Copyright
©The Author(s) 2025.
World J Gastroenterol. Jun 28, 2025; 31(24): 108021
Published online Jun 28, 2025. doi: 10.3748/wjg.v31.i24.108021
Published online Jun 28, 2025. doi: 10.3748/wjg.v31.i24.108021
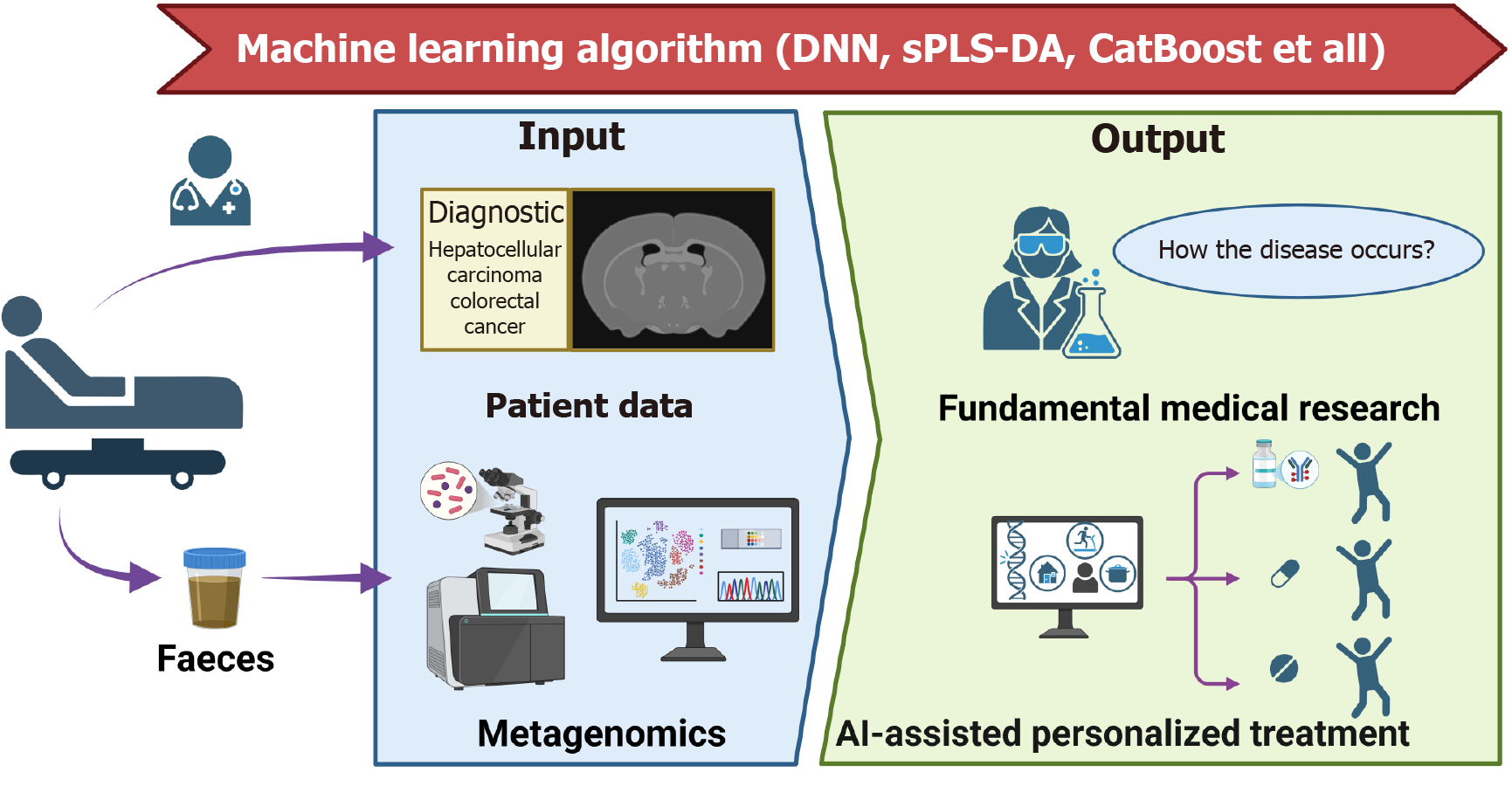
Figure 2 Schematic illustration of the deep integration between gut microbiome and machine learning for decoding cross-disease features to drive precision treatment.
In the “Input” section, patient data (e.g., diagnostic information on hepatocellular carcinoma and colorectal cancer) and metagenomics data (faeces samples) are utilized. Various machine learning algorithms, such as deep neural network, sparse partial least squares-discriminant analysis, and CatBoost-process these inputs. The “Output” involves two key dimensions: One is fundamental medical research exploring “How the Disease Occurs?” through gut microbiome pattern analysis; the other is artificial intelligence-assisted personalized treatment, which leverages the integration of microbiomics and artificial intelligence to formulate tailored therapeutic strategies. This synergy between metagenomics and artificial intelligence significantly promotes in-depth disease mechanism research and personalized patient treatment. sPLS-DA: Sparse partial least squares-discriminant analysis; AI: Artificial intelligence; DNN: Deep neural network.
- Citation: Chen ZL, Wang C, Wang F. Revolutionizing gastroenterology and hepatology with artificial intelligence: From precision diagnosis to equitable healthcare through interdisciplinary practice. World J Gastroenterol 2025; 31(24): 108021
- URL: https://www.wjgnet.com/1007-9327/full/v31/i24/108021.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i24.108021