Published online Oct 15, 2003. doi: 10.3748/wjg.v9.i10.2226
Revised: September 3, 2003
Accepted: September 10, 2003
Published online: October 15, 2003
AIM: To express all three HCV structural proteins in the presence or absence of HCV 5’NCR to investigate the requirement of 5’NCR for the assembly of HCV-like particles in insect cells.
METHODS: HCV structural protein encoding sequences CE1E2 and 5’NCR-CE1E2 were amplified with PCR. Recombinant baculovirus were constructed with recombinant DNA techniques. HCV structural proteins expressed in insect cells were analyzed by immunofluorescence and SDS-PAGE. Immunoprecipitation experiment of insect cell lysates with anti-E2 monoclonal antibody (MAb) was carried out and the immunoprecipitated proteins were subjected to SDS-PAGE and immunoblotting with anti-C, anti-E2 MAbs and HCV positive serum. The virus-like particles in insect cells were visualized by electron microscopy (EM). The HCV-like particles were purified by sucrose gradient centrifugation and identified by EM and immune aggregation EM.
RESULTS: The recombinant baculovirus reBV/CE1E2 containing HCV C, E1, E2 genes and reBV/CS containing the same structural protein genes plus 5’NCR were constructed. The insect cells infected with either reBV/CE1E2 or reBV/CS expressed HCV C, E1 and E2 proteins with a molecular weight of 20 kD, 35 kD and 66 kD respectively. The results of immunoprecipitation and the immunoblotting revealed the coimmunoprecipitation of C, E1, and E2 proteins, indicating the interaction of HCV structural proteins expressed in insect cells. Electron microscopy of insect cells infected with reBV/CE1E2 or reBV/CS demonstrated spherical particles (40 to 60 nm in diameter) similar to the HCV virions from sera or hepatic tissues of HCV infected humans. The HCV-like particles were partially purified by sucrose gradient centrifugation, and the purified VLPs showed immuno-reactivity with anti-HCV antibodies.
CONCLUSION: HCV 5’NCR is not required for the assembly of HCV-like particles in insect cells, HCV core and envelope proteins are sufficient for viral particle formation.
- Citation: Zhao W, Liao GY, Jiang YJ, Jiang SD. No requirement of HCV 5’NCR for HCV-like particles assembly in insect cells. World J Gastroenterol 2003; 9(10): 2226-2231
- URL: https://www.wjgnet.com/1007-9327/full/v9/i10/2226.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i10.2226
Hepatitis C virus (HCV) is the major causative agent of posttransfusion and sporadic non-A, non-B hepatitis. It is estimated that 170 million people worldwide are infected with HCV[1], more than 75% of infected individuals develop a chronic infection, frequently with severe long-term pathologies such as cirrhosis and hepatocellular carcinoma[2]. Neither an effective treatment for chronic HCV infection nor a vaccine to prevent HCV infection is available at the present time.
HCV belongs to the genus hepacivirus of the flaviviridae family. Its genome is a 9.6-kb single-stranded RNA of positive polarity with a 5’ noncoding region (5’NCR) that functions as an internal ribosome entry site, a single long open reading frame encoding a polyprotein of approximately 3000 amino acids (aa) and a 3’NCR. This polyprotein is posttranslationally cleaved by host cell peptidases to yield structural proteins and by viral proteases, which generate nonstructural proteins. The three structural proteins, namely core (C) and envelope glycoproteins E1 and E2, are located within the amino-terminal region of the polyprotein. The nonstructural proteins (NS) 2 to 5B reside within the carboxyl-terminal part. By analogy with other flaviviruses, HCV virion is presumed to consist of a nucleocapsid or core protein and a viral genome coated by a lipid envelope containing glycoproteins E1 and E2. The study of HCV has been hampered by the low level of viral particles in infected individuals, the inability to propagate efficiently the virus in cultured cells, and the lack of a convenient animal model. Due to these obstacles, neither the structure of the virus nor the prerequisites for its assembly have been clearly defined. Synthesis of virus like particles (VLPs) in eukaryotic cells has opened up new possibilities for defining the structural requirements for viral particle assembly under natural intracellular conditions. In this way, by manipulating the sequence of the different proteins involved, it will be possible to define experimentally the essential morphogenic interactions required for the assembly process.
The assembly of VLPs has been reported for a number of viruses[3,4] with the most successful example of human papillomavirus (HPV). Baumert et al[5] have reported the recombinant baculovirus containing entire structural protein encoding sequences plus part of 5’NCR led to the expression and assembly of HCV-like particles in insect cells.
In the present study we reported the expression of HCV structural proteins in the presence or absence of HCV 5’NCR to investigate the requirement of 5’NCR for the assembly of HCV-like particles in insect cells.
HCV cDNA was isolated from a HCV patient from Hebei Province, China, as previously described[6], and used as the amplifying template. cDNA fragments encoding HCV structural proteins were generated by PCR with the following primers: P1: 5’-ACAGATCTACCATGAGCACGAATCCTAAACC-3’, P2: 5’-ACAGATCTACTCCACCATAGATCACTCCCC-3’, P3: 5’-ATCAAGCTTACGCGTCTGCTAGTAGAAGGA-3’, P1 and P3, P2 and P3 were for CE1E2 and 5’NCR-CE1E2 respectively. A Bgl II site was introduced in P1 and P2 primers separately, a stop codon and a Hind III site were introduced in P3 primer. The correct sequences of CE1E2 and 5’NCR-CE1E2 were confirmed by DNA sequencing.
For the construction of recombinant baculoviruses, a Bac-to-Bac baculovirus expression system (Gibco-BRL/Life Technologies) was applied. The Bgl II-Hind III digestion products of PCR fragments were subcloned into BamHI-Hind III site (multiple cloning site) of baculovirus donor plasmid pFastBacI. After identification by restriction digestion and PCR, each of the recombinant plasmids was used to transform DH10Bac. Through Tn7 transposon-mediated site-specific transposition foreign gene expression cassette was integrated into a baculovirus shuttle vector (bacmid). The size of inserts was confirmed by PCR with the pUC/M13 amplification primers, which were directed at sequences on either side of the mini-attTn7 site in the bacmid. The recombinant bacmids were used for transfection of Sf9 cells (Spodoptera frugiperda). Recombinant baculoviruses were harvested thereafter and purified by plaque screening. The recombinant baculoviruses were verified by PCR with CE1E2 and 5’NCR-CE1E2 gene specific primers and amplified by subsequent rounds of Sf9 cell infection until a final titer of 5 × 107 PFU/mL was achieved. Detailed methods for baculovirus manipulation were referred to the instruction manual. Sf9 insect cells were maintained in spinner or monolayer cultures at 27 °C in Grace’s medium (Gibco-BRL/Life Technologies) supplemented with 10% fetal bovine serum.
For all protein expression experiments, Sf9 cells in mid-log growth in monolayer cultures were infected with a multiplicity of infection (MOI) of 10. Infection of insect cells with non-recombinant baculovirus served as a negative control in all experiments. The expression of HCV structural proteins was analyzed at 72 h postinfection by immunofluorescence and SDS-PAGE. For immunofluorescence, cells were fixed with -20 °C acetone for 5 min. The primary antibody used was anti-HCV serum, and the secondary antibody was fluorescein isothiocyanate (FITC)-conjugated goat anti-human IgG antibody. Detailed methods were referred to Ausubel[7]. For SDS-PAGE, the cells were lysed by ultrasonic, the cell lysate was briefly centrifuged to remove cell debris and nuclei, and the supernatant was subjected to electrophoresis on a SDS-12% polyacrylamide gel. Detailed methods were referred to Sambrook[8].
The Immunoprecipitation starter packthe (Amersham Pharmacia Biotech) was used for immunoprecipitation analysis, details of the experimental procedures were referred to the manufacturer’s protocols. Briefly, at 96 h postinfection, cells were lysed with RIPA buffer [50 mM Tris-HCl, 150 mM NaCl, 0.1% Nonidet P-40 (NP-40), 5 mM EDTA (pH7.5), 0.5% sodium deoxycholate (DOC), 1 mM phenylmethylsulfonyl fluoride (PMSF)]. The cell lysate was cleared of cell debris and nuclei by low-speed centrifugation (15 min at 15000 × g and 4 °C) and immunoprecipitated with anti-E2 monoclonal antibody (MAb). The immunoprecipitated proteins were analyzed by electrophoresis on a 12% polyacrylamide gel. After gel transfer to polyvinylidene difluoride membranes, the blots were probed with anti-C, anti-E2 MAb, or anti-HCV serum followed by horseradish peroxidase (HRP)-conjugated goat anti-mouse (for anti-C and anti-E2 MAb) or anti-human (for serum) IgG antibody. Detailed methods for immunoblotting were referred to Sambrook[8].
A flask (25 cm2) of infected cells (MOI = 10, at 96 h postinfection) was harvested, washed in PBS and centrifuged at 1000 r/min for 5 min. The cell pellets were fixed with 3.8% glutaraldehyde in PBS for 24 h at 4 °C. After fixation, the cells were washed three times in PBS (8 h each time) at 4 °C and post-fixed with 1% osmium tetroxide for 2 h at 4 °C. The samples were then dehydrated with acetone of a series of graded concentration, and embedded in 1:1 acetone-Epon 812 for 30 min at room temperature, further embedded in Epon 812 overnight at 37 °C, and then polymerized for 24 h at 60 °C. Ultrathin sections were cut with LKBIII ultramicrotome, mounted on copper grids, stained with uranyl acetate and lead citrate, washed, dried and finally examined under transmission electron microscope.
Cells were harvested at 96 h postinfection, All purification steps were carried out on ice. Insect cells infected with reBV/CS or reBV/CE1E2 (approximately 5 × 107 cells, grown in suspension) were lysed in 50 mM Tris, 50 mM NaCl, 0.5 mM EDTA, 1 mM PMSF, 0.1% NP-40. Sonication of the lysate was performed. The lysate was homogenized and subjected to low-speed centrifugation (15 min at 4 °C and 15000 × g), and the supernatant was pelleted over a 30% (wt/vol) sucrose (in 20 mM Tris, 150 mM NaCl [pH7.4]) cushion (6 h at 4 °C and 150000 × g). The pellet was resuspended in 50 mM Tris, 100 mM NaCl (pH7.4), homogenized, and subjected to a second sucrose gradient centrifugation. The resuspended pellet was layered onto a 20% to 60% (wt/wt) sucrose (in 50 mM Tris, 100 mM NaCl [pH7.4]) gradient and centrifuged for 22 h at 4 °C and 150000 × g. Fractions (0.5 mL) were collected from the top and analyzed by immunoblotting.
After the sucrose gradient centrifugation, VLPs were diluted in PBS (1:10), centrifuged for 2 h at 4 °C and 150000 × g, the pellet was resuspended in PBS. Anti-HCV serum (diluted 1:200 in PBS) was homogenized, incubated at 37 °C for 1 h, then stored at 4 °C overnight. Samples were centrifuged again for 1h at 4 °C and 150000 × g, the pellet was resuspended in PBS. Samples (3 μL) were absorbed on the surface of carbon-coated 300 mesh copper grids, and stained negatively with 2% phosphotungustic acid for EM examination.
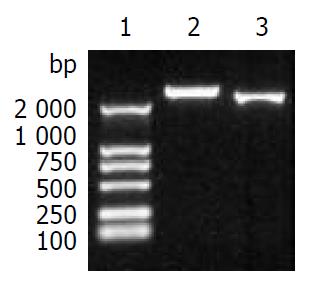
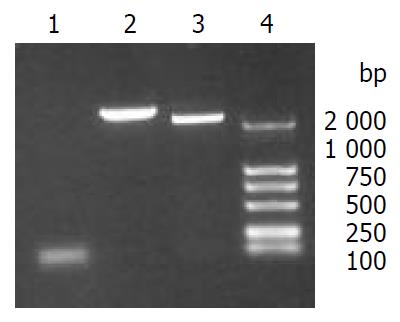
The fragment CE1E2 consisting of HCV C, E1, E2 encoding sequences and the fragment 5’NCR-CE1E2 containing the same structural protein genes plus 5’NCR were produced by PCR, with the size of 2578 bp and 2238 bp respectively (Figure 1). The correct sequences of CE1E2 and 5’NCR-CE1E2 were confirmed by DNA sequencing.
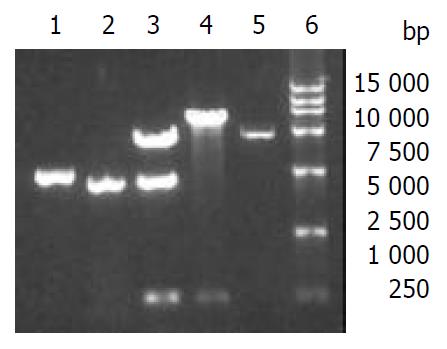
5’NCR-CE1E2 and CE1E2 were separately subcloned into baculovirus donor plasmid pFastBacI. The resulting recombinant plasmids pFB1-CS and pFB1-CE1E2 were verified (Figure 2) and used to transform DH10Bac. Through Tn7 transposon-mediated site-specific transposition foreign gene expression cassette was integrated into a baculovirus shuttle vector (bacmid). The result of PCR with M13/pUC forward and reverse primers indicted the correct insertion of 5’NCR-CE1E2 and CE1E2 fragments into bacmids (Figure 3), and the resulting recombinant bacmids were named as bacmid/CS and bacmid/CE1E2 respectively. The non-recombinant bacmid was named as bacmid/0.
Bacmid/CS and bacmid/CE1E2 were separately used to transfect Sf9 cells to generate recombinant baculoviruses, yielding reBV/CS and reBV/CE1E2, respectively. Bacmid/0 was used to generate non-recombinant baculovirus control, BV/Bac.
A large number of baculoviruses could be observed in nuclei of the Sf9 cells transfected with bacmid DNAs (Figure 4).
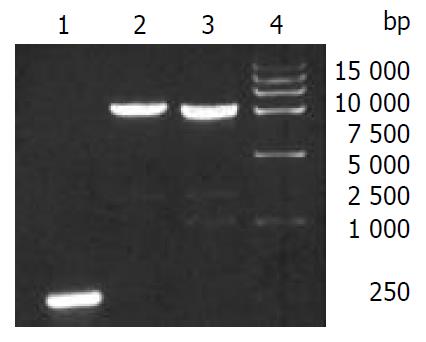
The recombinant baculoviruses reBV/CS and reBV/CE1E2 were confirmed by PCR with 5’NCR-CE1E2 and CE1E2 specific primers (Figure 5).
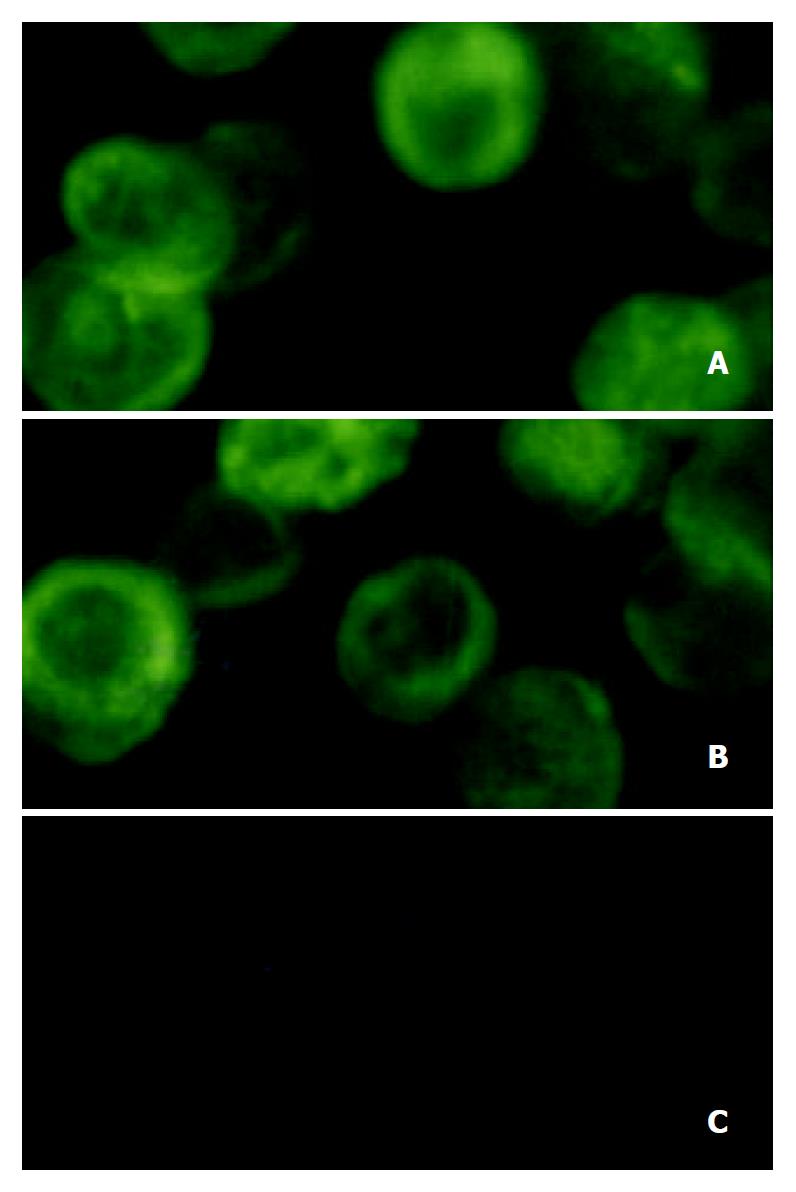
Either the recombinant baculovirus reBV/CS or reBV/CE1E2 directed the production of HCV structural proteins in insect cells, as demonstrated by immunofluorescence analysis of infected insect cells with anti-HCV antibodies (Figure 6). Immuno-staining was observed in the cells infected with recombinant baculovirus (Figure 6A and B). The anti-HCV antibodies used in this study did not display any cross-reactivity against insect cell or baculovirus proteins (Figure 6C).
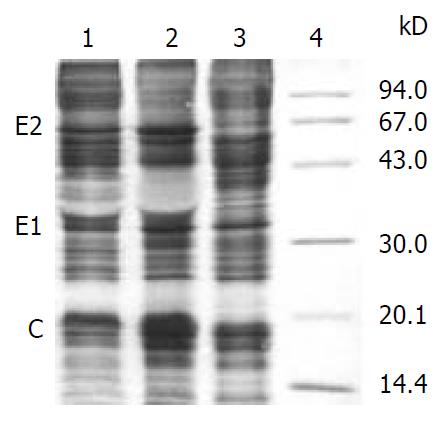
SDS-PAGE analysis of lysates from Sf-9 cells infected with either reBV/CS or reBV/CE1E2 demonstrated 3 novel bands of the size of expected HCV E2, E1 and C proteins which were 66 kD, 35 kD and 20 kD respectively, which were not present in cells infected with non-recombinant baculovirus BV/Bac (Figure 7).
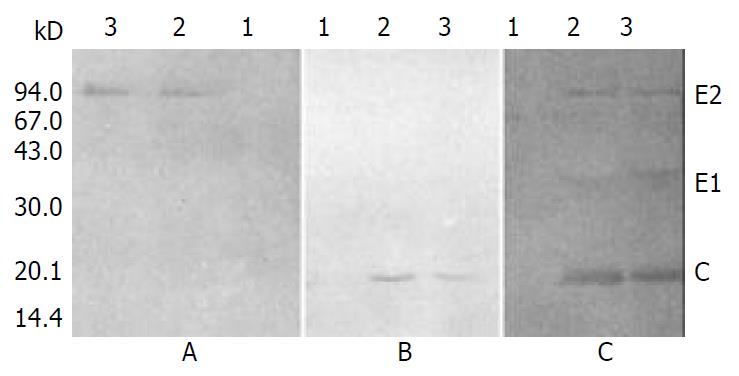
To further investigate the association of C, E1, and E2 proteins expressed in insect cells, cell extracts were immunoprecipitated with anti-E2 monoclonal antibody (MAb), and the precipitated proteins were separately probed with anti-C, anti-E2 MAb, or anti-HCV serum.
As shown in Figure 8, E2 protein was precipitated by anti-E2 MAb (Figure 8A), C protein could be detected in the immunoprecipitation complex when probed with anti-C MAb (Figure 8B), while C and E1 proteins could be detected together with E2 protein when probed with anti-HCV serum (Figure 8C). These results revealed the coimmunoprecipitation of C, E1 and E2 proteins.
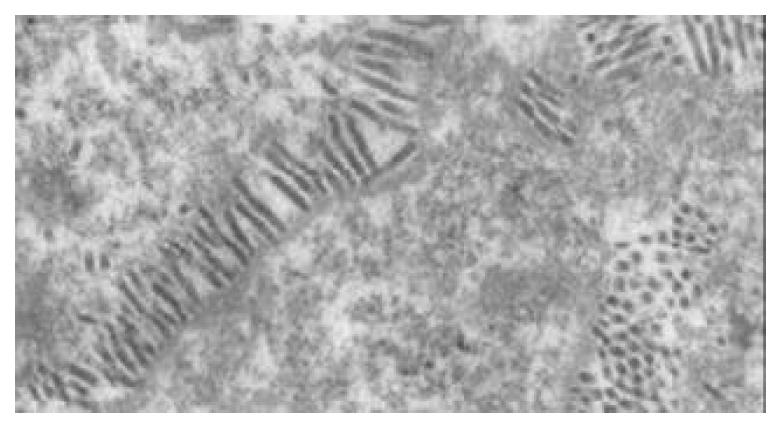
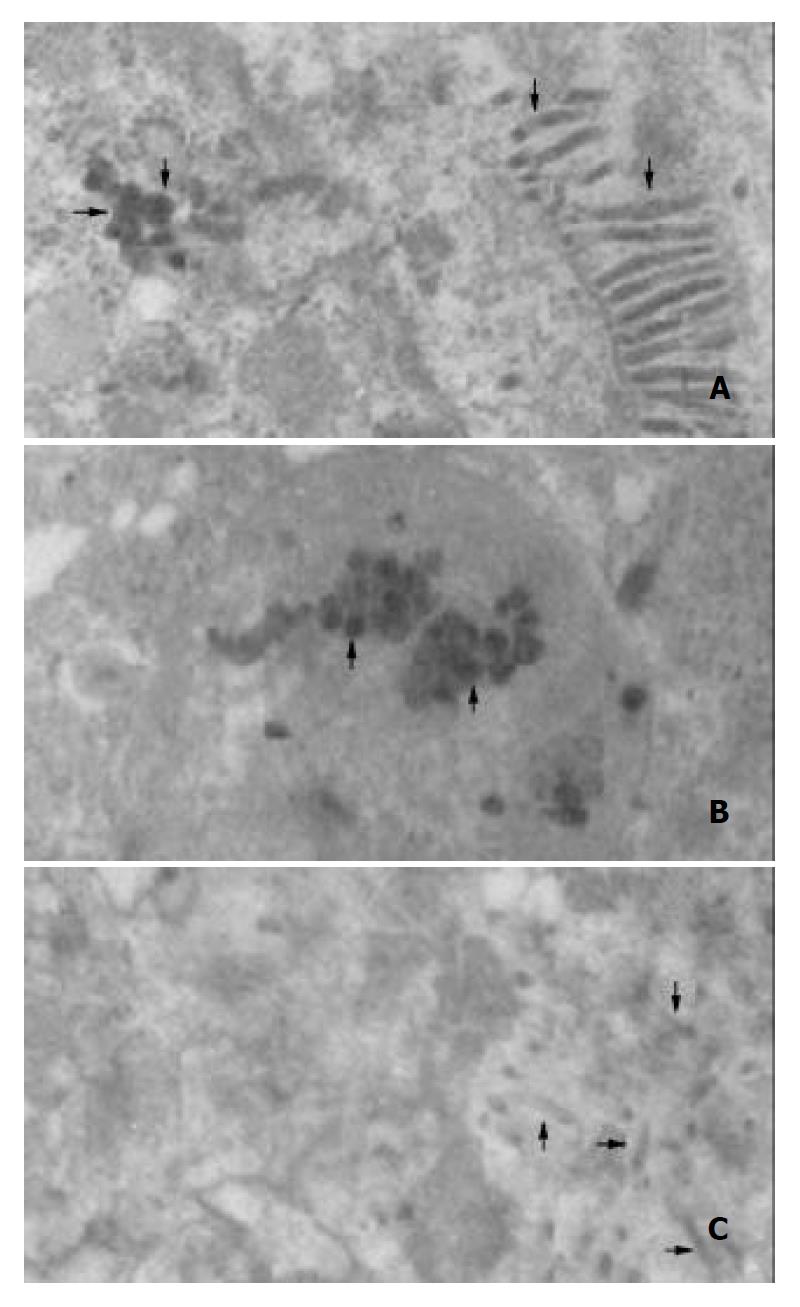
Transmission electron microscopy of the cells infected with either reBV/CS or reBV/CE1E2 revealed abundant virus-like particles in cytoplasm (Figure 9A and B). These particles (indicated by arrow “→”), 40 to 60 nm in diameter, were polymorphic in appearance, many of them had unevenly distributed electron-dense structures suggestive of possible nucleocapsids. No such structures were observed in the control cells infected with BV/Bac (Figure 9C) in spite of the existence of abundant baculoviruses in the nuclei (indicated by arrow “→”).
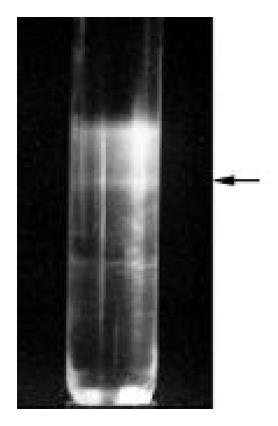
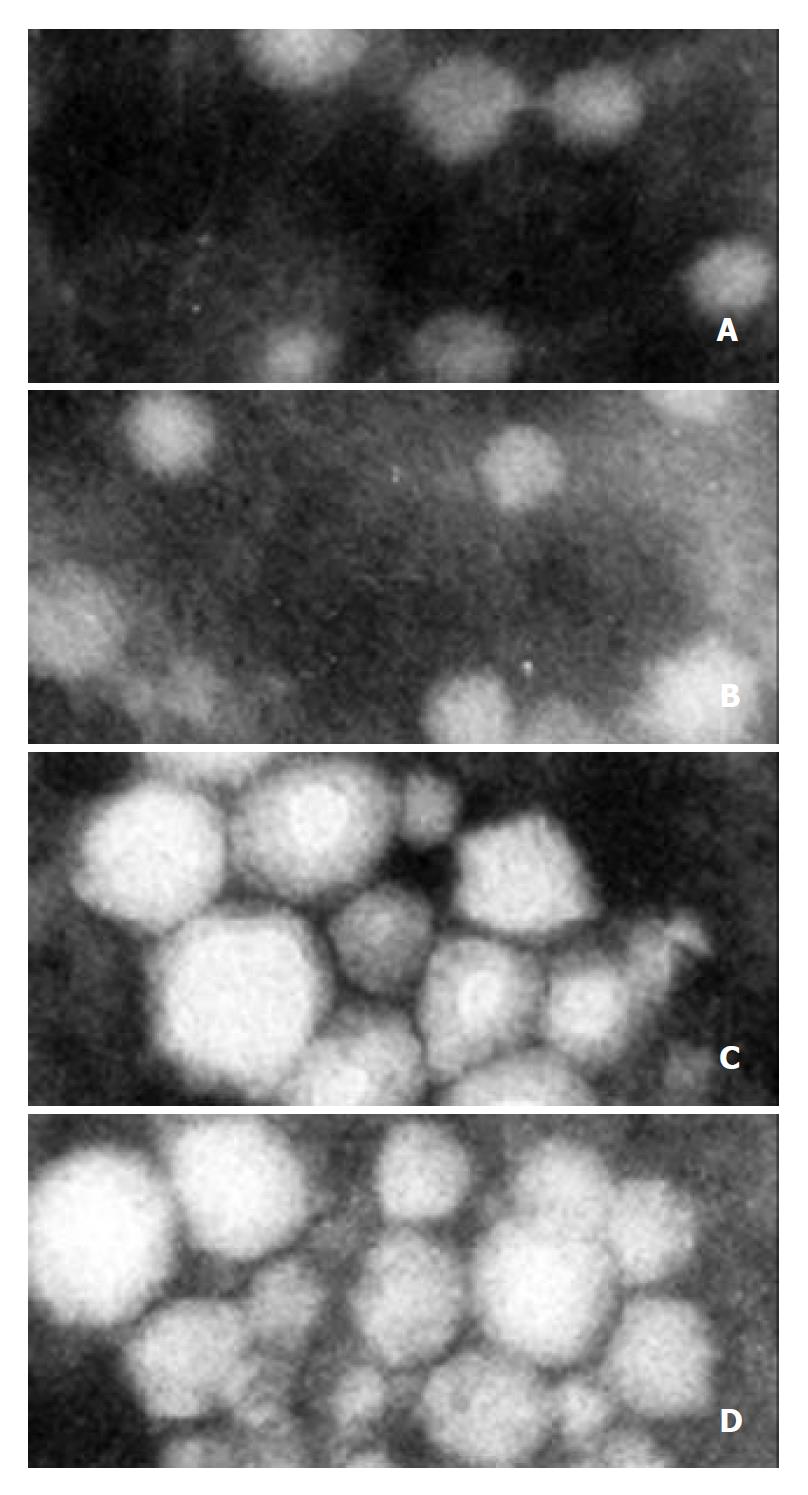
VLPs were purified from large-scale cell cultures. The band corresponding to VLPs in sucrose gradient centrifugation was indicated in Figure 10. Portions of the VLPs were examined by EM which showed separate spherical particles similar to those seen in Sf9 cells infected with recombinant baculoviruses (Figure 11A and B), other portions of VLPs were examined by immune aggregation EM which revealed aggregated VLPs by anti-HCV serum (Figure 11C and D), indicating the immuno-reactivity of VLPs with anti-HCV antibodies.
In this study, either in the presence or absence of HCV 5’NCR, the recombinant baculovirus (reBV/CS or reBV/CE1E2) efficiently expressed all the three HCV structural proteins, indicating the efficient cleavage of the polyproteins into individual proteins. The sizes of C, E1 and E2 proteins were 20 kD, 35 kD and 66 kD respectively, similar to those of HCV structural proteins expressed in mammalian cells[9], suggesting similar posttranslational processing of HCV structural proteins in insect and mammalian expression systems.
The interactions of HCV structural proteins expressed in insect cells were investigated by means of immunoprecipitation and immunoblotting. In addition to E2 protein, C and E1 proteins were also present in the immunoprecipitation complex (Figure 8B and C), indicating that C and E1 proteins were precipitated together with E2 protein by anti-E2 MAb. The result of immunoblotting of the immunoprecipitated proteins with anti-E2 MAb (Figure 8A) showed that the coimmunoprecipitation of the three structural proteins was not precipitated because there existed cross-reactivity between E2 MAb and C or E1 protein. So, we could conclude that the coimmunoprecipitation of C, E1 and E2 proteins was due to the association of the three structural proteins. These results extended the findings of previous studies which demonstrated interactions of E1 and E2 proteins[10] as well as C and E1 proteins[11], and provided immunological and biochemical evidences for the assembly of HCV structural proteins into VLPs in insect cells.
Detail ultrastructural features of HCV virions remain elusive since direct visualization of virus particles from infected serum and tissues has proven to be difficult. Filtration studies have estimated the virion particle size was 30 to 60 nm in diameter[12], Shimizu et al[13] detected VLPs with a diameter of approximately 50 nm in cytoplasm of the liver cells obtained during the acute phase of hepatitis C from a chimpanzee. Kaito et al[14] reported visualization of 55 to 65 nm VLPs by immunogold electron microscopy with antibodies to HCV envelope protein. Takahashi et al[15] observed by EM 55 nm VLPs in a gradient fraction when human plasma containing HCV was separated by potassium bromide density gradient centrifugation.
In our study, abundant spherical particles with the size of 40-60 nm in diameter were observed in the cytoplasm of Sf9 cells infected with either reBV/CS or reBV/CE1E2. Such particles could not be found in the cells infected with nonrecombinant baculovirus although the existence of abundant baculoviruses in the nuclei, indicating that VLPs in the infected cells were not correlated with the propagation of baculovirus in insect cells, but were resulted from the expression of HCV structural proteins in insect cells. Furthermore, VLPs isolated from recombinant baculovirus infected cells showed immuno-reactivity with anti-HCV antibodies, indicating that VLPs were derived from HCV proteins. Our data suggest that the morphology of HCV-like particles synthesized in insect cells is similar to the features described for HCV virions isolated from HCV-infected humans and chimpanzees. These results provided the morphological evidence for the assembly of HCV structural proteins into HCV-like particles.
In conclusion, either in the presence or absence of 5’NCR, the recombinant baculovirus containing HCV structural protein encoding sequences can direct the expression of correctly processed individual HCV structural proteins, which assemble into VLPs similar to HCV particles from sera or hepatic tissues of HCV infected humans or chimpanzees. HCV core and envelope proteins are sufficient for viral particle formation, 5’NCR is not required for the assembly of VLPs in insect cells.
Edited by Wang XL
| 1. | Cohen J. The scientific challenge of hepatitis C. Science. 1999;285:26-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 284] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 2. | Shimotohno K. Hepatitis C virus and its pathogenesis. Semin Cancer Biol. 2000;10:233-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 43] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 3. | Schiller JT, Lowy DR. Papillomavirus-like particle based vaccines: cervical cancer and beyond. Expert Opin Biol Ther. 2001;1:571-581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 45] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 4. | Latham T, Galarza JM. Formation of wild-type and chimeric influenza virus-like particles following simultaneous expression of only four structural proteins. J Virol. 2001;75:6154-6165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 192] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 5. | Baumert TF, Ito S, Wong DT, Liang TJ. Hepatitis C virus structural proteins assemble into viruslike particles in insect cells. J Virol. 1998;72:3827-3836. [PubMed] |
| 6. | Bi SL, Bai XH, Cong ME, Tian HW, Sun DG, Margolis HS, Liu CB. Primary Structure and Variation of Chinese Hepatitis C Vi-rus Genome. Bingdu Xuebao. 1993;9:114-127. |
| 7. | Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short Protocols in Molecular Biology. 3rd ed. John Wiley Sons, Inc. 1995;23-29. |
| 8. | Sambrook J, Fritsch EF, Maniatis T. Molecular cloning, A Labo-ratory Manual. 2nd ed. Cold Spring Harbor Laboratory Press. 1989;620-665. |
| 9. | Moradpour D, Wakita T, Wands JR, Blum HE. Tightly regulated expression of the entire hepatitis C virus structural region in continuous human cell lines. Biochem Biophys Res Commun. 1998;246:920-924. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 10. | Patel J, Patel AH, McLauchlan J. Covalent interactions are not required to permit or stabilize the non-covalent association of hepatitis C virus glycoproteins E1 and E2. J Gen Virol. 1999;80:1681-1690. [PubMed] |
| 11. | Ma HC, Ke CH, Hsieh TY, Lo SY. The first hydrophobic domain of the hepatitis C virus E1 protein is important for interaction with the capsid protein. J Gen Virol. 2002;83:3085-3092. [PubMed] |
| 12. | He LF, Alling D, Popkin T, Shapiro M, Alter HJ, Purcell RH. Determining the size of non-A, non-B hepatitis virus by filtration. J Infect Dis. 1987;156:636-640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 98] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 13. | Shimizu YK, Feinstone SM, Kohara M, Purcell RH, Yoshikura H. Hepatitis C virus: detection of intracellular virus particles by electron microscopy. Hepatology. 1996;23:205-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 131] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 14. | Kaito M, Watanabe S, Tsukiyama-Kohara K, Yamaguchi K, Kobayashi Y, Konishi M, Yokoi M, Ishida S, Suzuki S, Kohara M. Hepatitis C virus particle detected by immunoelectron microscopic study. J Gen Virol. 1994;75:1755-1760. [PubMed] |
| 15. | Takahashi K, Kishimoto S, Yoshizawa H, Okamoto H, Yoshikawa A, Mishiro S. p26 protein and 33-nm particle associated with nucleocapsid of hepatitis C virus recovered from the circulation of infected hosts. Virology. 1992;191:431-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 57] [Article Influence: 1.7] [Reference Citation Analysis (0)] |