Published online Feb 15, 2002. doi: 10.3748/wjg.v8.i1.135
Revised: August 6, 2001
Accepted: August 15, 2001
Published online: February 15, 2002
AIM: To establish an experimental model for exploring the role of hepatitis C virus (HCV) in the development of cholangiocarcinoma.
METHODS: Recombinant plasmid of HCV-core gene was constructed with molecular cloning technique and transfected into QBC939 cells with lipofection. After it was selected with G418, resistant colonies were obtained. The colonies were analysed by immunocytochemistry and Western blotting.The morphology was observed under transmission electron microscope(TEM) and microscope.
RESULTS: The recombinant plasmid was proved to carry the target gene by PCR and restriction enzymed mapping. Moreover, it could express HCV-C protein efficiently in QBC939 cells. The HCV-like particles were found in the cytoplasm by electron microscope, which were spherical with a diameter of 50 nm-80 nm possessing outer membrane.The transfected cells had lower differentiation and higher malignant degree under microscope.
CONCLUSION: Because HCV-core gene could express steadily in cholangiocarcinoma cells,the transfected tumor cells(QBC939-HCVC) could be used to study the effect of HCV in the development of cholangiocarcinoma.
- Citation: Liu XF, Zou SQ, Qiu FZ. Construction of HCV-core gene vector and its expression in cholangiocarcinoma. World J Gastroenterol 2002; 8(1): 135-138
- URL: https://www.wjgnet.com/1007-9327/full/v8/i1/135.htm
- DOI: https://dx.doi.org/10.3748/wjg.v8.i1.135
Cholangiocar cinoma is the second cancer of hepatobiliary system. The incidence and mortality of cholangiocarcinoma are increasing yearly.There are 3000 new patients in America each year. Recent reports showing the expression of HCVRNA and HCV antigents in cholangiocarcinoma[1-8], have provided new insights into the pathogenesis of cholangiocarcinoma. Hepatitis C virus(HCV) is recognized as a kind of serious infectious source to harm the health of the humans.It could lead to cancer[9-17].Moreover,its core protein could act as a transcriptional regulator of various viral and cellular promoters to potentially disrupt normal cellular functions[18-21]. Thus, the function of core protein is important for carcinogenesis.We constructed the recombinant plasmid of HCV core gene with molecular cloning technique and transfected into cholangiocarcinoma cells with lipofection, and established an experimental model for exploring the role of HCV in the development of cholangiocarcinoma.
Plasmids and bactrium strain PBK-HCV encompassing the core and envelope genomic regions of HCV,containing 330 nt-2020 nt of HCV II strain, was provided as a gift by Dr.Chen (Institute of Infectious Disease, Zhejiang University), whose restriction sites were PstI and EcoRI.The prokaryotic expressing vector of PBK-CMV containing MCS(multiple cloning site),the neomycin- and kanamycin-resistance gene, SV40 poly(A), was purchased from Stratagene Co. E.coli JM109 was obtained from the collection kept in our research group.
Cells The QBC939 cells(a cholangiocarcinoma cell line)were a generous gift from Dr. Wang Shuguang(Third Military Medical University,China). QBC939 cells were cultured in RPMI 1640 supplemented with 100 mL·L-1 FBS and incubated at 37 °C in a 50 mL·L-1 CO2 atomosphere.
Reagents Restriction enzymes(SalI and BamHI), Taq DNA polymerase, T4DNA ligase,were purchased from Hua Mei Co. Lipofection was provided by Boehringer Mannheim Co. Mouse anti-HCV C protein monoclonal antibody was purchased from Chemicon Co. House anti-mouse IgG(mouse)-AP, NBT/BICP were purchased from Zhong Shan Co. Biotinylated-conjugated sheep anti-mouse IgG was purchased from Boster Co.
PCR primers Two primers were designed according to the sequence of HCV core genomic regions and synthesized by the Shanghai GeneCore Bio Technologies Co. Primer 1:5’-CTCGTCGACCATGAGCACAAATCCTAA-3’; Primer 2:5’-CTCGGATCCTAAGCGGAAGCTGGGATG-3’. Primer 1 was 5’primer containing SalI site and Primer 2 was 5’ primer containing BamHI site.
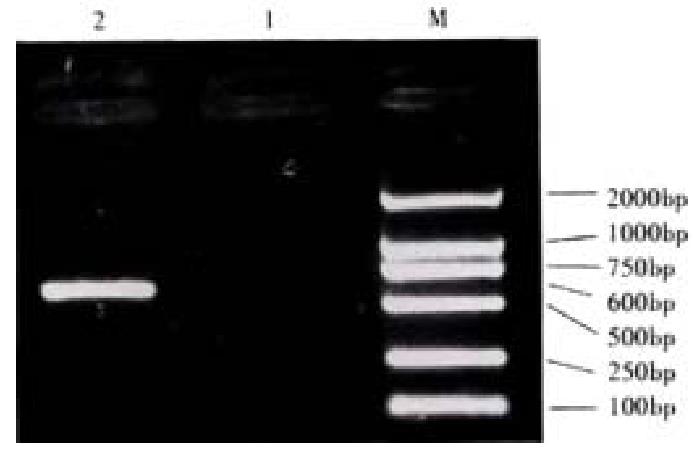
PCR amplification PCR amplification was done using the PBK-HCV as a template and primers 1 and 2 as primers.The reaction was performed according to the parameters of 94 °C 3 min, 94 °C 1 min, 55 °C 30 s and 72 °C 1 min for 35 cycles.Then it was extended 10 min at 72 °C. The amplified fragment (approx 600 bp)was analyzed by 9 g·L-1 agarose-gel electrophoresis.The product was purified and used for DNA recombinction.
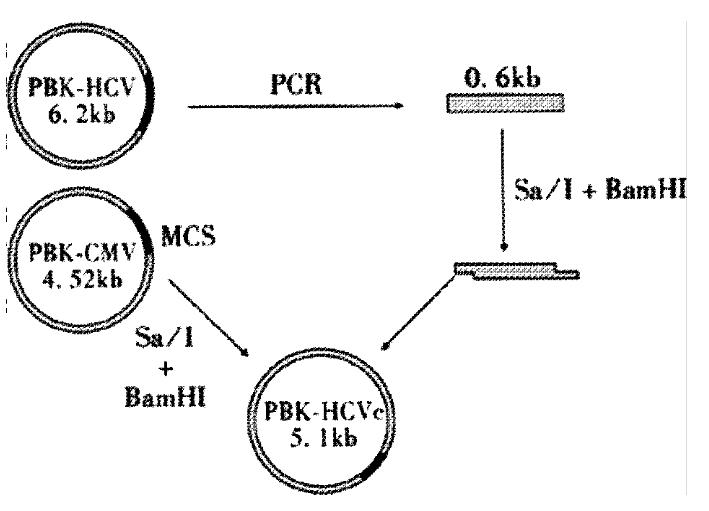
DNA recombination DNA recombination was performed according to the methods described in reference[22]. A 0.6 kb fragment,containing HCV core gene,was obtained from the PCR product after digestion with SalI and BamHI.The fragment was recombinated into plasmid PBK-CMV and the resulted recombinant plasmid was designated as PBK-HCVC (Figure 1).
Transfection of cells Transfection was performed with lipofection.The constructed vector and control plasmid were used to transfect QBC939 cells in culture. Seven-two h after transfection, they were selected with G418. Then the cells were harvested and used for the detections.
Immunocytochemistry for HCV core protein Detection of HCV core protein expression was performed using a mouse anti-HCV core protein monoclonal antibody(1:50) on cell sections of transfected QBC939 and control plasmid.
Western blotting Detection of HCV core protein by immunoblotting was performed. Briefly,cells (1 × 106) were scraped,centrifuged briefly,and lysed for 30 min on ice in 50 mmol·L-1 Tris-cl(pH 7.5), 150 mmol·L-1 NaCl, 0.2 mmol·L-1 EDTA, 1 mmol·L-1 PMSF and 10 g·L-1 NP-40. The samples were cleared by centrifugation(140000 r·min-1,30 min,4 °C),and assessed for protein concentration.SDS-PAGE was performed,and proteins were electroblotted onto nitrocellulose membranes.After 1h incubation in blocking solution,the membrane was exposed to the primary antiobody 2 h at 4 °C.After washing in PBS,the secondary AP-labeled antibody was added for 2 h at room temperature.The proteins were visualized with NBT-BICP.
Transmission electron microscope A pellet of the transfected cells was fixed in 2.5 g·L-1 glutraraldehyde,postfixed with 10 g·L-1 osmium tetroxide,treated with 20 g·L-1 uramyl acctate,dehydrated in ethanol, infiltrated with propylene oxide,and embedded in Epon mixture.Ultrathin sections were observed under Opton EM 10C(German).
Microscope After selected with G418, the cell sections were stained with HE.The cell sections were observed under microscope.
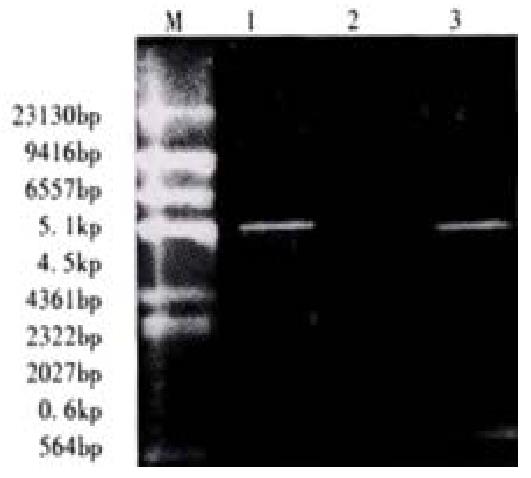
The reconstructed plasmid was amplified by PCR, using the PBK-HCVc as a template and primers 1 and 2 as primers, the reaction was performed according to the same parameters. PCR product was approx 600 bp(Figure 2). Then it was identified by the digestion with restriction enzymes(SalI and BamHI). Fragments of 4500 bp and 600 bp were produced from the digestion. It was proved to carry the target gene(Figure 3).
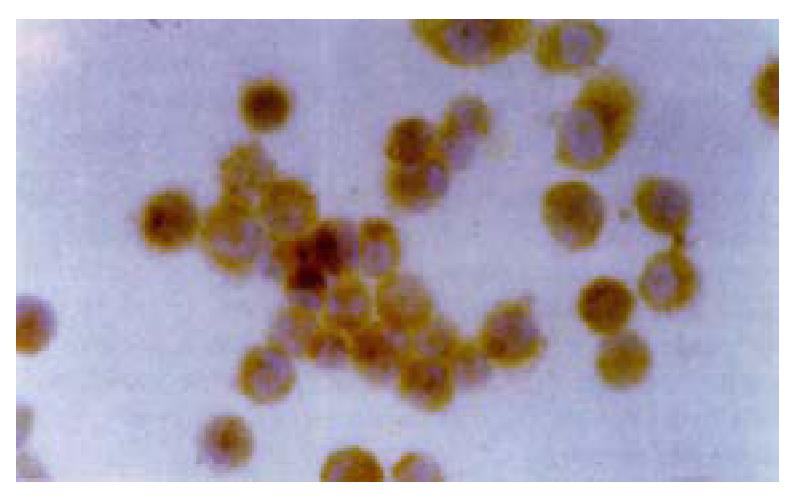
Cell sections of transfected with PBK-HCVC and control plasmid (PBK-CMV) were stained for HCV core protein. Staining for HCV core protein was seen in the PBK-HCVC transfected QBC939 cells. Positive staining was located to the cytoplasm(Figure 4).
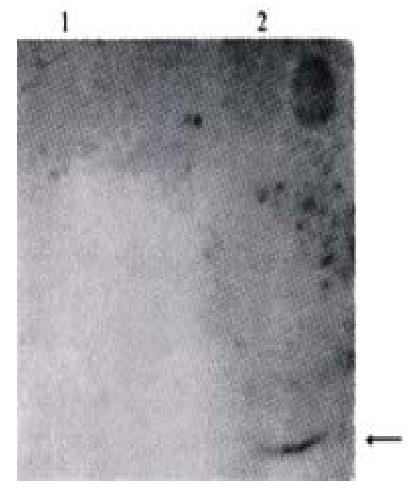
The expressed products in the supernatant which were transfected with PBK-HCVC and control plasmid(PBK-CMV) were identified by Western blotting. A appox 21 ku band,similar to the size of HCV core protein,was observed in the supernatant of transfection with PBK-HCVC(Figure 5). These results showed that the QBC939 cells transfected with PBK-HCVC could express HCV core protein
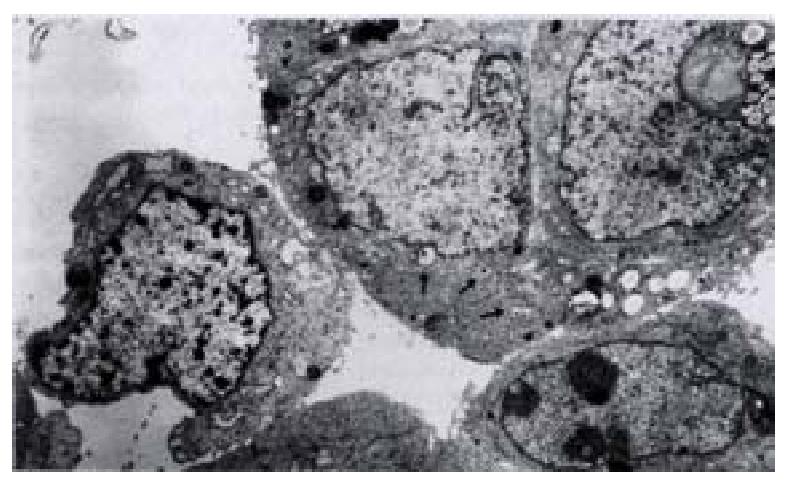
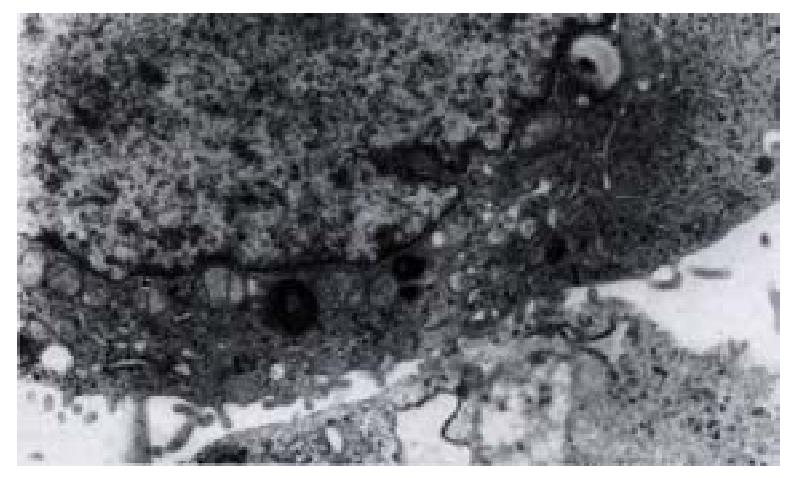
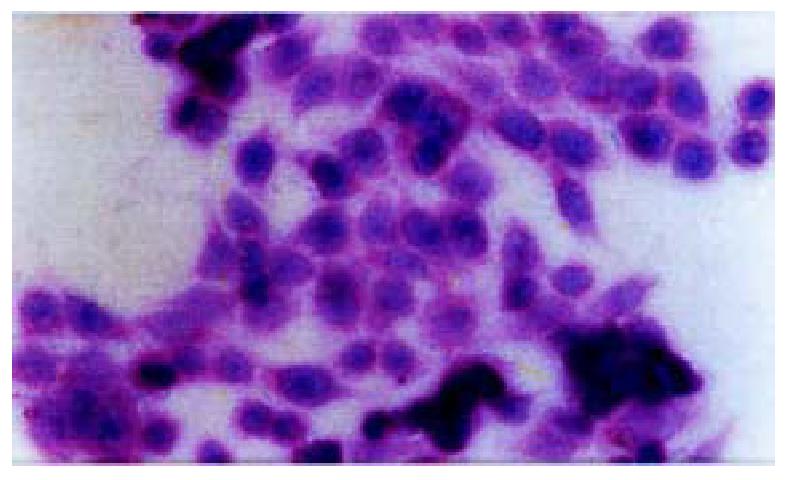
On day 15 after QBC939 cells were transfected with PBK-HCVC, the HCV-like particles were found in the cytoplasm under TEM, which were spherical with a diameter of 50 nm-80 nm possessing outer membrane(Figures 6 and 7).The structures were absent in the negative control.The results suggested that the recombinent plasmid could express HCV core protein efficiently in cholangiocarcinoma cell lines.The splits of nuclei were increased significantly in transfected QBC939 cells with PBK-HCVC than PBK-CMV and QBC939 cells (Figure 8). It suggested that the exent of the differentiation was reduced and malignant degree was enhanced after QBC939 cells were transfected with PBK-HCVC.
The pathogenesis of cholangiocarcinoma is not clear. Cholelithiasis,cystic dilation of the biliary system,ulcerative colitis and primary sclerosing cholangitis are thought to be the risk factors for cholangiocarcinoma.Recently, laboratory and epidemiological studies found and epidemic found the infection of HCV was related to the development of cholangiocarcinoma [1-8,23-25].But the mechanism of how they are related has not been studied deeply. HCV infection is an important cause of morbidity and mortality worldwide,causing a spectrum of liver diseases ranging from an asymptomative carrier state to end-stage liver disease.The most important feature of persistent HCV infection is the development of chronic hepatitis in half of the infected individuals and the potential for disease progression to hepatocellular carcinoma.In our country there are 30-40 million of HCV-carriers patients[26-28].
HCV are hepatotropic viruses.Viral replication and cellar injury are largely confined to the liver.Recent studies,however,have suggested that HCV may replicate in tissues other than in hepatocyte only. Many scholars have reported the presence of HCVRNA and HCV-antigens in lymph nodes, pancreas, ovary, kidney, heart and bile duct epithelial cells[29-35]. The infection of HCV could lead to bile duct damage and loss. Moreover,bile duct injury is more commonly associated with HCV than either HBV or autoimmune hepatitis,which is the characteristic histologic feature of HCV. The bile duct damage and loss are defined as variable epithelial, steatosis,lymphocytic infiltration of bile ducts[36-38].The more injury of HCV infection in bile duct provides new evidence that bile duct epithelial cells could be an important reservoir of HCV and might lead to the pathogenesis of cholangiocarcinoma. An HCV genome contains a linear,positive-strand RNA molecule of 9500 nucleotides encoding a single ployprotein precursor of 3000 amino acids.The polyprotein is cleaved by viral proteases to generate three putative structural proteins(C, E1 and E2) and at least six nonstructural proteins(NS2,NS3,NS4A, NS4B, NS5A and NS5B)[39,40]. The genomic region encoding the putative C protein is also called core protein. The HCV core protein could act as a transcriptional regulator of various viral and cellular promoters to potentially disrupt normal cellular functions[18-21]. The core protein may cooperate with ras oncogene and transform primary rat embryo fibroblasts to a tumorigenic phenotype[41]. It may also cause anti-apoptosis by ressive of tumor suppressor gene-p53 and activation of NF-κB and implicate a mechanism by which HCV may evade the host immune surveillance leading to viral persistence and possibly to carcinogenesis[42-51]. Thus,the HCV core protein plays a major role in the malignant transformation of cells. It is important to explore the role of HCV in the development of cholangiocarcinoma by establishing an experimential model which expresses efficiently HCV core protein in cholangiocarcinoma cell lines.
In our study, recombinant plasmid of HCV-core gene was constructed with molecular cloning technique,it was identified by restriction enzymes.Then it was transfected into QBC939 cells with lipofection.After it was selected with G418 made by the presence of the necomycin and kanamycin resistance gene, resistant colonies were obtained. The colonies were analysed by immunocytochemistry and Western blotting. The results suggest that the recombinant plasmid was proved to carry the target gene by PCR and restricted enzyme map, and it could express HCV core protein efficiently in QBC939 cells.The morphology was observed under transmission electron microscopy(TEM), and the HCV-like particles in the cytoplasm, were found spherical with a diameter of 50 nm-80 nm possessing outer membranes. Moreover, we also found that the splits of nuclei were increased in transfected QBC939 cells with PBK-HCVC, and the exent of differentiation was reduced. It suggested that the malignant degree was enhanced after QBC939 cells were transfected with PBK-HCVC. Because HCV-core gene could express steadily in cholangiocarcinoma cells, the transfected tumor cells(QBC939-HCVC) could be used to study the effect of HCV in the development of cholangiocarcinoma.
Edited by Ma JY
| 1. | Tomimatsu M, Ishiguro N, Taniai M, Okuda H, Saito A, Obata H, Yamamoto M, Takasaki K, Nakano M. Hepatitis C virus antibody in patients with primary liver cancer (hepatocellular carcinoma, cholangiocarcinoma, and combined hepatocellular-cholangiocarcinoma) in Japan. Cancer. 1993;72:683-688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 2. | Zhang H, Tang Y, Lu X. [Detection of hepatitis B virus DNA and hepatitis C virus RNA in human hepatocellular carcinoma by polymerase chain reaction]. Zhonghua Binglixue Zazhi. 1996;25:70-72. [PubMed] |
| 3. | Yin FZ, Chen BF, Xu CS. HCVRNA sequences in cholangiocarcinoma tissues. Zhonghua Shiyan Waike Zashi. 1998;15:109-110. |
| 4. | Chen RF, Zou SQ, Zhao XP. The expression of hepatitis C virus gene in hilar cholangiocarcinoma and implication. Zhonghua Shiyan Waike Zashi. 2000;17:223-224. |
| 5. | Lu H, Ye MQ, Thung SN, Dash S, Gerber MA. Detection of hepatitis C virus RNA sequences in cholangiocarcinomas in Chinese and American patients. Chin Med J (Engl). 2000;113:1138-1141. [PubMed] |
| 6. | Chen MY, Huang ZQ, Chen LZ, Gao YB, Peng RY, Wang DW. Detection of hepatitis C virus NS5 protein and genome in Chinese carcinoma of the extrahepatic bile duct and its significance. World J Gastroenterol. 2000;6:800-804. [PubMed] |
| 7. | Zhai SH, Liu JB, Liu YM, Zhang LL, Du ZP. Expression of HBsAg, HCV-Ag and AFP in liver cirrhosis and hepatocarcinoma. Shijie Huaren Xiaohua Zazhi. 2000;8:524-527. |
| 8. | Wang WL, Wang CJ, Wang BF. Significance of HCV gene and its antigen expression in human primary intrahepatic cholangiocarcinoma. Shijie Huaren Xiaohua Zazhi. 2001;9:542-545. |
| 9. | Hiramatsu N, Hayashi N, Haruna Y, Kasahara A, Fusamoto H, Mori C, Fuke I, Okayama H, Kamada T. Immunohistochemical detection of hepatitis C virus-infected hepatocytes in chronic liver disease with monoclonal antibodies to core, envelope and NS3 regions of the hepatitis C virus genome. Hepatology. 1992;16:306-311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 85] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 10. | Yan XB, Wu WY, Wei L. Clinical features of infection with different genotypes of hepatitis C virus. Huaren Xiaohua Zazhi. 1998;6:653-655. |
| 11. | Zhang LF, Peng WW, Yao JL, Tang YH. Immunohistochemical detection of HCV infection in patients with hepatocellular carcinoma and other liver diseases. World J Gastroenterol. 1998;4:64-65. [PubMed] |
| 12. | Sorensen HT, Friis S, Olsen JH, Thulstrup AM, Mellemkjaer L, Linet M, Trichopoulos D, Vilstrup H, Olsen J. Risk of liver and other types of cancer in patients with cirrhosis: a nationwide cohort study in Denmark. Hepatology. 1998;28:921-925. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 230] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 13. | Yang JM, Wang RQ, Bu BG, Zhou ZC, Fang DC, Luo YH. Effect of HCV infection on expression of several cancer-associated gene products in HCC. World J Gastroenterol. 1999;5:25-27. [PubMed] |
| 14. | Feng DY, Chen RX, Peng Y, Zheng H, Yan YH. Effect of HCV NS(3) protein on p53 protein expression in hep atocarcinogenesis. World J Gastroenterol. 1999;5:45-46. [PubMed] |
| 15. | Huang F, Zhao GZ, Li Y. HCV genotypes in hepatitis C patients and their clinical significances. World J Gastroenterol. 1999;5:547-549. [PubMed] |
| 16. | Rabe C, Pilz T, Klostermann C, Berna M, Schild HH, Sauerbruch T, Caselmann WH. Clinical characteristics and outcome of a cohort of 101 patients with hepatocellular carcinoma. World J Gastroenterol. 2001;7:208-215. [PubMed] |
| 17. | Tang ZY. Hepatocellular carcinoma--cause, treatment and metastasis. World J Gastroenterol. 2001;7:445-454. [PubMed] |
| 18. | Liu Q, Tackney C, Bhat RA, Prince AM, Zhang P. Regulated processing of hepatitis C virus core protein is linked to subcellular localization. J Virol. 1997;71:657-662. [PubMed] |
| 19. | Dai YM, Shou ZP, Ni CR, Wang NJ, Zhang SP. Localization of HCV RNA and capsid protein in human hepatocellular carcinoma. World J Gastroenterol. 2000;6:136-137. [PubMed] |
| 20. | Liu LH, Xiao WH, Liu WW. Effect of 5-Aza-2'-deoxycytidine on the P16 tumor suppressor gene in hepatocellular carcinoma cell line HepG2. World J Gastroenterol. 2001;7:131-135. [PubMed] |
| 21. | Ray RB, Meyer K, Ray R. Hepatitis C virus core protein promotes immortalization of primary human hepatocytes. Virology. 2000;271:197-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 134] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 22. | Sambrook J,Fritsch EF,Maniatis T(Translated by Jin DY,Liang MF). Molecular Cloning: A Laboratory Manal(2nd Ed). Beijing: Science Press. 1996;672-898. |
| 23. | Yamamoto M, Takasaki K, Nakano M, Saito A. Minute nodular intrahepatic cholangiocarcinoma. Cancer. 1998;82:2145-2149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 24. | Suriawinata A, Ivanov K, Ben Haim M, Schwartz ME. A 67-year-old man with hepatitis C virus infection and a liver tumor. Semin Liver Dis. 2000;20:227-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 25. | Kobayashi M, Ikeda K, Saitoh S, Suzuki F, Tsubota A, Suzuki Y, Arase Y, Murashima N, Chayama K, Kumada H. Incidence of primary cholangiocellular carcinoma of the liver in japanese patients with hepatitis C virus-related cirrhosis. Cancer. 2000;88:2471-2477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 26. | Zhang SL, Liang XS, Lin SM, Peng Chao Qiu. Relation between viremia level and liver disease in patients with chronic HCV infection. China Natl J New Gastroenterol. 1996;2:115-117. |
| 27. | Assy N, Minuk G. A comparison between previous and present histologic assessments of chronic hepatitis C viral infections in humans. World J Gastroenterol. 1999;5:107-110. [PubMed] |
| 28. | Li LF, Zhou Y, Xia S, Zhao LL, Wang ZX, Wang CQ. The epidemio-logic feature of HCV prevalence in Fujian. World J Gastroenterol. 2000;6:80. |
| 29. | Shimizu YK, Iwamoto A, Hijikata M, Purcell RH, Yoshikura H. Evidence for in vitro replication of hepatitis C virus genome in a human T-cell line. Proc Natl Acad Sci USA. 1992;89:5477-5481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 219] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 30. | Moldvay J, Deny P, Pol S, Brechot C, Lamas E. Detection of hepatitis C virus RNA in peripheral blood mononuclear cells of infected patients by in situ hybridization. Blood. 1994;83:269-273. [PubMed] |
| 31. | Zhou P, Cai Q, Chen YC, Zhang MS, Guan J, Li XJ. Hepatitis C virus RNA detection in serum and peripheral blood mononuclear cells of patients with hepatitis C. China Natl J New Gastroenterol. 1997;3:108-110. |
| 32. | Kato N, Nakazawa T, Mizutani T, Shimotohno K. Susceptibility of human T-lymphotropic virus type I infected cell line MT-2 to hepatitis C virus infection. Biochem Biophys Res Commun. 1995;206:863-869. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 84] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 33. | Fan XG, Tang FQ, Ou ZM, Zhang JX, Liu GC, Hu GL. Lymphoproliferative response to hepatitis C virus (HCV) antigens in patients with chronic HCV infection. Shijie Huaren Xiaohua Zazhi. 1999;7:1038-1040. |
| 34. | Yan FM, Chen AS, Hao F, Zhao XP, Gu CH, Zhao LB, Yang DL, Hao LJ. Hepatitis C virus may infect extrahepatic tissues in patients with hepatitis C. World J Gastroenterol. 2000;6:805-811. [PubMed] |
| 35. | Cheng JL, Liu BL, Zhang Y, Tong WB, Yan Z, Feng BF. Hepatitis C virus in human B lymphocytes transformed by Epstein-Barr virus in vitro by in situ reverse transcriptase-polymerase chain reaction. World J Gastroenterol. 2001;7:370-375. [PubMed] |
| 36. | Bach N, Thung SN, Schaffner F. The histological features of chronic hepatitis C and autoimmune chronic hepatitis: a comparative analysis. Hepatology. 1992;15:572-577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 359] [Cited by in RCA: 317] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 37. | Goldin RD, Patel NK, Thomas HC. Hepatitis C and bile duct loss. J Clin Pathol. 1996;49:836-838. [PubMed] |
| 38. | Giannini E, Ceppa P, Botta F, Fasoli A, Romagnoli P, Cresta E, Venturino V, Risso D, Celle G, Testa R. Steatosis and bile duct damage in chronic hepatitis C: distribution and relationships in a group of Northern Italian patients. Liver. 1999;19:432-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 39. | Zhang SZ, Liang JJ, Qi ZT, Hu YP. Cloning of the non-structural gene 3 of hepatitis C virus and its inducible expression in cultured cells. World J Gastroenterol. 1999;5:125-127. [PubMed] |
| 40. | Jiang RL, Lu QS, Luo KX. Cloning and expression of core gene cDNA of Chinese hepatitis C virus in cosmid pTM3. World J Gastroenterol. 2000;6:220-222. [PubMed] |
| 41. | Ray RB, Lagging LM, Meyer K, Ray R. Hepatitis C virus core protein cooperates with ras and transforms primary rat embryo fibroblasts to tumorigenic phenotype. J Virol. 1996;70:4438-4443. [PubMed] |
| 42. | Xiao WH, Liu WW, Lu YY, Li Z. Mutation of p53 tumor suppressor gene in hepatocellular carcinoma. Xin Xiaohuabingxue Zazhi. 1997;5:573-574. |
| 43. | Wei HS, Li DG, Lu HM. Hepatic cell apoptosis and fas gene. Shijie Huaren XiaohuaZazhi. 1999;7:531-532. |
| 44. | Li J, Chen YF, Wang WL, Lin SG. Translocated expression of HCV core protein inhibits apoptosis in the tissue of hepatocellular carcinoma. Shijie Huaren Xiaohua Zazhi. 1999;7:579-582. |
| 45. | Si XH, Yang LJ. Extraction and purification of TGFbeta and its effect on the induction of apoptosis of hepatocytes. World J Gastroenterol. 2001;7:527-531. [PubMed] |
| 46. | Ray RB, Meyer K, Ray R. Suppression of apoptotic cell death by hepatitis C virus core protein. Virology. 1996;226:176-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 188] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 47. | Marusawa H, Hijikata M, Chiba T, Shimotohno K. Hepatitis C virus core protein inhibits Fas- and tumor necrosis factor alpha-mediated apoptosis via NF-kappaB activation. J Virol. 1999;73:4713-4720. [PubMed] |
| 48. | Ray RB, Meyer K, Steele R, Shrivastava A, Aggarwal BB, Ray R. Inhibition of tumor necrosis factor (TNF-alpha)-mediated apoptosis by hepatitis C virus core protein. J Biol Chem. 1998;273:2256-2259. [PubMed] |
| 49. | Tai DI, Tsai SL, Chen YM, Chuang YL, Peng CY, Sheen IS, Yeh CT, Chang KS, Huang SN, Kuo GC. Activation of nuclear factor kappaB in hepatitis C virus infection: implications for pathogenesis and hepatocarcinogenesis. Hepatology. 2000;31:656-664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 151] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 50. | Hiramatsu N, Hayashi N, Katayama K, Mochizuki K, Kawanishi Y, Kasahara A, Fusamoto H, Kamada T. Immunohistochemical detection of Fas antigen in liver tissue of patients with chronic hepatitis C. Hepatology. 1994;19:1354-1359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 228] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 51. | Yen TS. Nuclear factor kappaB and hepatitis C--Is there a connection. Hepatology. 2000;31:785-787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |