Published online Jun 28, 2025. doi: 10.3748/wjg.v31.i24.108021
Revised: April 21, 2025
Accepted: June 4, 2025
Published online: June 28, 2025
Processing time: 84 Days and 18.5 Hours
Artificial intelligence (AI) is driving a paradigm shift in gastroenterology and hepa
Core Tip: This review highlights artificial intelligence (AI)-driven innovations in gastroenterology and hepatology, demonstrating breakthroughs in endoscopic/image analysis, multi-omics integration, and precision therapy. AI achieves diagnostic parity with experts in detecting early cancers and fibrosis, while addressing challenges like data fragmentation and bias through standardized databases, federated learning, and explainable systems. The study emphasizes interdisciplinary collaboration and ethical frameworks to advance equitable healthcare access and redefine digestive disease management.
- Citation: Chen ZL, Wang C, Wang F. Revolutionizing gastroenterology and hepatology with artificial intelligence: From precision diagnosis to equitable healthcare through interdisciplinary practice. World J Gastroenterol 2025; 31(24): 108021
- URL: https://www.wjgnet.com/1007-9327/full/v31/i24/108021.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i24.108021
The development of artificial intelligence (AI) applications in medicine dates back to the mid-20th century, and the com
The current medical AI technology system covers machine learning, deep learning and natural language processing technologies, and the algorithms include SVM, decision trees, random forests, logistic regression, CNN, recurrent neural networks, long and short-term memory networks, transformer, generative adversarial networks (GAN) and other algorithms. Machine learning algorithms have played an important role in medical image analysis and disease diagnosis. For example, algorithms such as SVM and random forests excel at processing structured data and can be used for disease risk prediction and diagnostic support. In the lung nodule segmentation task, the dice similarity coefficient of the fully automated lung nodule segmentation model constructed on the basis of random forest can reach 0.986[5]. Deep learning models, especially CNN and Transformer, have made significant progress in medical image analysis and multimodal data fusion. Among them, CNNs still dominate the field of medical image parsing[6]. In 2017, deep CNNs can reach a level comparable to experts in skin cancer classification[7]. Vision transformer achieves global feature extraction through the self-attention mechanism and demonstrates performance beyond traditional CNNs in analysis such as computed tomography three-dimensional (3D) reconstruction and full-slice pathology images[8-11]. Natural language processing technologies are important in medical text processing and information extraction, which cover several healthcare scenarios such as clinical practice, hospital management, personal care, public health and drug development[12]. Natural language processing facilitates the translation of cancer treatment from laboratory to clinic by mining unstructured text data in electronic medical records. It empowers oncologists to construct evidence-based research frameworks in case identification, staging, and outcome quantification. In this way, natural language processing lays a technological foundation for the advancement of an efficient and precise cancer diagnosis and treatment system[13].
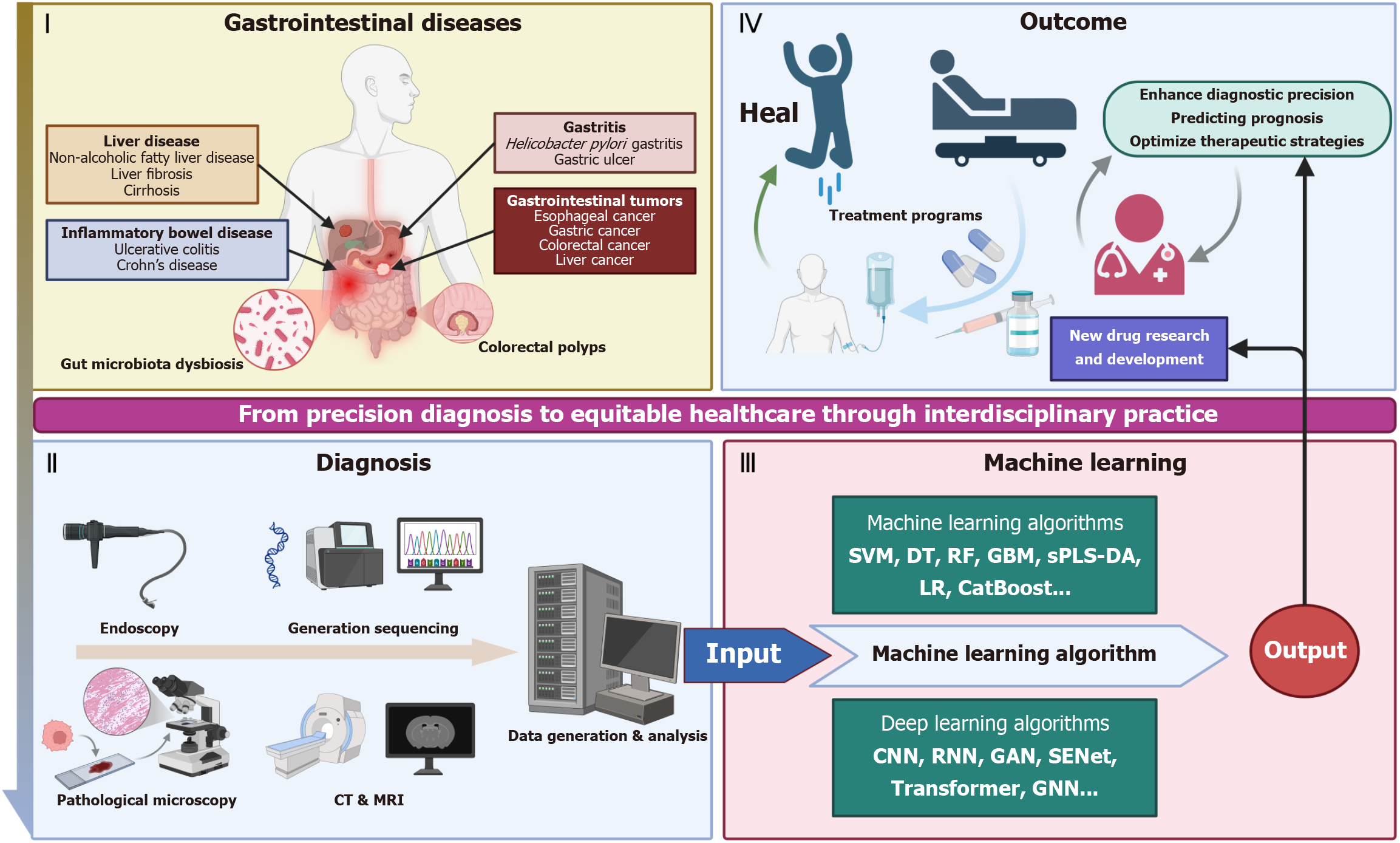
Global cancer statistics for 2022 show that cancers of the digestive tract are one of the major threats to human life and health, with esophageal, gastric, liver, and colorectal cancers having the seventh, fifth, third, and second highest mortality rates in the world, respectively[14,15]. And due to the lack of specific symptoms and early diagnostic markers in the early stages, there is a large number of potential patients who remain undiagnosed, resulting in the majority of patients often being diagnosed at a late stage[16-18]. Improving early detection of gastroenterological cancers is urgently needed. Benign ulcerative colorectal diseases, such as ulcerative colitis (UC), Crohn’s disease, ischemic colitis, and intestinal tuber
Recent advancements in AI have catalyzed transformative applications across gastrointestinal disease diagnostics and therapeutics, with machine learning and deep learning emerging as distinct yet complementary paradigms. As summarized in Table 1[21-51], machine learning algorithms including SVM, random forests, and gradient boosting have demonstrated robust performance in structured data analysis, particularly for risk stratification and treatment response prediction. For instance, SVM-based models achieved 98.5% accuracy in differentiating celiac disease from non-celiac duodenitis by analyzing duodenal histopathological features, while ensemble methods improved recurrence risk stratification in Crohn’s disease [area under the curve (AUC) = 0.84].
| AI algorithm | Parameters employed/study design | Sample size/control group/ validation | Outcomes | Performance | Ref. |
| SVM | Multi-center data + TCGA validation | Total n = 255 (training 212 + internal validation 43); external: 4 centers + TCGA | OS/DFS risk stratification (low/moderate/high); high-risk stage II/III chemotherapy benefit | AUC training 0.773 (OS)/0.751 (DFS); validation 0.852 (OS)/0.837 (DFS) | Li et al[21] |
| SVM + APINet/TransFG | Tongue features (color/morphology/coating) + microbiome (16S rDNA); multicenter prospective study | Cohort 1: GC = 328 vs NGC = 304; cohort 2: GC = 937 vs NGC = 1911 (10 centers); external: GC = 294 vs NGC = 521 (7 centers) | Distinguish GC/early GC/precancerous lesions (e.g., AG); superior to 8 blood biomarkers | Tongue model AUC: 0.89 (initial); 0.88-0.92 (internal); 0.83-0.88 (external); microbiome AUC: 0.94 (genus)/0.95 (species) | Yuan et al[22] |
| SVM/LR/kNN + feature selection | Liver/PBMC RNA-seq data | Liver = 67; PBMC = 137; external public dataset; controls: Healthy + AH/AC/MASLD/HCV | Precise differentiation of AH/AC/MASLD/HCV; minimal gene sets (33-75 genes) | Liver accuracy: 90% (AH/AC vs healthy), 91% (4-class); External 82%; PBMC accuracy: 75% (4-class) | Listopad et al[23] |
| SVM/LR/RF | Multiphase CT radiomics (n = 851) | Total n = 215 (training 150 + external 65) | Multiphase CT prediction (plain scan alternative) | Nomogram C-index 0.913 (95%CI: 0.878-0.956) | Liu et al[24] |
| SVM | Radiomics features extracted from CT images; integrated rad-score + clinicopathological characteristics | 693 GC patients (2 centers); training (n = 390), internal validation (n = 151), external validation (n = 152) cohorts | Rad-scores significantly associated with diffuse-type GC and SRCC (P < 0.001) | Lauren nomogram: AUC = 0.895 (training), 0.841 (internal), 0.893 (external). SRCC nomogram: AUC = 0.905 (training), 0.845 (internal), 0.918 (external) | Chen et al[25] |
| Counterfactual random forest + optimal policy trees | Imatinib duration inferred via counterfactual model; OPTs interpreted counterfactual predictions | Internal: 117 (MSKCC); external: 363 (polish) + 239 (spanish) | OPTs recommended no imatinib for low-risk subgroups: Gastric GIST < 15.9 cm + mitotic count < 11.5/5 mm². Any site GIST < 5.4 cm + mitotic count < 11.5/5 mm² | Sensitivity: 92.7% (internal), 95.4% (Spanish), 92.4% (Polish). Specificity: 33.9% (internal) | Bertsimas et al[26] |
| Markov decision tree model | Input variables from systematic review/meta-analysis of RCTs comparing DS, EUS-GE, and GJ; prospective cohort study for EUS-GE | 15 studies in Markov model | 1-month survival: DS (81.2%), EUS-GE (80.4%) > GJ (75.5%). 6-month survival: GJ (25.2%), EUS-GE (23.8%) > DS (21.3%) | EUS-GE and GJ outperformed DS for long-term palliation (6 months) | Chue et al[27] |
| Decision trees, LASSO, kNN, random forests | Pathomics features extracted from HE-stained WSIs; multicenter retrospective study | 584 gastric cancer patients (training: 325, internal validation: 113, external validation 1:73, external validation 2:73) | Pathomics signature independently predicted progression-free survival (P < 0.001, HR = 0.34) | Training: AUC = 0.985; Internal validation: AUC = 0.921 | Han et al[28] |
| Optimal classification trees | Input variables: Tumour size, mitotic count, tumour site | Internal: 395 patients (MSKCC + Spanish consortium); external: 556 patients (polish registry) | OCT significantly improved calibration compared to MSK nomogram | Higher C-index for OCT (0.805 vs 0.788); slope = 1.041 (OCT) vs 0.681 (MSK); no significant calibration error for OCT | Bertsimas et al[29] |
| Gradient-boosting decision tree | Baseline characteristics, endoscopic atrophy | Total: 1099 chronic gastritis patients, training: 879, test: 220 | Key predictors: Age, OLGIM/OLGA stage, endoscopic atrophy, history of other malignancies | Harrell’s c-index: 0.84 (test set). Stratified risk into 3 categories | Arai et al[30] |
| GBM | Prospective cohort study (15-year follow-up); 70% training and 30% validation split | FINRISK 2002 cohort: 7115 individuals (103 incident liver disease, 41 alcoholic liver disease) | Gut microbiome and conventional factors showed comparable predictive power | Liver disease: AUROC = 0.834 (microbiome + conventional) vs 0.768 (conventional); alcoholic liver disease: AUROC = 0.956 (microbiome + conventional) vs 0.875 (conventional) | Liu et al[31] |
| XGBoost (pre/delta-radiomics) + SMOTE | Pre/post-treatment MRI radiomics (n = 105); multisequence MRI integration | LARC patients n = 84; validation: 5/10-fold CV + independent; no control | Delta-radiomics > pre-radiomics | Pre-model: AUC 0.93 ± 0.06 (train)/0.79 (test); delta-model: AUC 0.96 ± 0.03 (train)/0.83 (test) | Wang et al[32] |
| sPLS-DA | Multi-site microbiome (saliva/esophagus/stomach); 16S rRNA analysis | EoE: Saliva = 29, biopsy = 25; controls: Non-EoE = 20 (saliva)/5 (biopsy) | Saliva model distinguishes EoE/non-EoE; esophageal microbiota detects disease activity | Saliva: CE 24%, validation Acc 78.6% (sensitivity 80%/specificity 75%); esophagus: CE 8% (activity detection) | Facchin et al[33] |
| sPLS-DA + LR | Genome-wide 5hmC features (n = 64); protein biomarkers | Healthy = 165; LC = 62; HCC = 135; longitudinal cohort | HCC diagnosis/recurrence prediction; tumor burden monitoring | Wild-score AUC = 93.24% (HCC vs non-HCC); HCC score AUC = 92.75% (HCC vs LC) | Cai et al[34] |
| LR + mixed-effects model | Multicohort clinical/serologic/genetic data; JAK-STAT/IL6 pathway | IBD patients = 12083 (4 cohorts); within-case design | Female/CD colonic location/surgery linked to EIMs; MHC/CPEB4 associations; therapeutic targets (TNF/JAK-STAT) | MHC OR = 2.5 (P = 1.4E-15); CPEB4 OR = 1.5 (P = 2.7 × 10-8); serologic panel OR = 1.7 (P = 3.6× 10-19) | Khrom et al[35] |
| LR + RF + kNN + SVM + NN | Recursive feature elimination; single-center retrospective | Total n = 864 (IIIa + n = 457 vs low-risk n = 407); 3-fold imputation/CV | NN outperforms others (Acc 68.8%); best in medical complications (AUC = 0.695) | NN: Overall Acc 0.688/AUC = 0.672; medical AUC = 0.695; surgical AUC = 0.653; cologne score Acc 0.510 | Jung et al[36] |
| RF vs cv-Enet/glmboost/ensemble | Multicenter preoperative features; elastic-net regularization | Development = 3182 (39 centers); validation = 260; no control | RF optimal prediction; surgical decision support | RF AUC = 0.844 (0.841-0.848) (development); similar in validation | Pera et al[37] |
| LR + Cox regression models | Endoscopic features (whitish/irregular) + Histology (marked IM); retrospective multicenter | Total n = 182 (malignant = 48); progression cohort = 98; ROC/KM validation | Misdiagnosis predictors (single/large/IM); progression predictors (whitish/margin/multi-diagnosis) | AUC 0.871 (sensitivity 68.7%/specificity 92.5%) | Zou et al[38] |
| RF + Swin transformer tongue model | Questionnaire features (n = 10) + tongue images; multicenter | Total n = 2229 (9 centers); validation AUC > 0.8 | Key factors: Age/TCM constitution/tongue features/diet/anxiety; dynamic nomogram | RF Acc 85.65%; tongue model Acc 73.33% (validation) | Yu et al[39] |
| LR | Tumor location/ulceration/biopsy features; H-L test/DCA validation | Training = 516; validation = 220 (7:3 split); no control | 4 fibrosis predictors; severe fibrosis prediction model | Raining AUC = 0.819; validation AUC = 0.812; DCA clinical benefit | Zeng et al[40] |
| Stepwise logistic regression | Demographics/history/Lab markers (AFP/AST/albumin); prospective multicenter | Total n = 1723; HCC events = 109; median follow-up 2.2 years; no control | Key factors: Male/cirrhosis duration/family history/age/obesity/AFP/AST | Incidence 24/100 person-years; multivariate OR 1.08-2.73 (P < 0.05) | Reddy et al[41] |
| Multivariate logistic regression | Radiomic features (peritumoral enhancement/necrosis); transcriptomic sequencing | Development = 470; validation: Control = 145 + HAIC = 143; multicenter | Imaging subtypes guide HAIC benefit; immune pathway correlation | Training AUC = 0.83; control AUC = 0.84; HAIC AUC = 0.73 | Ma et al[42] |
| LR | Multiphase CT radiomics (peritumoral); RNA sequencing | Total n = 773 (training 334 + internal 142 + external 141 + survival 121 + RNA35); 4 centers | MVI prediction + survival stratification (early recurrence/OS); glucose metabolism genes | Hybrid model AUC: 0.86 (internal)/0.84 (external); survival P < 0.01 | Xia et al[43] |
| Multivariate logistic regression | LI-RADS visualization score (A/B/C); obesity class II-III | Total n = 2053 (A = 1685, B = 262, C = 106); longitudinal = 1546; multicenter | Alcohol/MASLD cirrhosis + obesity linked to limited visualization; 19.6% worsened/53.1% improved | Baseline limited rate 18%; obesity OR = 2.1 (P < 0.001) | Schoenberger et al[44] |
| Regularized LR + GBM | RCT secondary analysis; mailed outreach; prior screening behavior | Total n = 1200 (training 960 + test 240); 3 screening rounds; no control | Surveillance adherence stratification; key variables: Prior screening/primary care contact | AUROC 0.66-0.77 (increasing); 41%-47% completion rate | Singal et al[45] |
| LASSO logistic regression | Pre/intraoperative variables; multicenter international | Total n = 2192 (train 70% + valid 30%); 12 centers | Dual prediction (PHLF/CCI > 40); online risk calculators | PHLF AUC = 0.80 (calib. slope = 0.95); CCI AUC = 0.76 | Wang et al[46] |
| LDpred2 PRS + QCancer-10 integration | Genetic/non-genetic factors; Cox proportional hazards | United Kingdom Biobank n = 434587; case-control/survival validation | C-index improvement (M + 7.3%/F + 6.5%); high-risk group 3.47 × (M)/2.77 × (F) | Integrated C-index: 0.730 (M)/0.687 (F); sensitivity/specificity: 47.8%/80.3% (M), 42.7%/80.1% (F) | Briggs et al[47] |
| Multivariable logistic + Cox regression | Multicenter FS screening; long-term follow-up (median 17 years) | Intervention = 40085 (13 centers) | High-ADR group: Distal CRC HR = 0.34 (incidence)/0.22 (mortality); all-site CRC HR = 0.58/0.52 | High vs low-ADR: Distal CRC HR 0.34 vs 0.55 (incidence), 0.22 vs 0.54 (mortality) | Cross et al[48] |
| RRR + elastic net models | Inflammatory markers (CRP/IL6/GDF15) + metabolic markers (BMI/waist/C-peptide); case-control | Total n = 1368 (cases 684 +controls 684); NHS = 818F + HPFS = 550M | Sex-specific: Median OR = 1.34 (inflammation)/1.25 (metabolic); NS in F; 11 key metabolites | Variance explained: 24% (inflammation)/27% (metabolic) | Bever et al[49] |
| RSF/GBM/Deep hit | Multivariable analysis + clinical feature selection; time-dependent C-index | CRC patients = 2157; stratified 5-fold CV (5 repeats) | Deep hit best discrimination; RSF best calibration; SHAP key factors (R0 resection/TNM) | Deep hit C-index 0.789 (0.779-0.799); RSF brier 0.096 (0.094-0.099) | Yang et al[50] |
| Multivariable logistic regression | Cell search CTCs detection; prospective CTCs + retrospective HGP; excluded neoadjuvant/extrahepatic | Total n = 177 (dHGP = 34, 19%); multivariable validation; no external cohort | CTC-negativity predicts dHGP (OR = 2.7); dHGP better survival | OR = 2.7 (1.1-6.8), P = 0.028 | Meyer et al[51] |
In parallel, Table 2[52-77] highlights the ascendancy of deep learning architectures, notably CNNs and vision trans
| AI algorithm | Parameters employed/study design | Sample size, control group, validation | Outcomes | Performance | Ref. |
| CNN | 14 EUS anatomical sites; multicenter validation | Training: 1812 patients/6230 images; internal: 47 patients/1569 images; external: 131 patients/85322 images | Outperformed novices in 11 sites; high expert agreement (kappa 084-0.98) | Internal Acc 92.1-100%; external sensitivity 89.45%-99.92%/specificity 93.35%-99.79% | Tian et al[52] |
| NNLS deconvolution + GCNN | Methylation atlas (TSMA) + genome-wide density; multi-modal strategy | 5 tumor types + WBC training; validation = 239 low-depth cfDNA | Multi-modal improves TOO in low-depth cfDNA | Validation Acc 69% | Nguyen et al[53] |
| CNN + survival MLP | CT + clinical multimodal data; 5-fold CV | GC patients = 1061; vs 3 SOTA methods; no control | Multimodal > single-modality; optimal OS/PFS prediction | OS C-index 0.849; PFS 0.783 (surpass SOTA) | Hao et al[54] |
| CNN | HE features for HER2 status; trastuzumab response | Surgical = 300; biopsy = 101; treated = 41; no control | HER2 amplification prediction; treatment response (CR + PR vs SD + PD) | Surgical AUC 0.847 (amplification)/0.903 (2 +); biopsy 0.723; treatment 0.833 | Wu et al[55] |
| DCNN | HE whole-slide imaging; fibrosis stage comparison | Non-HCC = 639; HCC = 46; paired training/unpaired validation | Detect HCC risk in mild fibrosis; saliency maps reveal nuclear atypia/immune infiltration | Training Acc 81.0% (AUC = 0.80); validation 82.3% (AUC = 0.84) | Nakatsuka et al[56] |
| Faster R-CNN model | Preoperative CT/MRI analysis; multicenter retrospective cohort (2012-2020) | Total n = 1141 (PCCCL = 62, CHCC = 1079); 4:1 split (train-val vs test); CHCC cases (n = 1079) as negative control | Differential diagnosis of rare PCCCL | Accuracy: 0.962 (95%CI: 0.931-0.992); AP: PCCCL 0.908, CHCC 0.907; Recall: 0.95 | Liu et al[57] |
| Transformer | End-to-end biomarker prediction; multicenter validation | Total n > 13k (16 CRC cohorts); resection training/biopsy validation | Solved biopsy MSI diagnosis; improved interpretability | MSI detection: Sensitivity 0.99/NPV > 0.99 | Wagner et al[58] |
| CNN + SMOTE/SVM | Pathomics/radiomics/immune score (CD3 +/CD8 +)/clinical; digital pathology | Lung metastasis = 103; internal validation | Path/radio features vs immunoscore (neg); triple independent prognosis | Integrated model: OS = 0.860/DFS = 0.875; Calib/DCA validated | Wang et al[59] |
| INSIGHT (CNN) + wise MSI (self-attention) two-stage | Tumor tile classification + ResNet pre-trained + attention pooling; multicenter | Chinese multicenter cohort; vs 5 DL methods | Outperforms SOTA in MSI prediction; high pathologist consistency | Wise MSI AUC 0.954 (0.948-0.960) | Chang et al[60] |
| CNN + RNN | Multicenter blinded trial; real-time monitoring + second observer | Total n = 946 (adenomas = 989); multicenter | CADe > human in adenoma detection (sensitivity 94.6% vs 96.0%); changed 2.3% follow-up | ADR + 1.1%/case; Non-neoplastic + 4.9%; time + 42.6% (6.6 minutes) | Sinonquel et al[61] |
| ANN | Pathological image analysis; retrospective multicenter | Training = 496 (GDPH); external validation = 150 (SYSMH) | Avoided 34.9% unnecessary surgeries; outperformed United States guidelines | Training AUC = 0.979; validation AUC = 0.978 | Su et al[62] |
| Multitask transformer | Preop MRI multiparametric features; 7-center retrospective | Total n = 725 (train 234 + internal 58); external = 212/111/110 | PA-TACE benefit in high-MVI/low-survival group (P < 0.001) | RFS C-index: Training 0.763/validation 0.628-0.728 | Wang et al[63] |
| Multistage DL models | Longitudinal MRI (pre/post-TA) + clinical variables; multicenter retrospective | Total n = 289 (train 254 + external 35); 3 hospitals | DL clinical improved ER prediction (AUC = 0.740); High/low-risk RFS P = 0.04 | DL clinical AUC: 0.740 vs 0.571/0.648/0.689 | Kong and Li[64] |
| CNN | Clinical data + MRI radiomics; 6 time-frame prediction | Early HCC = 120 (recurrence = 44); retrospective (2005-2018) | Imaging model > clinical (AUC 0.76 vs 0.68, P = 0.03) | Imaging model AUC 0.71-0.85; KM P < 0.05 (2-6 years) | Iseke et all[65] |
| RSF/ANN/decision tree | Inflammatory markers + ALBI + AFP + tumor size + INR; single-center retrospective | Total n = 808 (train 2:1 split) | ANN optimal (5 years AUC = 0.85); High-risk OS HR = 7.98 (5.85-10.93) | Training AUC 0.85 (0.82-0.88); validation 0.82 (0.74-0.85); P < 0.0001 | Zhang et al[66] |
| DL | DCE-MRI + clinical/radiologic features; retrospective multicenter | Total n = 355 (train 251 + internal 62 + external 42); 2 centers | Proliferative HCC prediction; fusion model improves recurrence stratification | DL + clinical + radiologic model AUC: Training 0.99/internal 0.87/external 0.80 | Qu et al[67] |
| DenseNet169 + MLP | Multiphase 25D CT + clinical features + RNA-seq; multicenter retrospective | Total n = 620 (TCIA + 3 centers); internal + 2 external test sets | Stratified RFS/OS (P < 0.001); high score links WNT/MYC/KRAS activation | DLER MLP 0.891 vs DLER 0.797 vs clinical model 0.752 | Guo et al[68] |
| scSE-CatBoost | Multi-site endoscopic images; CNN + scSE feature extraction | Total n = 302 (An Nan Hospital); RUT validation | Real-time Helicobacter pylori detection; NPV 100% | Acc 0.90; sensitivity 1.00/specificity 0.81; AUC = 0.88 | Lin et al[69] |
| Transformer + MIL | HE WSIs; dual-task (subtype + TMB prediction) | EC = 529/918; CRC = 594/1495; vs 7 SOTA methods | Strong subtype-TMB association (fisher P < 0.001); guides immunotherapy | Outperformed SOTA in both tasks | Wang et al[70] |
| GAN + ViT distillation | HE/HPS staining; multi-task prognosis (OS/TTR/TRG) | Internal = 258 CLM; two public datasets | TRG dichotomization. Acc 86.9-90.3%; 3-class Acc 78.5-82.1% | OS C-index 0.804 (± 0.014); TTR C-index 0.735 (± 0.016) | Elforaici et al[71] |
| Transfer learning | HE WSIs analysis | Segmentation = 100 WSI; validation: 4 cohorts (3 internal +1 external) + 6-month series = 217 | Fine-tuning improved F1 0.797-0.949 (P < 0.00001); 100% visual overlay accuracy | Detection model AUC 0.959-0.978 (P < 0.00001) | Khan et al[72] |
| DBMIA-Net | GIA + EIA modules; adaptive channel graph convolution | 5 public datasets (CVC-Clinic DB); vs SOTA methods | Enhanced generalization | 94.12% dice (vs PraNet + 4.22%); leading in 6 metrics | Zhang et al[73] |
| UC-former vision transformer | Multicenter retrospective study; mayo endoscopic score prediction | Total n = 768 UC patients/15120 images; internal + 3 external validations | Surpassed senior endoscopists; strong multicenter stability | Internal Acc 90.8%; external Acc 82.4%-85.0% | Qi et al[74] |
| MIST | Self-supervised contrastive learning + dual-stream MIL | Total n = 480/666 WSI (Drum Tower); external = 273 WSI (Nanjing First) | Acc comparable to pathologists (0.784 vs 0.806) | External Acc 0.784 | Cai et al[75] |
| ResTransUNet | Global context (transformer) + local features (CNN); LiTS2017/3Dircadb/Chaos/Sliver07 | LiTS2017/3Dircadb/Chaos/Sliver07 | Solved small/discontinuous region segmentation; outperformed SOTA | LiTS2017 dice 09535/VOE 0.0804/RVD -0.0007 | Ou et al[76] |
| GCN | Pathological micronecrosis analysis + multicenter datasets; GCN feature fusion | Total n = 752/3622 slides; internal (FAH-ZJUMS) + external (TCGA-LIHC) | Improved prognostic stratification; precise necrosis localization | Internal + 8.18%; External + 9.02%; superior C-index vs baseline | Deng et al[77] |
AI demonstrates multidimensional value in the management of UC and Crohn’s disease. In the field of UC, deep lear
In addition, the UC-severity classification and localized extent (SCALE) algorithm enables topological visualization of disease severity by automating the assessment of inflammation distribution in full-length colonoscopy videos (86.5% accuracy, κ = 0.813)[83].
For Crohn’s disease, a multimodal machine learning model integrating magnetic resonance small bowel imaging and biochemical markers noninvasively assessed terminal ileal endoscopic activity with an AUC of 0.84[84]; and a deep learning model (SA-AbMILP) predicted Global Histologic Disease Activity scores from histologic images (65%-89% accu
AI significantly improves the efficiency and accuracy of inflammatory gastrointestinal disease screening. In capsule endoscopy, CNN models can quickly identify ulcers, erosions, and vascular malformations (with 96.9%-99% sensitivity) with processing speeds up to 90 frames/second[87,88]; a sequential CNN model for Crohn’s disease ulcer severity has an accuracy of 91% in identifying grade 3 ulcers[89]. In the field of colonoscopy, the FRCNN-AA-CIF model based on the attention mechanism and context fusion has a leakage rate of only 4.22% and the mean of average precision of all categories of detection of 0.817[90]; the ResNet50 migration learning model has an accuracy of 99.8% in the polyp classification task, which is superior to the traditional methods[91,92]. In imaging, 3D-CNN distinguished colon cancer from acute diverticulitis by computed tomography images (sensitivity 83.3%, specificity 86.6%), and AI support enabled radiologists to increase the sensitivity to 85.6%[93]. For data-scarce scenarios, computed tomography colon segmentation adopts a guided sequential scenario training strategy, which requires only 10 cases of annotation to achieve a dice coefficient of 97.15%, and its cross-layer feature comparison learning mechanism enables polyp detail retention to exceed 98.28%[94]. The most clinically translatable value is the hybrid architecture of 3D-CNN and random forest: By modeling spatial continuity, this model achieves a physician-level agreement of κ = 0.83 for the severity classification of 7.5-mm-level lesions in computed tomography enterography images, with a 91.51% accuracy in localizing moderate-to-severe lesions[95].
AI has also made breakthroughs in gastritis, celiac disease and small bowel disease. Multi-stage semantic segmentation model (AI-G) has an accuracy of 91% in gastric biopsy diagnosis, with significant cross-institutional validation robustness[96]; CNN can differentiate between Helicobacter pylori gastritis and autoimmune gastritis (with 100% diagnostic concordance)[97]. For celiac disease, the SVM model distinguishes between celiac disease and non-celiac duodenitis based on images of the duodenal lamina propria with 98.5% accuracy[98]. These innovations signal that AI is breaking through the traditional qualitative diagnostic framework and driving the transition of the diagnostic paradigm from qualitative detection to accurate quantitative assessment through interpretable feature engineering and anatomical constraint algo
Deep learning-based endoscopic image analysis system significantly improves the efficiency and detection rate of upper gastrointestinal tumors. The study showed that the sensitivity of the GRAIDS system in diagnosing upper gastrointestinal cancer by analyzing more than 1 million endoscopic images (0.942) was comparable to that of expert endoscopists (0.945) and significantly better than that of non-experts (P < 0.0001), and its negative predictive value (0.978) was close to the level of experts (0.980), which demonstrated that AI can effectively assist grassroots hospitals in improving diagnostic capabilities[99]. For the determination of the depth of infiltration of early gastric cancer, the diagnostic model F1 value of AI collaborating with endoscopists (0.776) was higher than that of AI alone (0.768) or physician majority vote (0.662), confirming that human-machine synergy can compensate for the limitations of a single method[100]. In addition, AI-assisted white light endoscopy reduced the leakage rate of gastric tumors from 27.3% to 6.1% (P = 0.015), which significantly improved the efficiency of identifying microscopic lesions[101]. In the evaluation of Barrett’s esophagus for heterozygous hyperplasia, the AI system surpassed most pathologists in diagnostic accuracy (76.4%) of whole slide images, with an AUC of 0.94 and a sensitivity of 0.92 for the prediction of heterozygous hyperplasia[102]. For the prediction of peritoneal recurrence in gastric cancer, the deep learning model based on preoperative computed tomogra
AI fused with multi-omics further breaks through the limits of molecular typing, metastatic mechanisms and immunotherapy response prediction in gastric and esophageal cancer. Based on the dynamic features of mitochondrial adenosine triphosphate metabolism, membrane potential and lactate/pyruvate/glucose metabolism, the researchers constructed a MitoScore quantitative assessment system by integrating 10 machine learning algorithms, which can accurately differentiate the immune-metabolic subtypes of gastric cancer. Data analysis showed that high MitoScore subgroups presented abnormal activation of glycolytic pathways and were significantly associated with tumor aggressiveness phenotype and poor patient prognosis[105]. A SVM model was constructed to predict the response of gastric cancer patients to Sintilimab combined with SOX treatment based on the screening of six key markers (DUOX2, HSPB1, S100A14, C1QA, TGFB1, and LTF) by combining the multiplex immunohistochemical data and the deep-learning feature extraction technique, which The AUC values were 0.93 and 0.84 in the exploratory cohort (n = 107) and validation cohort (n = 46), respectively, showing good predictive efficacy[106]. In addition, the MetImage technology based on metabolomics encoded liquid chromatograph mass spectrometer data into images, and the AI model constructed using the convolutional neuron network algorithm screened esophageal squamous carcinoma with a sensitivity of 85%, specificity of 92%, and AUC of 0.95[107].
AI plays an important role in the optimization of treatment strategies for upper gastrointestinal tumors. A study based on a counterfactual random forest model using predictors of recurrence (mitotic count, tumor size, and tumor site) and imatinib duration to infer the likelihood of recurrence in a given patient at 7 years for each imatinib treatment duration suggested that gastric-derived gastrointestinal stromal tumors (< 15.9 cm and low mitotic count) does not require ima
AI becomes a real-time aid in colonoscopy traditional colonoscopy suffers from the limitation of a high rate of adenoma leakage (15%-30%), which has been significantly improved by deep learning-based computer-aided detection (CADe) systems through real-time polyp identification. For example, a multicenter randomized controlled trial (RCT) showed an approximately twofold reduction in adenoma missed diagnosis (AMR) in the AI-assisted colonoscopy group compared to the control group (15.5% vs 32.4%), with a particularly significant advantage in the detection of tiny polyps (≤ 5 mm) and nonpolypoid lesions [odds ratio (OR) = 0.34-0.24][109]. Another United States multicenter study (CADeT-CS trial) further validated that the AMR and SSL leakage rates in the AI-assisted group decreased to 20.12% and 7.14%, respectively, while the number of first-pass adenomas detected increased significantly (1.19 vs 0.90)[110]. In addition, the gastrointestinal Genius system improved adenoma detection rate by 8.3% in a large RCT (COLO-DETECT) without the need for extended operating time[111]. These results suggest that AI has become a central tool for optimizing the quality of colonoscopy by reducing the number of perceptual errors.
Digital pathology combined with AI algorithms demonstrated high accuracy in tissue classification and tumor de
Imaging and multimodal data integration, AI in imaging analysis optimizes treatment prediction by integrating multimodal data. Multimodal AI models integrating imaging, pathology and genomic data provide new tools for individualized treatment of colorectal tumors. The radio pathomics integrated prediction system combines magnetic resonance imaging radiomics with HE pathologic features to predict pathological complete remission from neoadjuvant radiotherapy for rectal cancer, with an AUC of 0.872 in the validation cohort, which was significantly better than the unimodal model (P < 0.0001)[119]. AI has significantly improved the prediction of treatment response by integrating imaging histology and clinical data. One study utilized random forest and gradient boosting algorithms to predict response to radiotherapy for colorectal cancer with an accuracy of 93.8%[120]. Another multi-omics analysis combined with spatial interaction mapping developed the CCIM-Net model, which effectively predicts chemosensitivity and guides combination therapy targeting FOLR2 macrophages[121].
The application of AI technology to surgical planning for colorectal tumors has also seen breakthroughs. An automated computed tomography-based tumor segmentation model accurately assessed the total tumor volume of colorectal liver metastases with an intraclass correlation coefficient of 0.98[122]. For colorectal cancer liver metastases, the game-theory-based Shapley’s additive interpretation AI model recommended individualized margin widths (6 mm-12 mm), with an AUC of 0.78 in the validation cohort, confirming the association between an optimal margin width of 7 mm and a significant prolongation of survival in an external cohort[123]. In addition, the multilayer perceptron model for predicting lymph node metastasis in the inferior mesenteric artery (AUC = 0.873) was significantly better than expert judgment (AUC = 0.509), which may reduce unnecessary clearance[124]. Machine learning algorithms performed well in survival prediction. logistic regression models combined with clinical variables (e.g., distant metastases, number of lymph nodes) predicted 1- and 5-year survival with AUCs of 0.850 and 0.872, respectively[125]. Genomic profiling identified 32 key genes by interpretable AI and predicted stage II colorectal cancer recurrence with an AUC of 0.952[126].
The degree of liver fibrosis is the most sensitive clinical warning sign for metabolic dysfunction-associated steatotic liver disease (MASLD)-associated hepatocellular carcinoma, and significant and advanced liver fibrosis not only increases the risk of hepatic and extra-hepatic complications, but is also significantly associated with liver-related mortality[127,128]. Therefore, dynamic monitoring of liver fibrosis progression (e.g., liver stiffness value testing) has become a core strategy for early screening of hepatocellular carcinoma, and how to achieve accurate assessment of fibrosis staging remains an urgent clinical challenge. In recent years, AI technologies have significantly improved the objectivity and sensitivity of assessment through multimodal innovations: The performance of deep learning models in ultrasound steatosis grading (AUC = 0.85) is not unlike the judgment of radiologists[129]; at the histological level, an AI-based measurement tool effectively predicts the survival of patients with fibrosis through reproducible necroinflammation grading (κ = 1) and pathologists’ consensus highly concordant (κ = 0.62-0.74), effectively predicted progression-free survival of fibrosis patients (P < 0.05), providing a highly sensitive method for clinical trial endpoint assessment[130]; in the analysis of pathomechanisms, AI-based digital pathology combined with second harmonic generation/two-photon excited fluore
AI has shown no less ability than human experts in diagnostic imaging of liver tumors. Ultrasound-based deep learning models significantly improve the accuracy of benign and malignant liver tumors identification by integrating patient background and blood biomarkers. For example, a multimodal deep learning model combining B-mode ultrasound images with clinical data improved the diagnostic accuracy from 68.52% to 96.30% (AUC = 0.994) in unimodal mode[134]. In addition, the application of AI in computed tomography and magnetic resonance imaging image analysis is equally prominent. A GAN-based model effectively mitigates the data imbalance problem by synthesizing high-fidelity computed tomography images of liver tumors, improving the accuracy of classification models by 21%-34%[135]. In the magnetic resonance imaging segmentation task, the 3D CNN performs well in semi-automatic segmentation of hepatocellular carcinoma tumors, and especially performs best in diffusion-weighted imaging and T1-weighted imaging, with dice similarity coefficients up to 0.778[136]. The AI-driven software also detects liver metastases missed by radiologists on contrast-enhanced computed tomography with a sensitivity of 70.8% and a false-positive rate of only 0.48/case[137]. These techniques not only improve the efficiency of the diagnosis, but also reduce human error.
AI has also demonstrated strong classification and prediction capabilities in liver cancer histopathology image analysis. Deep learning models based on the attention mechanism (e.g., SENet) achieved 95.27% accuracy in the task of classifying the degree of differentiation of hepatocellular carcinoma, which is significantly better than traditional manual reading[138]. The clustering-constrained attention multiple instance learning model predicted immunogenetic signature activation by whole slide images with AUCs of 0.78-0.91, and pathologic analysis showed that the predicted areas were enriched with lymphocytes and neutrophils[139]. Microvascular infiltration (MVI) is a major risk factor for overall postoperative mortality and recurrence in hepatocellular carcinoma. Models combining imaging histology and deep learning perform well in MVI prediction. A deep learning model based on image-pathology fusion (Swin Transformer) was effective in predicting the vessels encapsulating tumor clusters (VETC) patterns of perivascular envelope tumor clusters, and its radiological-pathological histology column-line diagrams had a C-index of 0.67 in the external test set[140]. The random forest model based on preoperative computed tomography had an accuracy of 96.8% (sensitivity 95.2%) in the test set[141], while the column-line graph model combining clinical features with deep learning features had an AUC of up to 0.940[142]. In addition, the multitask learning framework significantly improved prognostic stratification by simultaneously predicting MVI and VETC via 3D CNN with AUCs of 0.917 and 0.860, respectively[143].
AI shows potential to outperform traditional statistical methods in prognostic modeling. The machine-learning-based smart-hepatocellular carcinoma score integrates nine clinical features, including liver stiffness, and its 5-year predictive AUC is ≥ 0.89, which is superior to existing scoring systems[144]. Deep learning column line drawings combined with magnetic resonance imaging histologic features and clinical variables had an AUC of 0.949 for predicting early recurrence after hepatocellular carcinoma[145]. The randomized survival forest model, by incorporating risk factors such as MVI and satellite nodules, predicted early recurrence with a C-index of 0.896 in the training group 0.798 in the validation group, which was significantly better than the Cox proportional risk model[146]. In addition, the knowledge-enhanced dual-style visual transformer improves the interpretability and performance of recurrence prediction by fusing multi-period computed tomography images with domain knowledge[147]. Migration learning and multimodal strategies were used to optimize hepatocellular carcinoma risk prediction in patients with MASLD, significantly alleviating the problem of data scarcity and gender bias[148]. These models provide precise tools for individualized treatment and follow-up.
AI plays a key role in mining emerging molecular markers for liver cancer and optimizing immunotherapy strategies. Based on single-cell transcriptome analysis and machine learning, S100A10 was identified as a core gene for hepatocellular carcinoma diagnosis and immunotherapy response, and its expression was positively correlated with the stem cell marker POU5F1[149]. Stem-related classifiers (9-gene model, PPARGC1A, FTCD, CFHR3, MAGEA6, CXCL8, CABYR, EPO, HMMR, and UCK2) predicted the state of the tumor immune microenvironment and the efficacy of immune checkpoint inhibitors, with a significant enrichment of Treg cells and immune-suppressing pathways in high-risk patients[150]. These findings provide new ideas for targeting and stratifying immunotherapy in hepatocellular carcinoma.
Using AI to plan radiotherapy and manage post-liver transplantation is also maturing. The hierarchical feature fusion network generated radiotherapy dose distributions close to the clinical standard (homogeneity index = 0.31, conformation index = 0.87) by integrating computed tomography images with organ contours[151]. For recurrence prediction after liver transplantation, the deep learning model had an AUC of 0.86 based on preoperative factors (e.g., tumor diameter and alpha fetoprotein level), with a 5-year recurrence-free survival rate of 92.6% in the low-risk group[152].
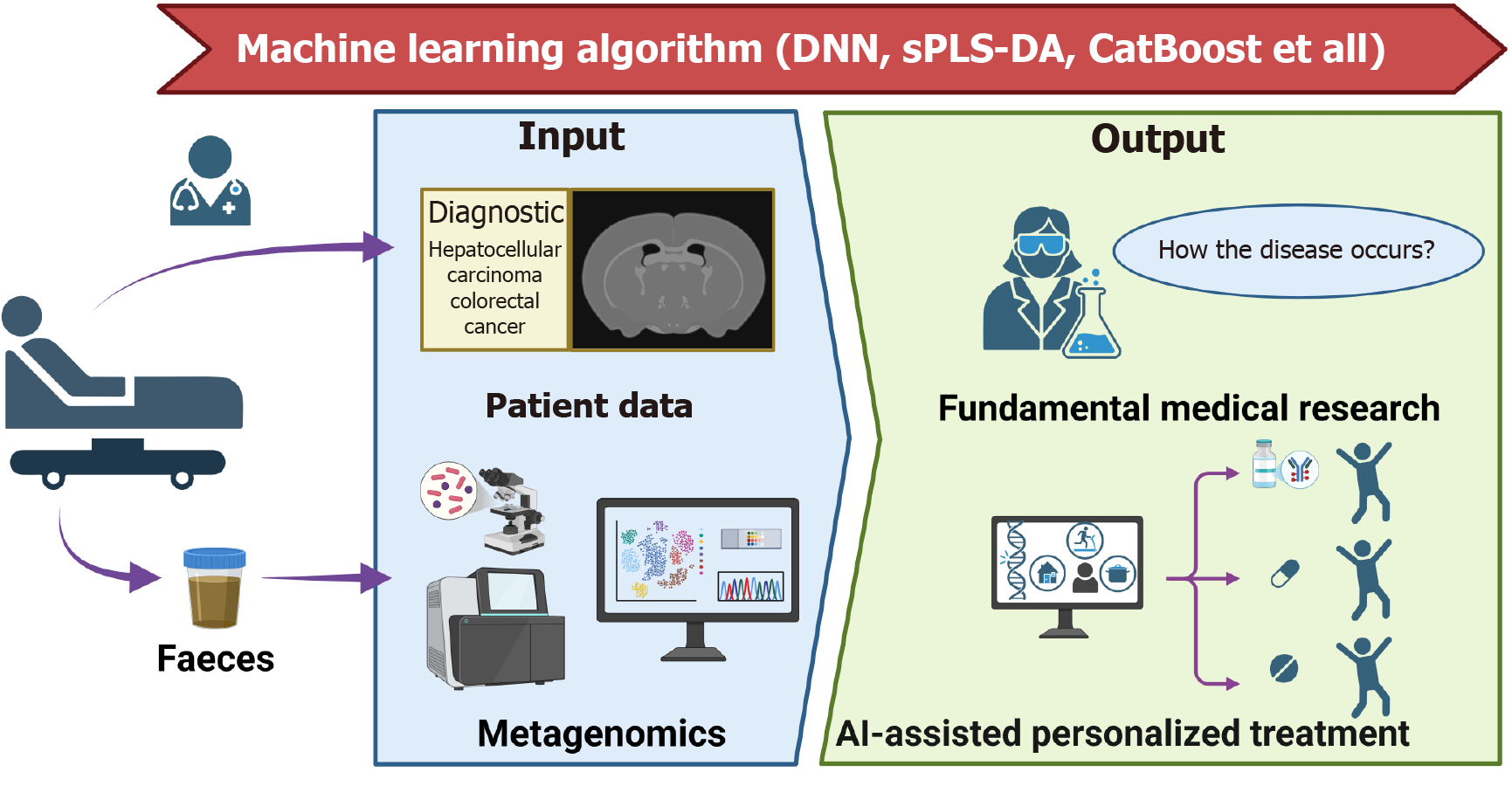
In the study of gut microbiology in inflammatory bowel disease, machine learning techniques have systematically deconstructed the gut-type-specific pathogenicity network of UC. Cluster analysis of 16S rRNA data from 11 cohorts by a deep neural network defined for the first time the three enterotypes of Enterobacteriaceae, Trichoderma, and Clostridiales: Among them, patients with Enterobacteriaceae had abnormally elevated abundance of Rumatococcus, which was positively correlated with the proliferation of Clostridium difficile (P < 0.01), and the machine learning-guided metabolic pathway analysis revealed that Odoribacter splanchnicus and Bacteroides uniformis exerted a protective effect by activating the adenosine 5’-monophosphate-activated protein kinase signaling pathway[153]. A sparse partial least squares discriminant analysis was further used to construct a prediction model for active UC, which maintained more than 90% accuracy even when only 5% of the feature volume was used, and to establish Bifidobacterium bifidum/Haemophilus parainfluenzae as a negative/positive marker of disease activity[154]. Machine learning algorithm-based analysis of multicohort fecal macrogenomic data revealed that microbial gene markers in Crohn’s disease patients had significantly better diagnostic performance than species and single nucleotide variants. The gene diagnostic model constructed by machine learning performed optimally in cross-geographic validation (mean AUC = 0.91) and targeted the key gene (celB/manY) of the phosphotransferase system, whose specificity was experimentally confirmed, revealing the potential of microbial functional genes for AI-driven precision diagnosis[155].
In terms of diagnostic technology innovation, CatBoost algorithm increased the colorectal precancer detection accuracy to 87.27% by integrating bacterial-viral two-dimensional features, identified Prevotella sp900557255 and phage Felixounavirus as early warning markers, and the combined typing strategy enabled the prediction accuracy to exceed 98%[156]; while the iterative random forest model revealed the characteristics of pancreatic cancer metastasis-associated flora, and found that changes in the abundance of six genera, including Anaero stipes hadrus, were significantly correlated with the enrichment of Gram-negative bacteria (OR = 2.34, P = 0.007)[157]. A neural network model based on 20 characteristic microorganisms combined with nanopore sequencing technology enabled rapid detection of hepatic encephalopathy (84% specificity), which drove the optimization of diagnostic and treatment protocols in 40% of cases[158]; while hepatocellular carcinoma studies constructed a prognostic model with an AUC of 81% by integrating microbiome-transcriptome data through randomized forests, which reveals that Mycobacterium anisopliae spp. mediated tumor through bile acids key mechanism of immune microenvironment remodeling[159]. It is worth emphasizing that machine learning-enabled cross-disease meta-analysis identified significant overlap of flora characteristics between Crohn’s disease and colorectal cancer, and between Parkinson’s disease and type 2 diabetes mellitus (Jaccard’s index > 0.65), which provides a new paradigm for cross-disease therapeutic target discovery[160]. This paradigm-shifting integration of gut microbiome-AI symbiosis, as exemplified in Figure 2, not only deciphers conserved microbial signatures across diseases (e.g., bile acid-immune axis remodeling in hepatocellular carcinoma) but also establishes a computational framework for translating multi-omics interactions into clinically actionable biomarkers and therapeutic blueprints.
The threat to human health posed by AMR has evolved from a theoretical warning to an urgent public health crisis. The Lancet research team modeled that by 2050, 1.91 million deaths globally will be directly attributed to AMR, with another 8.22 million deaths significantly associated with it[161]. However, in the face of “key priority pathogens”[162] repre
Excitingly, driven by AI, researchers have constructed numerous ways to quickly find new and efficient antibiotics. In response to the threat of multidrug-resistant Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter species pathogens, traditional high-throughput screening (HTS) is becoming insufficient due to the limitations of bacterial penetration barriers and resistance mechanisms[165]. In this context, the integration of deep learning and big data technologies significantly improves the efficiency of antibiotic discovery. For example, virtual screening of 1.4 billion compounds by out-of-distribution generalization techniques in the GNEprop model identified 82 anti-microbially active molecules with a 90-fold increase in hit rate compared to conventional HTS, and most of the structures were significantly different from those of known antibiotics[166]. Similarly, the AMPSphere platform, constructed based on global microbiome data, utilized machine learning to mine 863000 non-redundant antimicrobial peptides, of which 79% of the synthetically validated peptides target drug-resistant bacteria through membrane disruption mechanisms, providing an open repository for antibiotic development[167].
It is clear that deep learning models show unique advantages in discovering novel antibiotic structures. The research team screened structurally unique halicin from Drug Repurposing Hub, which showed potent bactericidal activity against both carbapenem-resistant Enterobacteriaceae and Acinetobacter baumannii[168]. To further break through the limitations of the “black box model”, interpretable graphical neural networks were used to resolve chemical substructures related to antibiotic activity, successfully targeting lead compounds from 283 candidate molecules to inhibit methicillin-resistant Staphylococcus aureus and Vancomycin-Resistant Enterococcus, which significantly reduced pathogen load in a mouse infection model[169]. Innovative strategies such as “molecular extinction” have further expanded antibiotic sources. Ancient proteome mining techniques based on the panCleave random forest model screened stable, non-toxic antimicrobial peptides from extinct molecules and validated their efficacy in a model of Acinetobacter baumannii infection[170]. Against this persistent drug-resistant bacterium, machine learning has screened for the narrow-spectrum com
AI technology has shown remarkable potential in the diagnosis and treatment of gastrointestinal diseases, especially in the field of gastrointestinal and liver tumors, where several major breakthroughs have been achieved. However, technology diffusion still faces common challenges: First, the lack of interpretability caused by the black-box nature of the algorithms undermines clinical trust; Second, the lack of data and heterogeneity (including endoscopic device differences and cross-center data bias) lead to limitations in model generalization; and Third, the chain of evidence for clinical translation is still incomplete as existing studies generally lack large-scale RCTs and prospective validation[112,172-176]. In addition, the validation of molecular markers discovered by AI technology needs to expand the sample size, and the technology implementation needs to address the issue of geographic medical appropriateness[149,150]. The breakthrough of these bottlenecks will determine the process of AI technology leapfrogging from experimental results to clinical routine applications.
In response to current challenges in model interpretability, for Gradient-weighted Class Activation Mapping (Grad-CAM) and Shapley value analysis were integrated into diagnostic systems to assist clinicians in understanding the AI decision basis[177]. Grad-CAM generates a heat map to visualize the basis of model decision-making by quantifying the gradient of the target category probability relative to the final convolutional layer feature map. Its implementation is divided into three steps: First, calculate the gradient of the target category scores with respect to a specific convolutional layer feature map to obtain the importance weights of each channel; Second, perform channel-weighted summation of the feature map to generate a coarse-grained heatmap; and finally, up sample the heatmap to the input image size by bilinear interpolation to highlight the key regions. The method does not require modification of the model structure and is suitable for all types of CNNs. In biomedical research, Grad-CAM can localize molecular pathways driving classification
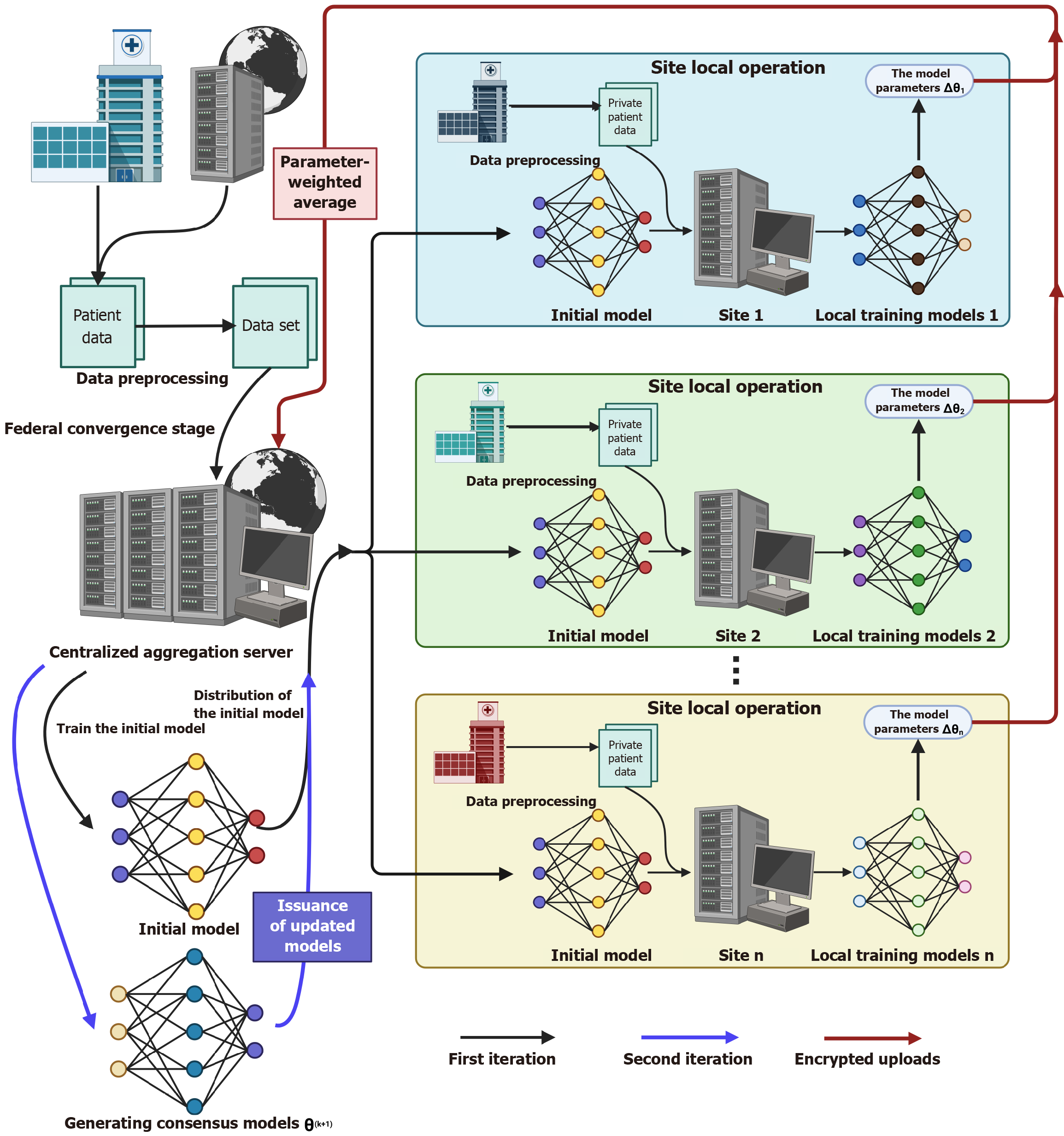
In addressing the lack of data and heterogeneity, multi-task networks (e.g., TransMT-Net) combined with active learning can maintain high accuracy with small samples (96.94% classification accuracy and 77.76% segmentation dice similarity coefficient[181]; and the course self-supervised learning framework utilizes unlabeled images to improve classification performance (73.39% F1 score)[182]. In addition, the migration learning strategy significantly alleviates the dilemma of annotated data scarcity in the medical field. In a deep migration learning study for catheter-dependent congenital heart disease (CHD) screening, the duct dependent congenital heart diseases-DenseNet model based on a two-stage migration strategy achieves the highest sensitivity for critical CHD detection by integrating the multi-center data of 6698 images and 48 videos 0.973 and specificity 0.985, and its hierarchical architectural design significantly improves cross-center generalization, providing an innovative solution for computer-aided hierarchical diagnosis of fetal heart defects in low-resource areas and model scalability[183]. And the breakthrough application of GANs is reflected in two aspects: On the one hand, it can synthesize augmented training data, and on the other hand, it can realize cross-modality image conversion (e.g., magnetic resonance imaging to computed tomography) by CycleGAN, which can effectively support multi-center studies[184,185]. In one of the largest federated machine learning studies to date, researchers integrated data from 6314 patients from 71 healthcare sites across 6 continents to construct an automated tumor boundary detection model. The framework effectively integrates heterogeneous datasets from multiple sources without sharing the original data by weighting and averaging the encrypted model parameters across sites via a central aggregation server. Compared with the model trained only on publicly available datasets, the federated learning model achieves significant breakthroughs in surgical target area detection (33% enhancement) and overall tumor extent identification (23% enhancement), which directly corroborates the critical role of distributed data aggregation in breaking through the data size limitations of a single center. In the study, a pre-training model initialization strategy was adopted to accelerate convergence, and data diversity was gradually improved through phased training (from the initial model at 16 sites to the final consensus model at 71 sites), successfully constructing a cross-regional privacy-preserving collaborative network in rare disease scenarios, which provides a scalable technological paradigm for solving the problem of scarcity of diagnostic resources in low-resource regions[186]. In a multicenter study of prostate magnetic resonance imaging segmentation, three institutions (University of California, LA, State University of New York Upstate Medical Center, and National Cancer Institute) integrated their respective private datasets (100 cases of T2-weighted magnetic resonance imaging images each) through federated learning, and aggregated the model weights by weighted averaging without sharing the original data, so that the federated model on the external dataset (343 cases) demonstrated significantly higher perfor
With the clinical validation and scale deployment of AI-assisted decision-making tools (e.g., the UC-SCALE standar
AI has demonstrated revolutionary potential in the fields of gastroenterology and hepatology, significantly improving the diagnostic accuracy and personalized treatment of gastrointestinal diseases through image analysis, pathology, multi
| 1. | Shortliffe EH, Davis R, Axline SG, Buchanan BG, Green CC, Cohen SN. Computer-based consultations in clinical therapeutics: explanation and rule acquisition capabilities of the MYCIN system. Comput Biomed Res. 1975;8:303-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 155] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 2. | Listgarten J, Damaraju S, Poulin B, Cook L, Dufour J, Driga A, Mackey J, Wishart D, Greiner R, Zanke B. Predictive models for breast cancer susceptibility from multiple single nucleotide polymorphisms. Clin Cancer Res. 2004;10:2725-2737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 102] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 3. | Lecun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proc IEEE. 1998;86:2278-2324. [DOI] [Full Text] |
| 4. | Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Commun ACM. 2017;60:84-90. [DOI] [Full Text] |
| 5. | Liu C, Zhao R, Pang M. A fully automatic segmentation algorithm for CT lung images based on random forest. Med Phys. 2020;47:518-529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 6. | Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, Dehghani M, Minderer M, Heigold G, Gelly S, Uszkoreit J, Houlsby N. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. 2021 Preprint. Available from: arXiv:2010.11929. [DOI] [Full Text] |
| 7. | Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5683] [Cited by in RCA: 5360] [Article Influence: 670.0] [Reference Citation Analysis (0)] |
| 8. | Naseem U, Khushi M, Kim J. Vision-Language Transformer for Interpretable Pathology Visual Question Answering. IEEE J Biomed Health Inform. 2023;27:1681-1690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 15] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 9. | Wu J, Hu R, Xiao Z, Chen J, Liu J. Vision Transformer-based recognition of diabetic retinopathy grade. Med Phys. 2021;48:7850-7863. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 10. | Ayana G, Dese K, Dereje Y, Kebede Y, Barki H, Amdissa D, Husen N, Mulugeta F, Habtamu B, Choe SW. Vision-Transformer-Based Transfer Learning for Mammogram Classification. Diagnostics (Basel). 2023;13:178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 27] [Reference Citation Analysis (0)] |
| 11. | Li J, Chen J, Tang Y, Wang C, Landman BA, Zhou SK. Transforming medical imaging with Transformers? A comparative review of key properties, current progresses, and future perspectives. Med Image Anal. 2023;85:102762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 77] [Article Influence: 38.5] [Reference Citation Analysis (0)] |
| 12. | Zhou B, Yang G, Shi Z, Ma S. Natural Language Processing for Smart Healthcare. IEEE Rev Biomed Eng. 2024;17:4-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 30] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 13. | Yim WW, Yetisgen M, Harris WP, Kwan SW. Natural Language Processing in Oncology: A Review. JAMA Oncol. 2016;2:797-804. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 186] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 14. | Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5690] [Cited by in RCA: 8169] [Article Influence: 8169.0] [Reference Citation Analysis (2)] |
| 15. | Wang J, Ni BY, Wang J, Han L, Ni X, Wang XM, Cao LC, Sun QH, Han XP, Cui HJ. Research progress of Paris polyphylla in the treatment of digestive tract cancers. Discov Oncol. 2024;15:31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 16. | Kobayashi T, Aikata H, Kobayashi T, Ohdan H, Arihiro K, Chayama K. Patients with early recurrence of hepatocellular carcinoma have poor prognosis. Hepatobiliary Pancreat Dis Int. 2017;16:279-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 17. | Ramachandran R, Grantham T, Parvataneni S, Budh D, Gollapalli S, Reddy M, Gaduputi V. Gastric Cancer: Clinical Features, Screening, Diagnosis, Treatment, and Prevention. J Community Hosp Intern Med Perspect. 2024;14:49-57. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 18. | Mukherjee S, Vagha S, Gadkari P. Navigating the Future: A Comprehensive Review of Artificial Intelligence Applications in Gastrointestinal Cancer. Cureus. 2024;16:e54467. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 19. | Luo X, Wang J, Tan C, Dou Q, Han Z, Wang Z, Tasnim F, Wang X, Zhan Q, Li X, Zhou Q, Cheng J, Liao F, Yip HC, Jiang J, Tan RT, Liu S, Yu H. Rapid Endoscopic Diagnosis of Benign Ulcerative Colorectal Diseases With an Artificial Intelligence Contextual Framework. Gastroenterology. 2024;167:591-603.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 20. | Zhou S, Xie Y, Feng X, Li Y, Shen L, Chen Y. Artificial intelligence in gastrointestinal cancer research: Image learning advances and applications. Cancer Lett. 2025;614:217555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 21. | Li X, Zhai Z, Ding W, Chen L, Zhao Y, Xiong W, Zhang Y, Lin D, Chen Z, Wang W, Gao Y, Cai S, Yu J, Zhang X, Liu H, Li G, Chen T. An artificial intelligence model to predict survival and chemotherapy benefits for gastric cancer patients after gastrectomy development and validation in international multicenter cohorts. Int J Surg. 2022;105:106889. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 22. | Yuan L, Yang L, Zhang S, Xu Z, Qin J, Shi Y, Yu P, Wang Y, Bao Z, Xia Y, Sun J, He W, Chen T, Chen X, Hu C, Zhang Y, Dong C, Zhao P, Wang Y, Jiang N, Lv B, Xue Y, Jiao B, Gao H, Chai K, Li J, Wang H, Wang X, Guan X, Liu X, Zhao G, Zheng Z, Yan J, Yu H, Chen L, Ye Z, You H, Bao Y, Cheng X, Zhao P, Wang L, Zeng W, Tian Y, Chen M, You Y, Yuan G, Ruan H, Gao X, Xu J, Xu H, Du L, Zhang S, Fu H, Cheng X. Development of a tongue image-based machine learning tool for the diagnosis of gastric cancer: a prospective multicentre clinical cohort study. EClinicalMedicine. 2023;57:101834. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 40] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 23. | Listopad S, Magnan C, Asghar A, Stolz A, Tayek JA, Liu ZX, Morgan TR, Norden-Krichmar TM. Differentiating between liver diseases by applying multiclass machine learning approaches to transcriptomics of liver tissue or blood-based samples. JHEP Rep. 2022;4:100560. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 24. | Liu Y, He C, Fang W, Peng L, Shi F, Xia Y, Zhou Q, Zhang R, Li C. Prediction of Ki-67 expression in gastrointestinal stromal tumors using radiomics of plain and multiphase contrast-enhanced CT. Eur Radiol. 2023;33:7609-7617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 8] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 25. | Chen T, Wu J, Cui C, He Q, Li X, Liang W, Liu X, Liu T, Zhou X, Zhang X, Lei X, Xiong W, Yu J, Li G. CT-based radiomics nomograms for preoperative prediction of diffuse-type and signet ring cell gastric cancer: a multicenter development and validation cohort. J Transl Med. 2022;20:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 26. | Bertsimas D, Margonis GA, Sujichantararat S, Koulouras A, Ma Y, Antonescu CR, Brennan MF, Martín-Broto J, Tang S, Rutkowski P, Kreis ME, Beyer K, Wang J, Bylina E, Sobczuk P, Gutierrez A, Jadeja B, Tap WD, Chi P, Singer S. Interpretable artificial intelligence to optimise use of imatinib after resection in patients with localised gastrointestinal stromal tumours: an observational cohort study. Lancet Oncol. 2024;25:1025-1037. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 12] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 27. | Chue KM, Douglass BR, Ong LWL, Tan JTH, Teh JGX, Putera M, Kwan CKW, Wong WK, Yeung BPM. Maximizing oral intake tolerance in malignant gastric outlet obstruction - a Markov decision tree analysis comparing duodenal stenting, endoscopic ultrasound-guided gastroenterostomy and surgical gastrojejunostomy based on a meta-analysis of randomized controlled trials. Int J Surg. 2025;111:3006-3019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 28. | Han Z, Zhang Z, Yang X, Li Z, Sang S, Islam MT, Guo AA, Li Z, Wang X, Wang J, Zhang T, Sun Z, Yu L, Wang W, Xiong W, Li G, Jiang Y. Development and interpretation of a pathomics-driven ensemble model for predicting the response to immunotherapy in gastric cancer. J Immunother Cancer. 2024;12:e008927. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 29. | Bertsimas D, Margonis GA, Tang S, Koulouras A, Antonescu CR, Brennan MF, Martin-Broto J, Rutkowski P, Stasinos G, Wang J, Pikoulis E, Bylina E, Sobczuk P, Gutierrez A, Jadeja B, Tap WD, Chi P, Singer S. An interpretable AI model for recurrence prediction after surgery in gastrointestinal stromal tumour: an observational cohort study. EClinicalMedicine. 2023;64:102200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 30. | Arai J, Aoki T, Sato M, Niikura R, Suzuki N, Ishibashi R, Tsuji Y, Yamada A, Hirata Y, Ushiku T, Hayakawa Y, Fujishiro M. Machine learning-based personalized prediction of gastric cancer incidence using the endoscopic and histologic findings at the initial endoscopy. Gastrointest Endosc. 2022;95:864-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 34] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 31. | Liu Y, Méric G, Havulinna AS, Teo SM, Åberg F, Ruuskanen M, Sanders J, Zhu Q, Tripathi A, Verspoor K, Cheng S, Jain M, Jousilahti P, Vázquez-Baeza Y, Loomba R, Lahti L, Niiranen T, Salomaa V, Knight R, Inouye M. Early prediction of incident liver disease using conventional risk factors and gut-microbiome-augmented gradient boosting. Cell Metab. 2022;34:719-730.e4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 44] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 32. | Wang L, Wu X, Tian R, Ma H, Jiang Z, Zhao W, Cui G, Li M, Hu Q, Yu X, Xu W. MRI-based pre-Radiomics and delta-Radiomics models accurately predict the post-treatment response of rectal adenocarcinoma to neoadjuvant chemoradiotherapy. Front Oncol. 2023;13:1133008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 33. | Facchin S, Calgaro M, Pandolfo M, Caldart F, Ghisa M, Greco E, Sattin E, Valle G, Dellon ES, Vitulo N, Savarino EV. Salivary microbiota composition may discriminate between patients with eosinophilic oesophagitis (EoE) and non-EoE subjects. Aliment Pharmacol Ther. 2022;56:450-462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 17] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 34. | Cai Z, Zhang J, He Y, Xia L, Dong X, Chen G, Zhou Y, Hu X, Zhong S, Wang Y, Chen H, Xie D, Liu X, Liu J. Liquid biopsy by combining 5-hydroxymethylcytosine signatures of plasma cell-free DNA and protein biomarkers for diagnosis and prognosis of hepatocellular carcinoma. ESMO Open. 2021;6:100021. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 35. | Khrom M, Long M, Dube S, Robbins L, Botwin GJ, Yang S, Mengesha E, Li D, Naito T, Bonthala NN, Ha C, Melmed G, Rabizadeh S, Syal G, Vasiliauskas E, Ziring D, Brant SR, Cho J, Duerr RH, Rioux J, Schumm P, Silverberg M, Ananthakrishnan AN, Faubion WA, Jabri B, Lira SA, Newberry RD, Sandler RS, Xavier RJ, Kugathasan S, Hercules D, Targan SR, Sartor RB, Haritunians T, McGovern DPB. Comprehensive Association Analyses of Extraintestinal Manifestations in Inflammatory Bowel Disease. Gastroenterology. 2024;167:315-332. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 36. | Jung JO, Pisula JI, Bozek K, Popp F, Fuchs HF, Schröder W, Bruns CJ, Schmidt T. Prediction of postoperative complications after oesophagectomy using machine-learning methods. Br J Surg. 2023;110:1361-1366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 37. | Pera M, Gibert J, Gimeno M, Garsot E, Eizaguirre E, Miró M, Castro S, Miranda C, Reka L, Leturio S, González-Duaigües M, Codony C, Gobbini Y, Luna A, Fernández-Ananín S, Sarriugarte A, Olona C, Rodríguez-Santiago J, Osorio J, Grande L; Spanish EURECCA Esophagogastric Cancer Group. Machine Learning Risk Prediction Model of 90-day Mortality After Gastrectomy for Cancer. Ann Surg. 2022;276:776-783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
| 38. | Zou L, Jiang Q, Guo T, Wu X, Wang Q, Feng Y, Zhang S, Fang W, Zhou W, Yang A. Endoscopic characteristics in predicting prognosis of biopsy-diagnosed gastric low-grade intraepithelial neoplasia. Chin Med J (Engl). 2022;135:26-35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 39. | Yu S, Jiang H, Xia J, Gu J, Chen M, Wang Y, Zhao X, Liao Z, Zeng P, Xie T, Sui X. Construction of machine learning-based models for screening the high-risk patients with gastric precancerous lesions. Chin Med. 2025;20:7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 40. | Zeng Y, Li J, Zheng Y, Zhang D, Zhong N, Zuo X, Li Y, Yu W, Lu J. Development and validation of a predictive model for submucosal fibrosis in patients with early gastric cancer undergoing endoscopic submucosal dissection: experience from a large tertiary center. Ann Med. 2024;56:2391536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 41. | Reddy KR, McLerran D, Marsh T, Parikh N, Roberts LR, Schwartz M, Nguyen MH, Befeler A, Page-Lester S, Tang R, Srivastava S, Rinaudo JA, Feng Z, Marrero JA. Incidence and Risk Factors for Hepatocellular Carcinoma in Cirrhosis: The Multicenter Hepatocellular Carcinoma Early Detection Strategy (HEDS) Study. Gastroenterology. 2023;165:1053-1063.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 27] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 42. | Ma L, Zhang C, Wen Y, Xing K, Li S, Geng Z, Liao S, Yuan S, Li X, Zhong C, Hou J, Zhang J, Gao M, Xu B, Guo R, Wei W, Xie C, Lu L. Imaging-based surrogate classification for risk stratification of hepatocellular carcinoma with microvascular invasion to adjuvant hepatic arterial infusion chemotherapy: a multicenter retrospective study. Int J Surg. 2025;111:872-883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 43. | Xia TY, Zhou ZH, Meng XP, Zha JH, Yu Q, Wang WL, Song Y, Wang YC, Tang TY, Xu J, Zhang T, Long XY, Liang Y, Xiao WB, Ju SH. Predicting Microvascular Invasion in Hepatocellular Carcinoma Using CT-based Radiomics Model. Radiology. 2023;307:e222729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 103] [Reference Citation Analysis (1)] |
| 44. | Schoenberger H, Chong N, Fetzer DT, Rich NE, Yokoo T, Khatri G, Olivares J, Parikh ND, Yopp AC, Marrero JA, Singal AG. Dynamic Changes in Ultrasound Quality for Hepatocellular Carcinoma Screening in Patients With Cirrhosis. Clin Gastroenterol Hepatol. 2022;20:1561-1569.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 81] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 45. | Singal AG, Chen Y, Sridhar S, Mittal V, Fullington H, Shaik M, Waljee AK, Tiro J. Novel Application of Predictive Modeling: A Tailored Approach to Promoting HCC Surveillance in Patients With Cirrhosis. Clin Gastroenterol Hepatol. 2022;20:1795-1802.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 46. | Wang JJ, Feng J, Gomes C, Calthorpe L, Ashraf Ganjouei A, Romero-Hernandez F, Benedetti Cacciaguerra A, Hibi T, Adam MA, Alseidi A, Abu Hilal M, Rashidian N; International Post-Hepatectomy Liver Failure Study Group. Development and Validation of Prediction Models and Risk Calculators for Posthepatectomy Liver Failure and Postoperative Complications Using a Diverse International Cohort of Major Hepatectomies. Ann Surg. 2023;278:976-984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 47. | Briggs SEW, Law P, East JE, Wordsworth S, Dunlop M, Houlston R, Hippisley-Cox J, Tomlinson I. Integrating genome-wide polygenic risk scores and non-genetic risk to predict colorectal cancer diagnosis using UK Biobank data: population based cohort study. BMJ. 2022;379:e071707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 24] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 48. | Cross AJ, Robbins EC, Saunders BP, Duffy SW, Wooldrage K. Higher Adenoma Detection Rates at Screening Associated With Lower Long-Term Colorectal Cancer Incidence and Mortality. Clin Gastroenterol Hepatol. 2022;20:e148-e167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 49. | Bever AM, Hang D, Lee DH, Tabung FK, Ugai T, Ogino S, Meyerhardt JA, Chan AT, Eliassen AH, Liang L, Stampfer MJ, Song M. Metabolomic signatures of inflammation and metabolic dysregulation in relation to colorectal cancer risk. J Natl Cancer Inst. 2024;116:1126-1136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 50. | Yang X, Qiu H, Wang L, Wang X. Predicting Colorectal Cancer Survival Using Time-to-Event Machine Learning: Retrospective Cohort Study. J Med Internet Res. 2023;25:e44417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 21] [Reference Citation Analysis (0)] |
| 51. | Meyer YM, Wilting SM, Kraan J, Olthof P, Vermeulen P, Martens J, Grünhagen DJ, Sleijfer S, Verhoef C. Circulating tumour cells are associated with histopathological growth patterns of colorectal cancer liver metastases. Clin Exp Metastasis. 2023;40:69-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 52. | Tian S, Shi H, Chen W, Li S, Han C, Du F, Wang W, Wen H, Lei Y, Deng L, Tang J, Zhang J, Lin J, Shi L, Ning B, Zhao K, Miao J, Wang G, Hou H, Huang X, Kong W, Jin X, Ding Z, Lin R. Artificial intelligence-based diagnosis of standard endoscopic ultrasonography scanning sites in the biliopancreatic system: a multicenter retrospective study. Int J Surg. 2024;110:1637-1644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 53. | Nguyen TH, Doan NNT, Tran TH, Huynh LAK, Doan PL, Nguyen THH, Nguyen VTC, Nguyen GTH, Nguyen HN, Giang H, Tran LS, Phan MD. Tissue of origin detection for cancer tumor using low-depth cfDNA samples through combination of tumor-specific methylation atlas and genome-wide methylation density in graph convolutional neural networks. J Transl Med. 2024;22:618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 54. | Hao D, Li Q, Feng QX, Qi L, Liu XS, Arefan D, Zhang YD, Wu S. SurvivalCNN: A deep learning-based method for gastric cancer survival prediction using radiological imaging data and clinicopathological variables. Artif Intell Med. 2022;134:102424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 55. | Wu Z, Wang T, Lan J, Wang J, Chen G, Tong T, Zhang H. Deep learning-based prediction of HER2 status and trastuzumab treatment efficacy of gastric adenocarcinoma based on morphological features. J Transl Med. 2025;23:13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 56. | Nakatsuka T, Tateishi R, Sato M, Hashizume N, Kamada A, Nakano H, Kabeya Y, Yonezawa S, Irie R, Tsujikawa H, Sumida Y, Yoneda M, Akuta N, Kawaguchi T, Takahashi H, Eguchi Y, Seko Y, Itoh Y, Murakami E, Chayama K, Taniai M, Tokushige K, Okanoue T, Sakamoto M, Fujishiro M, Koike K. Deep learning and digital pathology powers prediction of HCC development in steatotic liver disease. Hepatology. 2025;81:976-989. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 57. | Liu B, Li J, Yang X, Chen F, Zhang Y, Li H. Diagnosis of primary clear cell carcinoma of the liver based on Faster region-based convolutional neural network. Chin Med J (Engl). 2023;136:2706-2711. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 58. | Wagner SJ, Reisenbüchler D, West NP, Niehues JM, Zhu J, Foersch S, Veldhuizen GP, Quirke P, Grabsch HI, van den Brandt PA, Hutchins GGA, Richman SD, Yuan T, Langer R, Jenniskens JCA, Offermans K, Mueller W, Gray R, Gruber SB, Greenson JK, Rennert G, Bonner JD, Schmolze D, Jonnagaddala J, Hawkins NJ, Ward RL, Morton D, Seymour M, Magill L, Nowak M, Hay J, Koelzer VH, Church DN; TransSCOT consortium, Matek C, Geppert C, Peng C, Zhi C, Ouyang X, James JA, Loughrey MB, Salto-Tellez M, Brenner H, Hoffmeister M, Truhn D, Schnabel JA, Boxberg M, Peng T, Kather JN. Transformer-based biomarker prediction from colorectal cancer histology: A large-scale multicentric study. Cancer Cell. 2023;41:1650-1661.e4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 95] [Article Influence: 47.5] [Reference Citation Analysis (0)] |
| 59. | Wang R, Dai W, Gong J, Huang M, Hu T, Li H, Lin K, Tan C, Hu H, Tong T, Cai G. Development of a novel combined nomogram model integrating deep learning-pathomics, radiomics and immunoscore to predict postoperative outcome of colorectal cancer lung metastasis patients. J Hematol Oncol. 2022;15:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 145] [Article Influence: 48.3] [Reference Citation Analysis (0)] |
| 60. | Chang X, Wang J, Zhang G, Yang M, Xi Y, Xi C, Chen G, Nie X, Meng B, Quan X. Predicting colorectal cancer microsatellite instability with a self-attention-enabled convolutional neural network. Cell Rep Med. 2023;4:100914. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 61. | Sinonquel P, Eelbode T, Pech O, De Wulf D, Dewint P, Neumann H, Antonelli G, Iacopini F, Tate D, Lemmers A, Pilonis ND, Kaminski MF, Roelandt P, Hassan C, Ingrid D, Maes F, Bisschops R. Clinical consequences of computer-aided colorectal polyp detection. Gut. 2024;73:1974-1983. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 62. | Su J, Liu Z, Li H, Kang L, Huang K, Wu J, Huang H, Ling F, Yao X, Huang C. Artificial intelligence-based model to predict recurrence after local excision in T1 rectal cancer. Eur J Surg Oncol. 2025;51:109717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 63. | Wang F, Zhan G, Chen QQ, Xu HY, Cao D, Zhang YY, Li YH, Zhang CJ, Jin Y, Ji WB, Ma JB, Yang YJ, Zhou W, Peng ZY, Liang X, Deng LP, Lin LF, Chen YW, Hu HJ. Multitask deep learning for prediction of microvascular invasion and recurrence-free survival in hepatocellular carcinoma based on MRI images. Liver Int. 2024;44:1351-1362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 14] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 64. | Kong Q, Li K. Predicting early recurrence of hepatocellular carcinoma after thermal ablation based on longitudinal MRI with a deep learning approach. Oncologist. 2025;30:oyaf013. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 65. | Iseke S, Zeevi T, Kucukkaya AS, Raju R, Gross M, Haider SP, Petukhova-Greenstein A, Kuhn TN, Lin M, Nowak M, Cooper K, Thomas E, Weber MA, Madoff DC, Staib L, Batra R, Chapiro J. Machine Learning Models for Prediction of Posttreatment Recurrence in Early-Stage Hepatocellular Carcinoma Using Pretreatment Clinical and MRI Features: A Proof-of-Concept Study. AJR Am J Roentgenol. 2023;220:245-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 66. | Zhang Y, Shi K, Feng Y, Wang XB. Machine learning model using immune indicators to predict outcomes in early liver cancer. World J Gastroenterol. 2025;31:101722. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (4)] |
| 67. | Qu H, Zhang S, Guo M, Miao Y, Han Y, Ju R, Cui X, Li Y. Deep Learning Model for Predicting Proliferative Hepatocellular Carcinoma Using Dynamic Contrast-Enhanced MRI: Implications for Early Recurrence Prediction Following Radical Resection. Acad Radiol. 2024;31:4445-4455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 68. | Guo X, Chen M, Zhou L, Zhu L, Liu S, Zheng L, Chen Y, Li Q, Xia S, Lu C, Chen M, Chen F, Ji J. Predicting early recurrence in locally advanced gastric cancer after gastrectomy using CT-based deep learning model: a multicenter study. Int J Surg. 2025;111:2089-2100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 69. | Lin CH, Hsu PI, Tseng CD, Chao PJ, Wu IT, Ghose S, Shih CA, Lee SH, Ren JH, Shie CB, Lee TF. Application of artificial intelligence in endoscopic image analysis for the diagnosis of a gastric cancer pathogen-Helicobacter pylori infection. Sci Rep. 2023;13:13380. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 10] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 70. | Wang CW, Liu TC, Lai PJ, Muzakky H, Wang YC, Yu MH, Wu CH, Chao TK. Ensemble transformer-based multiple instance learning to predict pathological subtypes and tumor mutational burden from histopathological whole slide images of endometrial and colorectal cancer. Med Image Anal. 2025;99:103372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 71. | Elforaici MEA, Montagnon E, Romero FP, Le WT, Azzi F, Trudel D, Nguyen B, Turcotte S, Tang A, Kadoury S. Semi-supervised ViT knowledge distillation network with style transfer normalization for colorectal liver metastases survival prediction. Med Image Anal. 2025;99:103346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 72. | Khan A, Brouwer N, Blank A, Müller F, Soldini D, Noske A, Gaus E, Brandt S, Nagtegaal I, Dawson H, Thiran JP, Perren A, Lugli A, Zlobec I. Computer-Assisted Diagnosis of Lymph Node Metastases in Colorectal Cancers Using Transfer Learning With an Ensemble Model. Mod Pathol. 2023;36:100118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
| 73. | Zhang W, Lu F, Su H, Hu Y. Dual-branch multi-information aggregation network with transformer and convolution for polyp segmentation. Comput Biol Med. 2024;168:107760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 74. | Qi J, Ruan G, Ping Y, Xiao Z, Liu K, Cheng Y, Liu R, Zhang B, Zhi M, Chen J, Xiao F, Zhao T, Li J, Zhang Z, Zou Y, Cao Q, Nian Y, Wei Y. Development and validation of a deep learning-based approach to predict the Mayo endoscopic score of ulcerative colitis. Therap Adv Gastroenterol. 2023;16:17562848231170945. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 75. | Cai H, Feng X, Yin R, Zhao Y, Guo L, Fan X, Liao J. MIST: multiple instance learning network based on Swin Transformer for whole slide image classification of colorectal adenomas. J Pathol. 2023;259:125-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 76. | Ou J, Jiang L, Bai T, Zhan P, Liu R, Xiao H. ResTransUnet: An effective network combined with Transformer and U-Net for liver segmentation in CT scans. Comput Biol Med. 2024;177:108625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 77. | Deng B, Tian Y, Zhang Q, Wang Y, Chai Z, Ye Q, Yao S, Liang T, Li J. NecroGlobalGCN: Integrating micronecrosis information in HCC prognosis prediction via graph convolutional neural networks. Comput Methods Programs Biomed. 2024;257:108435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 78. | Gui X, Bazarova A, Del Amor R, Vieth M, de Hertogh G, Villanacci V, Zardo D, Parigi TL, Røyset ES, Shivaji UN, Monica MAT, Mandelli G, Bhandari P, Danese S, Ferraz JG, Hayee B, Lazarev M, Parra-Blanco A, Pastorelli L, Panaccione R, Rath T, Tontini GE, Kiesslich R, Bisschops R, Grisan E, Naranjo V, Ghosh S, Iacucci M. PICaSSO Histologic Remission Index (PHRI) in ulcerative colitis: development of a novel simplified histological score for monitoring mucosal healing and predicting clinical outcomes and its applicability in an artificial intelligence system. Gut. 2022;71:889-898. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 91] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 79. | Peyrin-Biroulet L, Adsul S, Stancati A, Dehmeshki J, Kubassova O. An artificial intelligence-driven scoring system to measure histological disease activity in ulcerative colitis. United European Gastroenterol J. 2024;12:1028-1033. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 14] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 80. | Turan M, Durmus F. UC-NfNet: Deep learning-enabled assessment of ulcerative colitis from colonoscopy images. Med Image Anal. 2022;82:102587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 81. | Kuroki T, Maeda Y, Kudo SE, Ogata N, Iacucci M, Takishima K, Ide Y, Shibuya T, Semba S, Kawashima J, Kato S, Ogawa Y, Ichimasa K, Nakamura H, Hayashi T, Wakamura K, Miyachi H, Baba T, Nemoto T, Ohtsuka K, Misawa M. A novel artificial intelligence-assisted "vascular healing" diagnosis for prediction of future clinical relapse in patients with ulcerative colitis: a prospective cohort study (with video). Gastrointest Endosc. 2024;100:97-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 20] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 82. | Ohara J, Nemoto T, Maeda Y, Ogata N, Kudo SE, Yamochi T. Deep learning-based automated quantification of goblet cell mucus using histological images as a predictor of clinical relapse of ulcerative colitis with endoscopic remission. J Gastroenterol. 2022;57:962-970. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 27] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 83. | Gutierrez-Becker B, Fraessle S, Yao H, Luscher J, Girycki R, Machura B, Czornik J, Goslinsky J, Pitura M, Levitte S, Arús-Pous J, Fisher E, Bojic D, Richmond D, Bigorgne AE, Prunotto M. Ulcerative Colitis Severity Classification and Localized Extent (UC-SCALE): An Artificial Intelligence Scoring System for a Spatial Assessment of Disease Severity in Ulcerative Colitis. J Crohns Colitis. 2025;19:jjae187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 84. | Guez I, Focht G, Greer MC, Cytter-Kuint R, Pratt LT, Castro DA, Turner D, Griffiths AM, Freiman M. Development of a multimodal machine-learning fusion model to non-invasively assess ileal Crohn's disease endoscopic activity. Comput Methods Programs Biomed. 2022;227:107207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 85. | Rymarczyk D, Schultz W, Borowa A, Friedman JR, Danel T, Branigan P, Chałupczak M, Bracha A, Krawiec T, Warchoł M, Li K, De Hertogh G, Zieliński B, Ghanem LR, Stojmirovic A. Deep Learning Models Capture Histological Disease Activity in Crohn's Disease and Ulcerative Colitis with High Fidelity. J Crohns Colitis. 2024;18:604-614. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 23] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 86. | Kiyokawa H, Abe M, Matsui T, Kurashige M, Ohshima K, Tahara S, Nojima S, Ogino T, Sekido Y, Mizushima T, Morii E. Deep Learning Analysis of Histologic Images from Intestinal Specimen Reveals Adipocyte Shrinkage and Mast Cell Infiltration to Predict Postoperative Crohn Disease. Am J Pathol. 2022;192:904-916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 87. | Ribeiro T, Mascarenhas M, Afonso J, Cardoso H, Andrade P, Lopes S, Ferreira J, Mascarenhas Saraiva M, Macedo G. Artificial intelligence and colon capsule endoscopy: Automatic detection of ulcers and erosions using a convolutional neural network. J Gastroenterol Hepatol. 2022;37:2282-2288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 88. | Mascarenhas Saraiva MJ, Afonso J, Ribeiro T, Ferreira J, Cardoso H, Andrade AP, Parente M, Natal R, Mascarenhas Saraiva M, Macedo G. Deep learning and capsule endoscopy: automatic identification and differentiation of small bowel lesions with distinct haemorrhagic potential using a convolutional neural network. BMJ Open Gastroenterol. 2021;8:e000753. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 89. | Barash Y, Azaria L, Soffer S, Margalit Yehuda R, Shlomi O, Ben-Horin S, Eliakim R, Klang E, Kopylov U. Ulcer severity grading in video capsule images of patients with Crohn's disease: an ordinal neural network solution. Gastrointest Endosc. 2021;93:187-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 65] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 90. | Gong R, He S, Tian T, Chen J, Hao Y, Qiao C. FRCNN-AA-CIF: An automatic detection model of colon polyps based on attention awareness and context information fusion. Comput Biol Med. 2023;158:106787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 91. | Sharma A, Kumar R, Garg P. Deep learning-based prediction model for diagnosing gastrointestinal diseases using endoscopy images. Int J Med Inform. 2023;177:105142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 21] [Reference Citation Analysis (0)] |
| 92. | Tang CP, Chen KH, Lin TL. Computer-Aided Colon Polyp Detection on High Resolution Colonoscopy Using Transfer Learning Techniques. Sensors (Basel). 2021;21:5315. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 93. | Ziegelmayer S, Reischl S, Havrda H, Gawlitza J, Graf M, Lenhart N, Nehls N, Lemke T, Wilhelm D, Lohöfer F, Burian E, Neumann PA, Makowski M, Braren R. Development and Validation of a Deep Learning Algorithm to Differentiate Colon Carcinoma From Acute Diverticulitis in Computed Tomography Images. JAMA Netw Open. 2023;6:e2253370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 94. | Harb SF, Ali A, Yousuf M, Elshazly S, Farag A. G-SET-DCL: a guided sequential episodic training with dual contrastive learning approach for colon segmentation. Int J Comput Assist Radiol Surg. 2025;20:279-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 95. | Enchakalody BE, Wasnik AP, Al-Hawary MM, Wang SC, Su GL, Ross B, Stidham RW. Local Assessment and Small Bowel Crohn's Disease Severity Scoring using AI. Acad Radiol. 2024;31:4045-4056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 96. | Abe H, Kurose Y, Takahama S, Kume A, Nishida S, Fukasawa M, Yasunaga Y, Ushiku T, Ninomiya Y, Yoshizawa A, Murao K, Sato S, Kitsuregawa M, Harada T, Kitagawa M, Fukayama M; Japan Pathology AI Diagnostics/National Institution of Informatics (JP-AID/NII) Study Group for Gastric Biopsy Pathology. Development and multi-institutional validation of an artificial intelligence-based diagnostic system for gastric biopsy. Cancer Sci. 2022;113:3608-3617. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 97. | Franklin MM, Schultz FA, Tafoya MA, Kerwin AA, Broehm CJ, Fischer EG, Gullapalli RR, Clark DP, Hanson JA, Martin DR. A Deep Learning Convolutional Neural Network Can Differentiate Between Helicobacter Pylori Gastritis and Autoimmune Gastritis With Results Comparable to Gastrointestinal Pathologists. Arch Pathol Lab Med. 2022;146:117-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 98. | Faust O, De Michele S, Koh JE, Jahmunah V, Lih OS, Kamath AP, Barua PD, Ciaccio EJ, Lewis SK, Green PH, Bhagat G, Acharya UR. Automated analysis of small intestinal lamina propria to distinguish normal, Celiac Disease, and Non-Celiac Duodenitis biopsy images. Comput Methods Programs Biomed. 2023;230:107320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 99. | Luo H, Xu G, Li C, He L, Luo L, Wang Z, Jing B, Deng Y, Jin Y, Li Y, Li B, Tan W, He C, Seeruttun SR, Wu Q, Huang J, Huang DW, Chen B, Lin SB, Chen QM, Yuan CM, Chen HX, Pu HY, Zhou F, He Y, Xu RH. Real-time artificial intelligence for detection of upper gastrointestinal cancer by endoscopy: a multicentre, case-control, diagnostic study. Lancet Oncol. 2019;20:1645-1654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 253] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 100. | Goto A, Kubota N, Nishikawa J, Ogawa R, Hamabe K, Hashimoto S, Ogihara H, Hamamoto Y, Yanai H, Miura O, Takami T. Cooperation between artificial intelligence and endoscopists for diagnosing invasion depth of early gastric cancer. Gastric Cancer. 2023;26:116-122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 22] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 101. | Wu L, Shang R, Sharma P, Zhou W, Liu J, Yao L, Dong Z, Yuan J, Zeng Z, Yu Y, He C, Xiong Q, Li Y, Deng Y, Cao Z, Huang C, Zhou R, Li H, Hu G, Chen Y, Wang Y, He X, Zhu Y, Yu H. Effect of a deep learning-based system on the miss rate of gastric neoplasms during upper gastrointestinal endoscopy: a single-centre, tandem, randomised controlled trial. Lancet Gastroenterol Hepatol. 2021;6:700-708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 77] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 102. | Botros M, de Boer OJ, Cardenas B, Bekkers EJ, Jansen M, van der Wel MJ, Sánchez CI, Meijer SL. Deep Learning for Histopathological Assessment of Esophageal Adenocarcinoma Precursor Lesions. Mod Pathol. 2024;37:100531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 103. | Jiang Y, Zhang Z, Yuan Q, Wang W, Wang H, Li T, Huang W, Xie J, Chen C, Sun Z, Yu J, Xu Y, Poultsides GA, Xing L, Zhou Z, Li G, Li R. Predicting peritoneal recurrence and disease-free survival from CT images in gastric cancer with multitask deep learning: a retrospective study. Lancet Digit Health. 2022;4:e340-e350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 86] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 104. | Liu L, Zhang R, Shi Y, Sun J, Xu X. Automated machine learning for predicting liver metastasis in patients with gastrointestinal stromal tumor: a SEER-based analysis. Sci Rep. 2024;14:12415. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 105. | Ma Y, Jin J, Xue Z, Zhao J, Cai W, Zhang W. Integrated multi-omics analysis and machine learning developed a prognostic model based on mitochondrial function in a large multicenter cohort for Gastric Cancer. J Transl Med. 2024;22:381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 106. | Che G, Yin J, Wang W, Luo Y, Chen Y, Yu X, Wang H, Liu X, Chen Z, Wang X, Chen Y, Wang X, Tang K, Tang J, Shao W, Wu C, Sheng J, Li Q, Liu J. Circumventing drug resistance in gastric cancer: A spatial multi-omics exploration of chemo and immuno-therapeutic response dynamics. Drug Resist Updat. 2024;74:101080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 23] [Reference Citation Analysis (0)] |
| 107. | Wang H, Yin Y, Zhu ZJ. Encoding LC-MS-Based Untargeted Metabolomics Data into Images toward AI-Based Clinical Diagnosis. Anal Chem. 2023;95:6533-6541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 108. | Liu L, Yu Z, Chen H, Gong Z, Huang X, Chen L, Fan Z, Zhang J, Yan J, Tian H, Zeng X, Chen Z, Zhang P, Zhou H. Imatinib adherence prediction using machine learning approach in patients with gastrointestinal stromal tumor. Cancer. 2025;131:e35548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 109. | Wallace MB, Sharma P, Bhandari P, East J, Antonelli G, Lorenzetti R, Vieth M, Speranza I, Spadaccini M, Desai M, Lukens FJ, Babameto G, Batista D, Singh D, Palmer W, Ramirez F, Palmer R, Lunsford T, Ruff K, Bird-Liebermann E, Ciofoaia V, Arndtz S, Cangemi D, Puddick K, Derfus G, Johal AS, Barawi M, Longo L, Moro L, Repici A, Hassan C. Impact of Artificial Intelligence on Miss Rate of Colorectal Neoplasia. Gastroenterology. 2022;163:295-304.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 142] [Article Influence: 47.3] [Reference Citation Analysis (0)] |
| 110. | Glissen Brown JR, Mansour NM, Wang P, Chuchuca MA, Minchenberg SB, Chandnani M, Liu L, Gross SA, Sengupta N, Berzin TM. Deep Learning Computer-aided Polyp Detection Reduces Adenoma Miss Rate: A United States Multi-center Randomized Tandem Colonoscopy Study (CADeT-CS Trial). Clin Gastroenterol Hepatol. 2022;20:1499-1507.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 115] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 111. | Seager A, Sharp L, Neilson LJ, Brand A, Hampton JS, Lee TJW, Evans R, Vale L, Whelpton J, Bestwick N, Rees CJ; COLO-DETECT trial team. Polyp detection with colonoscopy assisted by the GI Genius artificial intelligence endoscopy module compared with standard colonoscopy in routine colonoscopy practice (COLO-DETECT): a multicentre, open-label, parallel-arm, pragmatic randomised controlled trial. Lancet Gastroenterol Hepatol. 2024;9:911-923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 9] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 112. | Griem J, Eich ML, Schallenberg S, Pryalukhin A, Bychkov A, Fukuoka J, Zayats V, Hulla W, Munkhdelger J, Seper A, Tsvetkov T, Mukhopadhyay A, Sanner A, Stieber J, Fuchs M, Babendererde N, Schömig-Markiefka B, Klein S, Buettner R, Quaas A, Tolkach Y. Artificial Intelligence-Based Tool for Tumor Detection and Quantitative Tissue Analysis in Colorectal Specimens. Mod Pathol. 2023;36:100327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 113. | Song JH, Hong Y, Kim ER, Kim SH, Sohn I. Utility of artificial intelligence with deep learning of hematoxylin and eosin-stained whole slide images to predict lymph node metastasis in T1 colorectal cancer using endoscopically resected specimens; prediction of lymph node metastasis in T1 colorectal cancer. J Gastroenterol. 2022;57:654-666. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 31] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 114. | Yang J, Ye H, Fan X, Li Y, Wu X, Zhao M, Hu Q, Ye Y, Wu L, Li Z, Zhang X, Liang C, Wang Y, Xu Y, Li Q, Yao S, You D, Zhao K, Liu Z. Artificial intelligence for quantifying immune infiltrates interacting with stroma in colorectal cancer. J Transl Med. 2022;20:451. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 115. | Saillard C, Dubois R, Tchita O, Loiseau N, Garcia T, Adriansen A, Carpentier S, Reyre J, Enea D, von Loga K, Kamoun A, Rossat S, Wiscart C, Sefta M, Auffret M, Guillou L, Fouillet A, Kather JN, Svrcek M. Validation of MSIntuit as an AI-based pre-screening tool for MSI detection from colorectal cancer histology slides. Nat Commun. 2023;14:6695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 32] [Reference Citation Analysis (0)] |
| 116. | Gerwert K, Schörner S, Großerueschkamp F, Kraeft AL, Schuhmacher D, Sternemann C, Feder IS, Wisser S, Lugnier C, Arnold D, Teschendorf C, Mueller L, Timmesfeld N, Mosig A, Reinacher-Schick A, Tannapfel A. Fast and label-free automated detection of microsatellite status in early colon cancer using artificial intelligence integrated infrared imaging. Eur J Cancer. 2023;182:122-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 117. | Wang FA, Zhuang Z, Gao F, He R, Zhang S, Wang L, Liu J, Li Y. TMO-Net: an explainable pretrained multi-omics model for multi-task learning in oncology. Genome Biol. 2024;25:149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 118. | Liu Y, Ji Y, Chen J, Zhang Y, Li X, Li X. Pioneering noninvasive colorectal cancer detection with an AI-enhanced breath volatilomics platform. Theranostics. 2024;14:4240-4255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 119. | Feng L, Liu Z, Li C, Li Z, Lou X, Shao L, Wang Y, Huang Y, Chen H, Pang X, Liu S, He F, Zheng J, Meng X, Xie P, Yang G, Ding Y, Wei M, Yun J, Hung MC, Zhou W, Wahl DR, Lan P, Tian J, Wan X. Development and validation of a radiopathomics model to predict pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer: a multicentre observational study. Lancet Digit Health. 2022;4:e8-e17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 169] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 120. | Bahrambanan F, Alizamir M, Moradveisi K, Heddam S, Kim S, Kim S, Soleimani M, Afshar S, Taherkhani A. The development of an efficient artificial intelligence-based classification approach for colorectal cancer response to radiochemotherapy: deep learning vs. machine learning. Sci Rep. 2025;15:62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 121. | Bao X, Li Q, Chen D, Dai X, Liu C, Tian W, Zhang H, Jin Y, Wang Y, Cheng J, Lai C, Ye C, Xin S, Li X, Su G, Ding Y, Xiong Y, Xie J, Tano V, Wang Y, Fu W, Deng S, Fang W, Sheng J, Ruan J, Zhao P. A multiomics analysis-assisted deep learning model identifies a macrophage-oriented module as a potential therapeutic target in colorectal cancer. Cell Rep Med. 2024;5:101399. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Reference Citation Analysis (0)] |
| 122. | Wesdorp NJ, Zeeuw JM, Postma SCJ, Roor J, van Waesberghe JHTM, van den Bergh JE, Nota IM, Moos S, Kemna R, Vadakkumpadan F, Ambrozic C, van Dieren S, van Amerongen MJ, Chapelle T, Engelbrecht MRW, Gerhards MF, Grunhagen D, van Gulik TM, Hermans JJ, de Jong KP, Klaase JM, Liem MSL, van Lienden KP, Molenaar IQ, Patijn GA, Rijken AM, Ruers TM, Verhoef C, de Wilt JHW, Marquering HA, Stoker J, Swijnenburg RJ, Punt CJA, Huiskens J, Kazemier G. Deep learning models for automatic tumor segmentation and total tumor volume assessment in patients with colorectal liver metastases. Eur Radiol Exp. 2023;7:75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 123. | Bertsimas D, Margonis GA, Sujichantararat S, Boerner T, Ma Y, Wang J, Kamphues C, Sasaki K, Tang S, Gagniere J, Dupré A, Løes IM, Wagner D, Stasinos G, Macher-Beer A, Burkhart R, Morioka D, Imai K, Ardiles V, O'Connor JM, Pawlik TM, Poultsides G, Seeliger H, Beyer K, Kaczirek K, Kornprat P, Aucejo FN, de Santibañes E, Baba H, Endo I, Lønning PE, Kreis ME, Weiss MJ, Wolfgang CL, D'Angelica M. Using Artificial Intelligence to Find the Optimal Margin Width in Hepatectomy for Colorectal Cancer Liver Metastases. JAMA Surg. 2022;157:e221819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 27] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 124. | Wang X, Zheng Z, Xie Z, Yu Q, Lu X, Zhao Z, Huang S, Huang Y, Chi P. Development and validation of artificial intelligence models for preoperative prediction of inferior mesenteric artery lymph nodes metastasis in left colon and rectal cancer. Eur J Surg Oncol. 2022;48:2475-2486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 125. | Susič D, Syed-Abdul S, Dovgan E, Jonnagaddala J, Gradišek A. Artificial intelligence based personalized predictive survival among colorectal cancer patients. Comput Methods Programs Biomed. 2023;231:107435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 10] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 126. | Hummel JJ, Liu D, Tallon E, Snyder J, Warren W, Shyu CR, Mitchem J, Cortese R. Identification of Genomic Signatures for Colorectal Cancer Survival Using Exploratory Data Mining. Int J Mol Sci. 2024;25:3220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 127. | Taylor RS, Taylor RJ, Bayliss S, Hagström H, Nasr P, Schattenberg JM, Ishigami M, Toyoda H, Wai-Sun Wong V, Peleg N, Shlomai A, Sebastiani G, Seko Y, Bhala N, Younossi ZM, Anstee QM, McPherson S, Newsome PN. Association Between Fibrosis Stage and Outcomes of Patients With Nonalcoholic Fatty Liver Disease: A Systematic Review and Meta-Analysis. Gastroenterology. 2020;158:1611-1625.e12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 355] [Cited by in RCA: 739] [Article Influence: 147.8] [Reference Citation Analysis (0)] |
| 128. | Kanwal F, Shubrook JH, Adams LA, Pfotenhauer K, Wai-Sun Wong V, Wright E, Abdelmalek MF, Harrison SA, Loomba R, Mantzoros CS, Bugianesi E, Eckel RH, Kaplan LM, El-Serag HB, Cusi K. Clinical Care Pathway for the Risk Stratification and Management of Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology. 2021;161:1657-1669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 232] [Cited by in RCA: 380] [Article Influence: 95.0] [Reference Citation Analysis (0)] |
| 129. | Vianna P, Calce SI, Boustros P, Larocque-Rigney C, Patry-Beaudoin L, Luo YH, Aslan E, Marinos J, Alamri TM, Vu KN, Murphy-Lavallée J, Billiard JS, Montagnon E, Li H, Kadoury S, Nguyen BN, Gauthier S, Therien B, Rish I, Belilovsky E, Wolf G, Chassé M, Cloutier G, Tang A. Comparison of Radiologists and Deep Learning for US Grading of Hepatic Steatosis. Radiology. 2023;309:e230659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 130. | Iyer JS, Juyal D, Le Q, Shanis Z, Pokkalla H, Pouryahya M, Pedawi A, Stanford-Moore SA, Biddle-Snead C, Carrasco-Zevallos O, Lin M, Egger R, Hoffman S, Elliott H, Leidal K, Myers RP, Chung C, Billin AN, Watkins TR, Patterson SD, Resnick M, Wack K, Glickman J, Burt AD, Loomba R, Sanyal AJ, Glass B, Montalto MC, Taylor-Weiner A, Wapinski I, Beck AH. AI-based automation of enrollment criteria and endpoint assessment in clinical trials in liver diseases. Nat Med. 2024;30:2914-2923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 13] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 131. | Naoumov NV, Brees D, Loeffler J, Chng E, Ren Y, Lopez P, Tai D, Lamle S, Sanyal AJ. Digital pathology with artificial intelligence analyses provides greater insights into treatment-induced fibrosis regression in NASH. J Hepatol. 2022;77:1399-1409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 71] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 132. | Qiu QT, Zhang J, Duan JH, Wu SZ, Ding JL, Yin Y. Development and validation of radiomics model built by incorporating machine learning for identifying liver fibrosis and early-stage cirrhosis. Chin Med J (Engl). 2020;133:2653-2659. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 133. | Wen H, Zheng W, Li M, Li Q, Liu Q, Zhou J, Liu Z, Chen X. Multiparametric Quantitative US Examination of Liver Fibrosis: A Feature-Engineering and Machine-Learning Based Analysis. IEEE J Biomed Health Inform. 2022;26:715-726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 134. | Sato M, Kobayashi T, Soroida Y, Tanaka T, Nakatsuka T, Nakagawa H, Nakamura A, Kurihara M, Endo M, Hikita H, Sato M, Gotoh H, Iwai T, Tateishi R, Koike K, Yatomi Y. Development of novel deep multimodal representation learning-based model for the differentiation of liver tumors on B-mode ultrasound images. J Gastroenterol Hepatol. 2022;37:678-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 135. | Zheng Z, Wang M, Fan C, Wang C, He X, He X. Light&fast generative adversarial network for high-fidelity CT image synthesis of liver tumor. Comput Methods Programs Biomed. 2024;254:108252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 136. | Said D, Carbonell G, Stocker D, Hectors S, Vietti-Violi N, Bane O, Chin X, Schwartz M, Tabrizian P, Lewis S, Greenspan H, Jégou S, Schiratti JB, Jehanno P, Taouli B. Semiautomated segmentation of hepatocellular carcinoma tumors with MRI using convolutional neural networks. Eur Radiol. 2023;33:6020-6032. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 137. | Nakai H, Sakamoto R, Kakigi T, Coeur C, Isoda H, Nakamoto Y. Artificial intelligence-powered software detected more than half of the liver metastases overlooked by radiologists on contrast-enhanced CT. Eur J Radiol. 2023;163:110823. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 138. | Chen C, Chen C, Ma M, Ma X, Lv X, Dong X, Yan Z, Zhu M, Chen J. Classification of multi-differentiated liver cancer pathological images based on deep learning attention mechanism. BMC Med Inform Decis Mak. 2022;22:176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 13] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 139. | Zeng Q, Klein C, Caruso S, Maille P, Laleh NG, Sommacale D, Laurent A, Amaddeo G, Gentien D, Rapinat A, Regnault H, Charpy C, Nguyen CT, Tournigand C, Brustia R, Pawlotsky JM, Kather JN, Maiuri MC, Loménie N, Calderaro J. Artificial intelligence predicts immune and inflammatory gene signatures directly from hepatocellular carcinoma histology. J Hepatol. 2022;77:116-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 61] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 140. | Yu Y, Cao L, Shen B, Du M, Gu W, Gu C, Fan Y, Shi C, Wu Q, Zhang T, Zhu M, Wang X, Hu C. Deep Learning Radiopathomics Models Based on Contrast-enhanced MRI and Pathologic Imaging for Predicting Vessels Encapsulating Tumor Clusters and Prognosis in Hepatocellular Carcinoma. Radiol Imaging Cancer. 2025;7:e240213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 141. | Famularo S, Penzo C, Maino C, Milana F, Oliva R, Marescaux J, Diana M, Romano F, Giuliante F, Ardito F, Grazi GL, Donadon M, Torzilli G. Preoperative detection of hepatocellular carcinoma's microvascular invasion on CT-scan by machine learning and radiomics: A preliminary analysis. Eur J Surg Oncol. 2025;51:108274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 142. | Li X, Qi Z, Du H, Geng Z, Li Z, Qin S, Zhang X, Liang J, Zhang X, Liang W, Yang W, Xie C, Quan X. Deep convolutional neural network for preoperative prediction of microvascular invasion and clinical outcomes in patients with HCCs. Eur Radiol. 2022;32:771-782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 143. | Chu T, Zhao C, Zhang J, Duan K, Li M, Zhang T, Lv S, Liu H, Wei F. Application of a Convolutional Neural Network for Multitask Learning to Simultaneously Predict Microvascular Invasion and Vessels that Encapsulate Tumor Clusters in Hepatocellular Carcinoma. Ann Surg Oncol. 2022;29:6774-6783. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 26] [Reference Citation Analysis (0)] |
| 144. | Lin H, Li G, Delamarre A, Ahn SH, Zhang X, Kim BK, Liang LY, Lee HW, Wong GL, Yuen PC, Chan HL, Chan SL, Wong VW, de Lédinghen V, Kim SU, Yip TC. A Liver Stiffness-Based Etiology-Independent Machine Learning Algorithm to Predict Hepatocellular Carcinoma. Clin Gastroenterol Hepatol. 2024;22:602-610.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 17] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 145. | Yan M, Zhang X, Zhang B, Geng Z, Xie C, Yang W, Zhang S, Qi Z, Lin T, Ke Q, Li X, Wang S, Quan X. Deep learning nomogram based on Gd-EOB-DTPA MRI for predicting early recurrence in hepatocellular carcinoma after hepatectomy. Eur Radiol. 2023;33:4949-4961. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 22] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 146. | Zhang J, Chen Q, Zhang Y, Zhou J. Construction of a random survival forest model based on a machine learning algorithm to predict early recurrence after hepatectomy for adult hepatocellular carcinoma. BMC Cancer. 2024;24:1575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 147. | Gao Y, Yang X, Li H, Ding DW. A knowledge-enhanced interpretable network for early recurrence prediction of hepatocellular carcinoma via multi-phase CT imaging. Int J Med Inform. 2024;189:105509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 148. | Li Z, Lan L, Zhou Y, Li R, Chavin KD, Xu H, Li L, Shih DJH, Jim Zheng W. Developing deep learning-based strategies to predict the risk of hepatocellular carcinoma among patients with nonalcoholic fatty liver disease from electronic health records. J Biomed Inform. 2024;152:104626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 149. | Huang S, Tu T. Integrating single cell analysis and machine learning methods reveals stem cell-related gene S100A10 as an important target for prediction of liver cancer diagnosis and immunotherapy. Front Immunol. 2024;15:1534723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 150. | Chen E, Zou Z, Wang R, Liu J, Peng Z, Gan Z, Lin Z, Liu J. Predictive value of a stemness-based classifier for prognosis and immunotherapy response of hepatocellular carcinoma based on bioinformatics and machine-learning strategies. Front Immunol. 2024;15:1244392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 151. | Liao M, Di S, Zhao Y, Liang W, Yang Z. FA-Net: A hierarchical feature fusion and interactive attention-based network for dose prediction in liver cancer patients. Artif Intell Med. 2024;156:102961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 152. | Altaf A, Mustafa A, Dar A, Nazer R, Riyaz S, Rana A, Bhatti ABH. Artificial intelligence-based model for the recurrence of hepatocellular carcinoma after liver transplantation. Surgery. 2024;176:1500-1506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 153. | Wu X, Zhang T, Zhang T, Park S. The impact of gut microbiome enterotypes on ulcerative colitis: identifying key bacterial species and revealing species co-occurrence networks using machine learning. Gut Microbes. 2024;16:2292254. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 18] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 154. | Barberio B, Facchin S, Patuzzi I, Ford AC, Massimi D, Valle G, Sattin E, Simionati B, Bertazzo E, Zingone F, Savarino EV. A specific microbiota signature is associated to various degrees of ulcerative colitis as assessed by a machine learning approach. Gut Microbes. 2022;14:2028366. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 45] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 155. | Gao S, Gao X, Zhu R, Wu D, Feng Z, Jiao N, Sun R, Gao W, He Q, Liu Z, Zhu L. Microbial genes outperform species and SNVs as diagnostic markers for Crohn's disease on multicohort fecal metagenomes empowered by artificial intelligence. Gut Microbes. 2023;15:2221428. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 20] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 156. | Jing Z, Zheng W, Jianwen S, Hong S, Xiaojian Y, Qiang W, Yunfeng Y, Xinyue W, Shuwen H, Feimin Z. Gut microbes on the risk of advanced adenomas. BMC Microbiol. 2024;24:264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 157. | Villani A, Fontana A, Panebianco C, Ferro C, Copetti M, Pavlovic R, Drago D, Fiorentini C, Terracciano F, Bazzocchi F, Canistro G, Pisati F, Maiello E, Latiano TP, Perri F, Pazienza V. A powerful machine learning approach to identify interactions of differentially abundant gut microbial subsets in patients with metastatic and non-metastatic pancreatic cancer. Gut Microbes. 2024;16:2375483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 158. | Bajaj JS, O'Leary JG, Jakab SS, Fagan A, Sikaroodi M, Gillevet PM. Gut microbiome profiles to exclude the diagnosis of hepatic encephalopathy in patients with cirrhosis. Gut Microbes. 2024;16:2392880. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 159. | Huang H, Ren Z, Gao X, Hu X, Zhou Y, Jiang J, Lu H, Yin S, Ji J, Zhou L, Zheng S. Integrated analysis of microbiome and host transcriptome reveals correlations between gut microbiota and clinical outcomes in HBV-related hepatocellular carcinoma. Genome Med. 2020;12:102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 119] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 160. | Jin D-M, Morton JT, Bonneau R. Meta-analysis of the human gut microbiome uncovers shared and distinct microbial signatures between diseases. mSystems. 2024;9:e0029524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 161. | GBD 2021 Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance 1990-2021: a systematic analysis with forecasts to 2050. Lancet. 2024;404:1199-1226. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 621] [Cited by in RCA: 591] [Article Influence: 591.0] [Reference Citation Analysis (0)] |
| 162. | Melchiorri D, Rocke T, Alm RA, Cameron AM, Gigante V. Addressing urgent priorities in antibiotic development: insights from WHO 2023 antibacterial clinical pipeline analyses. Lancet Microbe. 2025;6:100992. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 163. | Solomkin JS, Gardovskis J, Lawrence K, Montravers P, Sway A, Evans D, Tsai L. IGNITE4: Results of a Phase 3, Randomized, Multicenter, Prospective Trial of Eravacycline vs Meropenem in the Treatment of Complicated Intraabdominal Infections. Clin Infect Dis. 2019;69:921-929. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 136] [Article Influence: 27.2] [Reference Citation Analysis (1)] |
| 164. | Wagenlehner FME, Cloutier DJ, Komirenko AS, Cebrik DS, Krause KM, Keepers TR, Connolly LE, Miller LG, Friedland I, Dwyer JP; EPIC Study Group. Once-Daily Plazomicin for Complicated Urinary Tract Infections. N Engl J Med. 2019;380:729-740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 149] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 165. | Tommasi R, Brown DG, Walkup GK, Manchester JI, Miller AA. ESKAPEing the labyrinth of antibacterial discovery. Nat Rev Drug Discov. 2015;14:529-542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 372] [Cited by in RCA: 459] [Article Influence: 45.9] [Reference Citation Analysis (0)] |
| 166. | Scalia G, Rutherford ST, Lu Z, Buchholz KR, Skelton N, Chuang K, Diamant N, Hütter J, Luescher J, Miu A, Blaney J, Gendelev L, Skippington E, Zynda G, Dickson N, Koziarski M, Bengio Y, Regev A, Tan M, Biancalani T. A high-throughput phenotypic screen combined with an ultra-large-scale deep learning-based virtual screening reveals novel scaffolds of antibacterial compounds. 2024 Preprint. [DOI] [Full Text] |
| 167. | Santos-Júnior CD, Torres MDT, Duan Y, Rodríguez Del Río Á, Schmidt TSB, Chong H, Fullam A, Kuhn M, Zhu C, Houseman A, Somborski J, Vines A, Zhao XM, Bork P, Huerta-Cepas J, de la Fuente-Nunez C, Coelho LP. Discovery of antimicrobial peptides in the global microbiome with machine learning. Cell. 2024;187:3761-3778.e16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 99] [Article Influence: 99.0] [Reference Citation Analysis (0)] |
| 168. | Stokes JM, Yang K, Swanson K, Jin W, Cubillos-Ruiz A, Donghia NM, MacNair CR, French S, Carfrae LA, Bloom-Ackermann Z, Tran VM, Chiappino-Pepe A, Badran AH, Andrews IW, Chory EJ, Church GM, Brown ED, Jaakkola TS, Barzilay R, Collins JJ. A Deep Learning Approach to Antibiotic Discovery. Cell. 2020;180:688-702.e13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 529] [Cited by in RCA: 952] [Article Influence: 190.4] [Reference Citation Analysis (0)] |
| 169. | Wong F, Zheng EJ, Valeri JA, Donghia NM, Anahtar MN, Omori S, Li A, Cubillos-Ruiz A, Krishnan A, Jin W, Manson AL, Friedrichs J, Helbig R, Hajian B, Fiejtek DK, Wagner FF, Soutter HH, Earl AM, Stokes JM, Renner LD, Collins JJ. Discovery of a structural class of antibiotics with explainable deep learning. Nature. 2024;626:177-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 144] [Article Influence: 144.0] [Reference Citation Analysis (0)] |
| 170. | Maasch JRMA, Torres MDT, Melo MCR, de la Fuente-Nunez C. Molecular de-extinction of ancient antimicrobial peptides enabled by machine learning. Cell Host Microbe. 2023;31:1260-1274.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 76] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 171. | Liu G, Catacutan DB, Rathod K, Swanson K, Jin W, Mohammed JC, Chiappino-Pepe A, Syed SA, Fragis M, Rachwalski K, Magolan J, Surette MG, Coombes BK, Jaakkola T, Barzilay R, Collins JJ, Stokes JM. Deep learning-guided discovery of an antibiotic targeting Acinetobacter baumannii. Nat Chem Biol. 2023;19:1342-1350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 144] [Article Influence: 72.0] [Reference Citation Analysis (0)] |
| 172. | Dilaghi E, Lahner E, Annibale B, Esposito G. Systematic review and meta-analysis: Artificial intelligence for the diagnosis of gastric precancerous lesions and Helicobacter pylori infection. Dig Liver Dis. 2022;54:1630-1638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 22] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 173. | Gao P, Xiao Q, Tan H, Song J, Fu Y, Xu J, Zhao J, Miao Y, Li X, Jing Y, Feng Y, Wang Z, Zhang Y, Yao E, Xu T, Mei J, Chen H, Jiang X, Yang Y, Wang Z, Gao X, Zheng M, Zhang L, Jiang M, Long Y, He L, Sun J, Deng Y, Wang B, Zhao Y, Ba Y, Wang G, Zhang Y, Deng T, Shen D, Wang Z. Interpretable multi-modal artificial intelligence model for predicting gastric cancer response to neoadjuvant chemotherapy. Cell Rep Med. 2024;5:101848. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 174. | Uema R, Hayashi Y, Kizu T, Igura T, Ogiyama H, Yamada T, Takeda R, Nagai K, Inoue T, Yamamoto M, Yamaguchi S, Kanesaka T, Yoshihara T, Kato M, Yoshii S, Tsujii Y, Shinzaki S, Takehara T. A novel artificial intelligence-based endoscopic ultrasonography diagnostic system for diagnosing the invasion depth of early gastric cancer. J Gastroenterol. 2024;59:543-555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 175. | Guo B, Li X, Yang M, Jonnagaddala J, Zhang H, Xu XS. Predicting microsatellite instability and key biomarkers in colorectal cancer from H&E-stained images: achieving state-of-the-art predictive performance with fewer data using Swin Transformer. J Pathol Clin Res. 2023;9:223-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 176. | van der Zander QEW, Roumans R, Kusters CHJ, Dehghani N, Masclee AAM, de With PHN, van der Sommen F, Snijders CCP, Schoon EJ. Appropriate trust in artificial intelligence for the optical diagnosis of colorectal polyps: the role of human/artificial intelligence interaction. Gastrointest Endosc. 2024;100:1070-1078.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 177. | Allgaier J, Mulansky L, Draelos RL, Pryss R. How does the model make predictions? A systematic literature review on the explainability power of machine learning in healthcare. Artif Intell Med. 2023;143:102616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 30] [Reference Citation Analysis (0)] |
| 178. | Ghassemi N, Shoeibi A, Khodatars M, Heras J, Rahimi A, Zare A, Zhang YD, Pachori RB, Gorriz JM. Automatic diagnosis of COVID-19 from CT images using CycleGAN and transfer learning. Appl Soft Comput. 2023;144:110511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 179. | Lin QH, Niu YW, Sui J, Zhao WD, Zhuo C, Calhoun VD. SSPNet: An interpretable 3D-CNN for classification of schizophrenia using phase maps of resting-state complex-valued fMRI data. Med Image Anal. 2022;79:102430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 180. | Troya J, Sudarevic B, Krenzer A, Banck M, Brand M, Walter BM, Puppe F, Zoller WG, Meining A, Hann A. Direct comparison of multiple computer-aided polyp detection systems. Endoscopy. 2024;56:63-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 181. | Tang S, Yu X, Cheang CF, Liang Y, Zhao P, Yu HH, Choi IC. Transformer-based multi-task learning for classification and segmentation of gastrointestinal tract endoscopic images. Comput Biol Med. 2023;157:106723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 182. | Kim JE, Choi YH, Lee YC, Seong G, Song JH, Kim TJ, Kim ER, Hong SN, Chang DK, Kim YH, Shin SY. Deep learning model for distinguishing Mayo endoscopic subscore 0 and 1 in patients with ulcerative colitis. Sci Rep. 2023;13:11351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 183. | Tang J, Liang Y, Jiang Y, Liu J, Zhang R, Huang D, Pang C, Huang C, Luo D, Zhou X, Li R, Zhang K, Xie B, Hu L, Zhu F, Xia H, Lu L, Wang H. A multicenter study on two-stage transfer learning model for duct-dependent CHDs screening in fetal echocardiography. NPJ Digit Med. 2023;6:143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 184. | You C, Li G, Zhang Y, Zhang X, Shan H, Li M, Ju S, Zhao Z, Zhang Z, Cong W, Vannier MW, Saha PK, Hoffman EA, Wang G. CT Super-Resolution GAN Constrained by the Identical, Residual, and Cycle Learning Ensemble (GAN-CIRCLE). IEEE Trans Med Imaging. 2020;39:188-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 191] [Article Influence: 38.2] [Reference Citation Analysis (0)] |
| 185. | Chartsias A, Joyce T, Papanastasiou G, Semple S, Williams M, Newby DE, Dharmakumar R, Tsaftaris SA. Disentangled representation learning in cardiac image analysis. Med Image Anal. 2019;58:101535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 74] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 186. | Pati S, Baid U, Edwards B, Sheller M, Wang SH, Reina GA, Foley P, Gruzdev A, Karkada D, Davatzikos C, Sako C, Ghodasara S, Bilello M, Mohan S, Vollmuth P, Brugnara G, Preetha CJ, Sahm F, Maier-Hein K, Zenk M, Bendszus M, Wick W, Calabrese E, Rudie J, Villanueva-Meyer J, Cha S, Ingalhalikar M, Jadhav M, Pandey U, Saini J, Garrett J, Larson M, Jeraj R, Currie S, Frood R, Fatania K, Huang RY, Chang K, Balaña C, Capellades J, Puig J, Trenkler J, Pichler J, Necker G, Haunschmidt A, Meckel S, Shukla G, Liem S, Alexander GS, Lombardo J, Palmer JD, Flanders AE, Dicker AP, Sair HI, Jones CK, Venkataraman A, Jiang M, So TY, Chen C, Heng PA, Dou Q, Kozubek M, Lux F, Michálek J, Matula P, Keřkovský M, Kopřivová T, Dostál M, Vybíhal V, Vogelbaum MA, Mitchell JR, Farinhas J, Maldjian JA, Yogananda CGB, Pinho MC, Reddy D, Holcomb J, Wagner BC, Ellingson BM, Cloughesy TF, Raymond C, Oughourlian T, Hagiwara A, Wang C, To MS, Bhardwaj S, Chong C, Agzarian M, Falcão AX, Martins SB, Teixeira BCA, Sprenger F, Menotti D, Lucio DR, LaMontagne P, Marcus D, Wiestler B, Kofler F, Ezhov I, Metz M, Jain R, Lee M, Lui YW, McKinley R, Slotboom J, Radojewski P, Meier R, Wiest R, Murcia D, Fu E, Haas R, Thompson J, Ormond DR, Badve C, Sloan AE, Vadmal V, Waite K, Colen RR, Pei L, Ak M, Srinivasan A, Bapuraj JR, Rao A, Wang N, Yoshiaki O, Moritani T, Turk S, Lee J, Prabhudesai S, Morón F, Mandel J, Kamnitsas K, Glocker B, Dixon LVM, Williams M, Zampakis P, Panagiotopoulos V, Tsiganos P, Alexiou S, Haliassos I, Zacharaki EI, Moustakas K, Kalogeropoulou C, Kardamakis DM, Choi YS, Lee SK, Chang JH, Ahn SS, Luo B, Poisson L, Wen N, Tiwari P, Verma R, Bareja R, Yadav I, Chen J, Kumar N, Smits M, van der Voort SR, Alafandi A, Incekara F, Wijnenga MMJ, Kapsas G, Gahrmann R, Schouten JW, Dubbink HJ, Vincent AJPE, van den Bent MJ, French PJ, Klein S, Yuan Y, Sharma S, Tseng TC, Adabi S, Niclou SP, Keunen O, Hau AC, Vallières M, Fortin D, Lepage M, Landman B, Ramadass K, Xu K, Chotai S, Chambless LB, Mistry A, Thompson RC, Gusev Y, Bhuvaneshwar K, Sayah A, Bencheqroun C, Belouali A, Madhavan S, Booth TC, Chelliah A, Modat M, Shuaib H, Dragos C, Abayazeed A, Kolodziej K, Hill M, Abbassy A, Gamal S, Mekhaimar M, Qayati M, Reyes M, Park JE, Yun J, Kim HS, Mahajan A, Muzi M, Benson S, Beets-Tan RGH, Teuwen J, Herrera-Trujillo A, Trujillo M, Escobar W, Abello A, Bernal J, Gómez J, Choi J, Baek S, Kim Y, Ismael H, Allen B, Buatti JM, Kotrotsou A, Li H, Weiss T, Weller M, Bink A, Pouymayou B, Shaykh HF, Saltz J, Prasanna P, Shrestha S, Mani KM, Payne D, Kurc T, Pelaez E, Franco-Maldonado H, Loayza F, Quevedo S, Guevara P, Torche E, Mendoza C, Vera F, Ríos E, López E, Velastin SA, Ogbole G, Soneye M, Oyekunle D, Odafe-Oyibotha O, Osobu B, Shu'aibu M, Dorcas A, Dako F, Simpson AL, Hamghalam M, Peoples JJ, Hu R, Tran A, Cutler D, Moraes FY, Boss MA, Gimpel J, Veettil DK, Schmidt K, Bialecki B, Marella S, Price C, Cimino L, Apgar C, Shah P, Menze B, Barnholtz-Sloan JS, Martin J, Bakas S. Federated learning enables big data for rare cancer boundary detection. Nat Commun. 2022;13:7346. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 123] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 187. | Sarma KV, Harmon S, Sanford T, Roth HR, Xu Z, Tetreault J, Xu D, Flores MG, Raman AG, Kulkarni R, Wood BJ, Choyke PL, Priester AM, Marks LS, Raman SS, Enzmann D, Turkbey B, Speier W, Arnold CW. Federated learning improves site performance in multicenter deep learning without data sharing. J Am Med Inform Assoc. 2021;28:1259-1264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 85] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 188. | Sharma J, Kumar D, Verma R. Secure and Collaborative Breast Cancer Detection Using Federated Learning. 2024 2nd World Conference on Communication & Computing (WCONF); 2024 July 12-14; RAIPUR, India. IEEE, 2024. [DOI] [Full Text] |
| 189. | Alsalman H, Al-rakhami MS, Alfakih T, Hassan MM. Federated Learning Approach for Breast Cancer Detection Based on DCNN. IEEE Access. 2024;12:40114-40138. [DOI] [Full Text] |
| 190. | Varkalaite G, Forster M, Franke A, Kupcinskas J, Skieceviciene J. Liquid Biopsy in Gastric Cancer: Analysis of Somatic Cancer Tissue Mutations in Plasma Cell-Free DNA for Predicting Disease State and Patient Survival. Clin Transl Gastroenterol. 2021;12:e00403. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 191. | Dai SL, Pan JQ, Su ZR. Multi-omics features of immunogenic cell death in gastric cancer identified by combining single-cell sequencing analysis and machine learning. Sci Rep. 2024;14:21751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 192. | Maeda Y, Kudo SE, Ogata N, Misawa M, Iacucci M, Homma M, Nemoto T, Takishima K, Mochida K, Miyachi H, Baba T, Mori K, Ohtsuka K, Mori Y. Evaluation in real-time use of artificial intelligence during colonoscopy to predict relapse of ulcerative colitis: a prospective study. Gastrointest Endosc. 2022;95:747-756.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 45] [Article Influence: 15.0] [Reference Citation Analysis (0)] |