs the short arm of chromosome 12, which carries the Nanog gene, is oriented more towards the center of the nucleus as in lymphoblastoid cells[106]. Although the distribution of autosomes is not significantly changed during ESC differentiation, as mentioned above. One exception is the transcriptionally inactive X chromosome in the female genome[28]. Moreover, the majority of centromeric heterochromatin is re-located closer to the nuclear periphery during differentiation of hESCs[107]. This suggests that chromosomal domains are relatively dynamic structures and individual chromosomal sub-regions can be moved during cellular processes, but always with respect to other chromosomal regions and related to general nuclear structures. Thus, the dynamics of chromosomal regions seems to be important for the regulation of expression of certain genes[106,108,109].
The spatial arrangement of specific genes in the nucleus also represents a very interesting phenomenon specific to ESCs. As mentioned above, in hESCs the short arm of chromosome 12 carrying the Nanog gene is oriented more towards the center of the nucleus compared with lymphoblastoid cells[106]. Similarly, human chromosome 6, which carries the major pluripotency gene Oct4, did not change its nuclear radial position during differentiation of hESCs[107]. Another aspect related to chromosome architecture is mapping of genes within a related chromosome territory. For example, the position of the Nanog and c-myc loci within their territories remained constant after differentiation. However, in pluripotent hESCs, the Oct4 locus was located outside of its chromosome territory on large de-condensed chromatin loops, but differentiation caused Oct4 repositioning in close proximity to the related chromosome territory[106]. This robust structural change did not affect the nuclear radial distribution of the Oct4 gene[107]. Interestingly to this fact, the transcriptionally active Oct4 locus is located more internally in mESCs than in hESCs. A possible explanation for this incongruity is the level of expression of neighboring genes or even whole chromosomes, which is significantly different in human and mouse cell nuclei and might influence the general nuclear architecture of mouse and human genomes[110].
Embryonic stem cells represent a promising tool for future regenerative medicine. Recently it becomes more evident that epigenetic process and chromatin plasticity are responsible for self-renewal and pluripotency of ESCs. Thus, genome-wide studies on histone signature, DNA methylation and cytosin hydroxymethylation enable us to better understand principles of stem cells pluripotency. It is important especially from the view of complete reprogramming of iPS cells that represent and advanced methodological tool how to get pluripotent cells with high potential as therapeutic tool. Also comma we must not forget the study performed on individual cellular level, including living cell studies. For example, in such experimental systems it possible to analyzed distribution and function of pluripotency factors within colonies of ESCs[100] or nuclear events, such as transcription regulation during specific ESC differentiation[111] or during DNA repair[55,112].
Linguistic revision of the manuscript was performed by BioScience Writers (Houston, TX, United States).
P- Reviewers Kiselev SL, Zhou FC S- Editor Zhai HH L- Editor A E- Editor Lu YJ
| 1. | Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145-1147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11399] [Cited by in RCA: 10418] [Article Influence: 385.9] [Reference Citation Analysis (0)] |
| 2. | Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17989] [Cited by in RCA: 18136] [Article Influence: 954.5] [Reference Citation Analysis (0)] |
| 3. | Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14327] [Cited by in RCA: 14287] [Article Influence: 840.4] [Reference Citation Analysis (0)] |
| 4. | Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917-1920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7589] [Cited by in RCA: 7234] [Article Influence: 401.9] [Reference Citation Analysis (0)] |
| 5. | Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell. 2012;10:678-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 543] [Cited by in RCA: 527] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 6. | Walczak P, Kedziorek DA, Gilad AA, Barnett BP, Bulte JW. Applicability and limitations of MR tracking of neural stem cells with asymmetric cell division and rapid turnover: the case of the shiverer dysmyelinated mouse brain. Magn Reson Med. 2007;58:261-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 124] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 7. | Walter J, Schermelleh L, Cremer M, Tashiro S, Cremer T. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J Cell Biol. 2003;160:685-697. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 233] [Cited by in RCA: 234] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 8. | Gerlich D, Beaudouin J, Kalbfuss B, Daigle N, Eils R, Ellenberg J. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell. 2003;112:751-764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 227] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 9. | Thomson I, Gilchrist S, Bickmore WA, Chubb JR. The radial positioning of chromatin is not inherited through mitosis but is established de novo in early G1. Curr Biol. 2004;14:166-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 136] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 10. | Cénas NK, Bironaité DA, Kulys JJ. On the mechanism of rotenone-insensitive reduction of quinones by mitochondrial NADH: ubiquinone reductase. The high affinity binding of NAD+ and NADH to the reduced enzyme form. FEBS Lett. 1991;284:192-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 21] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 11. | Strickfaden H, Zunhammer A, van Koningsbruggen S, Köhler D, Cremer T. 4D chromatin dynamics in cycling cells: Theodor Boveri’s hypotheses revisited. Nucleus. 2010;1:284-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 61] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 12. | Orlova DY, Stixová L, Kozubek S, Gierman HJ, Šustáčková G, Chernyshev AV, Medvedev RN, Legartová S, Versteeg R, Matula P. Arrangement of nuclear structures is not transmitted through mitosis but is identical in sister cells. J Cell Biochem. 2012;113:3313-3329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 13. | Brignier AC, Gewirtz AM. Embryonic and adult stem cell therapy. J Allergy Clin Immunol. 2010;125:S336-S344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 78] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 14. | Yamanaka Y, Ralston A. Early embryonic cell fate decisions in the mouse. Adv Exp Med Biol. 2010;695:1-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 15. | Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 2000;18:399-404. [PubMed] |
| 16. | Chui HC, Damasio AR. Progressive dialysis encephalopathy (“dialysis dementia”). J Neurol. 1980;222:145-157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3597] [Cited by in RCA: 3371] [Article Influence: 168.6] [Reference Citation Analysis (0)] |
| 17. | Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Schöler H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2554] [Cited by in RCA: 2499] [Article Influence: 92.6] [Reference Citation Analysis (0)] |
| 18. | Mitra J, Chowdhury M. Purification and characterization of rat uterine glycerylphosphorylcholine diesterase and its tissue-specific induction by 17 beta-estradiol. Endocrinology. 1991;129:1147-1154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1833] [Cited by in RCA: 1847] [Article Influence: 97.2] [Reference Citation Analysis (0)] |
| 19. | Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2725] [Cited by in RCA: 2668] [Article Influence: 106.7] [Reference Citation Analysis (0)] |
| 20. | Gu P, LeMenuet D, Chung AC, Mancini M, Wheeler DA, Cooney AJ. Orphan nuclear receptor GCNF is required for the repression of pluripotency genes during retinoic acid-induced embryonic stem cell differentiation. Mol Cell Biol. 2005;25:8507-8519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 238] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 21. | Wood HB, Episkopou V. Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech Dev. 1999;86:197-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 430] [Cited by in RCA: 426] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 22. | Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625-635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 856] [Cited by in RCA: 866] [Article Influence: 48.1] [Reference Citation Analysis (0)] |
| 23. | Adrian TE, Bloom SR, Bryant MG, Polak JM, Heitz Ph. Proceedings: Radioimmunoassay of a new gut hormone-human pancreatic polypeptide. Gut. 1976;17:393-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2410] [Cited by in RCA: 2328] [Article Influence: 105.8] [Reference Citation Analysis (0)] |
| 24. | Hamazaki T, Oka M, Yamanaka S, Terada N. Aggregation of embryonic stem cells induces Nanog repression and primitive endoderm differentiation. J Cell Sci. 2004;117:5681-5686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 89] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 25. | Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281-292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1593] [Cited by in RCA: 1548] [Article Influence: 73.7] [Reference Citation Analysis (0)] |
| 26. | Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2098] [Cited by in RCA: 1965] [Article Influence: 103.4] [Reference Citation Analysis (0)] |
| 27. | Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301-313. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1845] [Cited by in RCA: 1774] [Article Influence: 93.4] [Reference Citation Analysis (0)] |
| 28. | Bártová E, Galiová G, Krejcí J, Harnicarová A, Strasák L, Kozubek S. Epigenome and chromatin structure in human embryonic stem cells undergoing differentiation. Dev Dyn. 2008;237:3690-3702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 49] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 29. | Kuzmichev A, Jenuwein T, Tempst P, Reinberg D. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell. 2004;14:183-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 338] [Cited by in RCA: 337] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 30. | Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343-349. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2760] [Cited by in RCA: 2514] [Article Influence: 179.6] [Reference Citation Analysis (0)] |
| 31. | Smigová J, Juda P, Cmarko D, Raška I. Fine structure of the “PcG body” in human U-2 OS cells established by correlative light-electron microscopy. Nucleus. 2011;2:219-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 32. | Van Speybroeck L. From epigenesis to epigenetics: the case of C. H. Waddington. Ann N Y Acad Sci. 2002;981:61-81. [PubMed] |
| 33. | Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5074] [Cited by in RCA: 4872] [Article Influence: 211.8] [Reference Citation Analysis (0)] |
| 34. | Vaillant I, Paszkowski J. Role of histone and DNA methylation in gene regulation. Curr Opin Plant Biol. 2007;10:528-533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 35. | Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633-636. [PubMed] |
| 36. | Kouzarides T. SnapShot: Histone-modifying enzymes. Cell. 2007;131:822. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7446] [Cited by in RCA: 7655] [Article Influence: 425.3] [Reference Citation Analysis (0)] |
| 37. | Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol. 2001;13:263-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 499] [Cited by in RCA: 498] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 38. | Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025-1040. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1184] [Cited by in RCA: 1115] [Article Influence: 61.9] [Reference Citation Analysis (0)] |
| 39. | Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat Rev Genet. 2008;9:179-191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 291] [Cited by in RCA: 262] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 40. | Yuan X, Feng W, Imhof A, Grummt I, Zhou Y. Activation of RNA polymerase I transcription by cockayne syndrome group B protein and histone methyltransferase G9a. Mol Cell. 2007;27:585-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 129] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 41. | Krejcí J, Uhlírová R, Galiová G, Kozubek S, Smigová J, Bártová E. Genome-wide reduction in H3K9 acetylation during human embryonic stem cell differentiation. J Cell Physiol. 2009;219:677-687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 58] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 42. | Efroni S, Melcer S, Nissim-Rafinia M, Meshorer E. Stem cells do play with dice: a statistical physics view of transcription. Cell Cycle. 2009;8:43-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 43. | Mattout A, Meshorer E. Chromatin plasticity and genome organization in pluripotent embryonic stem cells. Curr Opin Cell Biol. 2010;22:334-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 98] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 44. | Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2091] [Cited by in RCA: 2127] [Article Influence: 88.6] [Reference Citation Analysis (0)] |
| 45. | Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 488] [Cited by in RCA: 492] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 46. | Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305-2312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 582] [Cited by in RCA: 541] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 47. | Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941-953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2977] [Cited by in RCA: 3210] [Article Influence: 160.5] [Reference Citation Analysis (0)] |
| 48. | Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters AH, Günther T, Buettner R, Schüle R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436-439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1269] [Cited by in RCA: 1367] [Article Influence: 68.4] [Reference Citation Analysis (0)] |
| 49. | Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 447] [Cited by in RCA: 467] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 50. | Reynolds N, Latos P, Hynes-Allen A, Loos R, Leaford D, O’Shaughnessy A, Mosaku O, Signolet J, Brennecke P, Kalkan T. NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell. 2012;10:583-594. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 193] [Cited by in RCA: 182] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 51. | Hayakawa T, Nakayama J. Physiological roles of class I HDAC complex and histone demethylase. J Biomed Biotechnol. 2011;2011:129383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 114] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 52. | Rayman JB, Takahashi Y, Indjeian VB, Dannenberg JH, Catchpole S, Watson RJ, te Riele H, Dynlacht BD. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev. 2002;16:933-947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 232] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 53. | Balciunaite E, Spektor A, Lents NH, Cam H, Te Riele H, Scime A, Rudnicki MA, Young R, Dynlacht BD. Pocket protein complexes are recruited to distinct targets in quiescent and proliferating cells. Mol Cell Biol. 2005;25:8166-8178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 94] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 54. | Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10:731-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 347] [Cited by in RCA: 344] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 55. | Bártová E, Šustáčková G, Stixová L, Kozubek S, Legartová S, Foltánková V. Recruitment of Oct4 protein to UV-damaged chromatin in embryonic stem cells. PLoS One. 2011;6:e27281. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 56. | HOTCHKISS RD. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J Biol Chem. 1948;175:315-332. [PubMed] |
| 57. | Clark SJ, Harrison J, Frommer M. CpNpG methylation in mammalian cells. Nat Genet. 1995;10:20-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 193] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 58. | Sakamoto H, Suzuki M, Abe T, Hosoyama T, Himeno E, Tanaka S, Greally JM, Hattori N, Yagi S, Shiota K. Cell type-specific methylation profiles occurring disproportionately in CpG-less regions that delineate developmental similarity. Genes Cells. 2007;12:1123-1132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 59. | Athanasiadou R, de Sousa D, Myant K, Merusi C, Stancheva I, Bird A. Targeting of de novo DNA methylation throughout the Oct-4 gene regulatory region in differentiating embryonic stem cells. PLoS One. 2010;5:e9937. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 55] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 60. | Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J, Deplus R, Fuks F, Shinkai Y, Cedar H. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 2008;15:1176-1183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 366] [Cited by in RCA: 333] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 61. | Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930-935. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4860] [Cited by in RCA: 4372] [Article Influence: 273.3] [Reference Citation Analysis (0)] |
| 62. | Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929-930. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2321] [Cited by in RCA: 2050] [Article Influence: 128.1] [Reference Citation Analysis (0)] |
| 63. | Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129-1133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2151] [Cited by in RCA: 1970] [Article Influence: 131.3] [Reference Citation Analysis (0)] |
| 64. | Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12:R54. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 359] [Cited by in RCA: 374] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 65. | Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394-397. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 703] [Cited by in RCA: 647] [Article Influence: 46.2] [Reference Citation Analysis (0)] |
| 66. | Szulwach KE, Li X, Li Y, Song CX, Han JW, Kim S, Namburi S, Hermetz K, Kim JJ, Rudd MK. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7:e1002154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 625] [Cited by in RCA: 640] [Article Influence: 45.7] [Reference Citation Analysis (0)] |
| 67. | James TC, Elgin SC. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol. 1986;6:3862-3872. [PubMed] |
| 68. | Jones DO, Cowell IG, Singh PB. Mammalian chromodomain proteins: their role in genome organisation and expression. Bioessays. 2000;22:124-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 69. | Nielsen AL, Oulad-Abdelghani M, Ortiz JA, Remboutsika E, Chambon P, Losson R. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol Cell. 2001;7:729-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 308] [Cited by in RCA: 312] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 70. | Fanti L, Pimpinelli S. HP1: a functionally multifaceted protein. Curr Opin Genet Dev. 2008;18:169-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 107] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 71. | Ball LJ, Murzina NV, Broadhurst RW, Raine AR, Archer SJ, Stott FJ, Murzin AG, Singh PB, Domaille PJ, Laue ED. Structure of the chromatin binding (chromo) domain from mouse modifier protein 1. EMBO J. 1997;16:2473-2481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 138] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 72. | Cowieson NP, Partridge JF, Allshire RC, McLaughlin PJ. Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr Biol. 2000;10:517-525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 197] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 73. | Smothers JF, Henikoff S. The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr Biol. 2000;10:27-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 209] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 74. | Cowell IG, Aucott R, Mahadevaiah SK, Burgoyne PS, Huskisson N, Bongiorni S, Prantera G, Fanti L, Pimpinelli S, Wu R. Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma. 2002;111:22-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 203] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 75. | Huang DW, Fanti L, Pak DT, Botchan MR, Pimpinelli S, Kellum R. Distinct cytoplasmic and nuclear fractions of Drosophila heterochromatin protein 1: their phosphorylation levels and associations with origin recognition complex proteins. J Cell Biol. 1998;142:307-318. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 92] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 76. | Eissenberg JC, James TC, Foster-Hartnett DM, Hartnett T, Ngan V, Elgin SC. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1990;87:9923-9927. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 391] [Cited by in RCA: 416] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 77. | Nielsen AL, Ortiz JA, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 1999;18:6385-6395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 297] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 78. | Hwang KK, Eissenberg JC, Worman HJ. Transcriptional repression of euchromatic genes by Drosophila heterochromatin protein 1 and histone modifiers. Proc Natl Acad Sci U S A. 2001;98:11423-11427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 88] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 79. | Ryan RF, Schultz DC, Ayyanathan K, Singh PB, Friedman JR, Fredericks WJ, Rauscher FJ. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Krüppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol Cell Biol. 1999;19:4366-4378. [PubMed] |
| 80. | Kwon SH, Workman JL. HP1c casts light on dark matter. Cell Cycle. 2011;10:625-630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 119] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 81. | Horáková AH, Bártová E, Galiová G, Uhlírová R, Matula P, Kozubek S. SUV39h-independent association of HP1 beta with fibrillarin-positive nucleolar regions. Chromosoma. 2010;119:227-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 28] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 82. | Piacentini L, Fanti L, Berloco M, Perrini B, Pimpinelli S. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J Cell Biol. 2003;161:707-714. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 174] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 83. | Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2:a003889. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 765] [Cited by in RCA: 818] [Article Influence: 54.5] [Reference Citation Analysis (0)] |
| 84. | Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006;4:e138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 568] [Cited by in RCA: 506] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 85. | Kozubek S, Lukásová E, Rýznar L, Kozubek M, Lisková A, Govorun RD, Krasavin EA, Horneck G. Distribution of ABL and BCR genes in cell nuclei of normal and irradiated lymphocytes. Blood. 1997;89:4537-4545. [PubMed] |
| 86. | Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 682] [Cited by in RCA: 658] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 87. | Dundr M. Nuclear bodies: multifunctional companions of the genome. Curr Opin Cell Biol. 2012;24:415-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 132] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 88. | Lanctôt C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 628] [Cited by in RCA: 595] [Article Influence: 33.1] [Reference Citation Analysis (0)] |
| 89. | Morris KJ, Chotalia M, Pombo A. Nuclear architecture in stem cells. The Cell Biology of Stem Cells. In: Meshorer E, Plath K, editors. Landes Bioscience and Springer Science Bussines media, LLC 2010; 14-25. |
| 90. | Dellaire G, Ching RW, Dehghani H, Ren Y, Bazett-Jones DP. The number of PML nuclear bodies increases in early S phase by a fission mechanism. J Cell Sci. 2006;119:1026-1033. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 74] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 91. | Krejcí J, Harnicarová A, Kůrová J, Uhlírová R, Kozubek S, Legartová S, Hájek R, Bártová E. Nuclear organization of PML bodies in leukaemic and multiple myeloma cells. Leuk Res. 2008;32:1866-1877. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 92. | Borden KL. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol Cell Biol. 2002;22:5259-5269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 235] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 93. | Lallemand-Breitenbach V, de Thé H. PML nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2:a000661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 432] [Cited by in RCA: 444] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 94. | Melcer S, Meshorer E. Chromatin plasticity in pluripotent cells. Essays Biochem. 2010;48:245-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 29] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 95. | Bhattacharya D, Talwar S, Mazumder A, Shivashankar GV. Spatio-temporal plasticity in chromatin organization in mouse cell differentiation and during Drosophila embryogenesis. Biophys J. 2009;96:3832-3839. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 91] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 96. | Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RD, Buetow KH. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 97. | Constantinescu D, Gray HL, Sammak PJ, Schatten GP, Csoka AB. Lamin A/C expression is a marker of mouse and human embryonic stem cell differentiation. Stem Cells. 2006;24:177-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 272] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 98. | Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105-116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 853] [Cited by in RCA: 785] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 99. | Stixová L, Matula P, Kozubek S, Gombitová A, Cmarko D, Raška I, Bártová E. Trajectories and nuclear arrangement of PML bodies are influenced by A-type lamin deficiency. Biol Cell. 2012;104:418-432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 100. | Sustáčková G, Legartová S, Kozubek S, Stixová L, Pacherník J, Bártová E. Differentiation-independent fluctuation of pluripotency-related transcription factors and other epigenetic markers in embryonic stem cell colonies. Stem Cells Dev. 2012;21:710-720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 101. | Arney KL, Fisher AG. Epigenetic aspects of differentiation. J Cell Sci. 2004;117:4355-4363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 127] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 102. | Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD. Molecular Biology of the Cell. 3rd edition. New York: Garland Science 1994; . |
| 103. | Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Müller S, Eils R, Cremer C, Speicher MR. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:e157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 587] [Cited by in RCA: 596] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 104. | Tanabe H, Müller S, Neusser M, von Hase J, Calcagno E, Cremer M, Solovei I, Cremer C, Cremer T. Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proc Natl Acad Sci U S A. 2002;99:4424-4429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 291] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 105. | Foster HA, Bridger JM. The genome and the nucleus: a marriage made by evolution. Genome organisation and nuclear architecture. Chromosoma. 2005;114:212-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 132] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 106. | Wiblin AE, Cui W, Clark AJ, Bickmore WA. Distinctive nuclear organisation of centromeres and regions involved in pluripotency in human embryonic stem cells. J Cell Sci. 2005;118:3861-3868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 130] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 107. | Bártová E, Krejcí J, Harnicarová A, Kozubek S. Differentiation of human embryonic stem cells induces condensation of chromosome territories and formation of heterochromatin protein 1 foci. Differentiation. 2008;76:24-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 57] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 108. | Kurz A, Lampel S, Nickolenko JE, Bradl J, Benner A, Zirbel RM, Cremer T, Lichter P. Active and inactive genes localize preferentially in the periphery of chromosome territories. J Cell Biol. 1996;135:1195-1205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 198] [Cited by in RCA: 200] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 109. | Bártová E, Kozubek S, Jirsová P, Kozubek M, Gajová H, Lukásová E, Skalníková M, Ganová A, Koutná I, Hausmann M. Nuclear structure and gene activity in human differentiated cells. J Struct Biol. 2002;139:76-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 49] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 110. | Hepperger C, Mannes A, Merz J, Peters J, Dietzel S. Three-dimensional positioning of genes in mouse cell nuclei. Chromosoma. 2008;117:535-551. [PubMed] |
| 111. | Masui O, Bonnet I, Le Baccon P, Brito I, Pollex T, Murphy N, Hupé P, Barillot E, Belmont AS, Heard E. Live-cell chromosome dynamics and outcome of X chromosome pairing events during ES cell differentiation. Cell. 2011;145:447-458. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 103] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 112. | Tichy ED, Stambrook PJ. DNA repair in murine embryonic stem cells and differentiated cells. Exp Cell Res. 2008;314:1929-1936. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 109] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 113. | Veselá I, Kotasová H, Jankovská S, Procházková J, Pacherník J. Leukaemia inhibitory factor inhibits cardiomyogenesis of mouse embryonic stem cells via STAT3 activation. Folia Biol (Praha). 2010;56:165-172. [PubMed] |
| 114. | Kotasová H, Veselá I, Kučera J, Houdek Z, Procházková J, Králičková M, Pacherník J. Phosphoinositide 3-kinase inhibition enables retinoic acid-induced neurogenesis in monolayer culture of embryonic stem cells. J Cell Biochem. 2012;113:563-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |