Published online Jan 28, 2024. doi: 10.3748/wjg.v30.i4.332
Peer-review started: September 29, 2023
First decision: November 6, 2023
Revised: December 4, 2023
Accepted: January 9, 2024
Article in press: January 9, 2024
Published online: January 28, 2024
Processing time: 118 Days and 23.4 Hours
Nonalcoholic fatty liver disease (NAFLD) is one of the most common chronic liver diseases in children and adolescents. NAFLD ranges in severity from isolated hepatic steatosis to nonalcoholic steatohepatitis (NASH), wherein hepatocellular inflammation and/or fibrosis coexist with steatosis. Circulating microRNA (miRNA) levels have been suggested to be altered in NAFLD, but the extent to which miRNA are related to NAFLD features remains unknown. This analysis tested the hypothesis that plasma miRNAs are significantly associated with histological features of NAFLD in adolescents.
To investigate the relationship between plasma miRNA expression and NAFLD features among adolescents with NAFLD.
This study included 81 adolescents diagnosed with NAFLD and 54 adolescents without NAFLD from the Teen-Longitudinal Assessment of Bariatric Surgery study. Intra-operative core liver biopsies were collected from participants and used to characterize histological features of NAFLD. Plasma samples were collected during surgery for miRNA profiling. A total of 843 plasma miRNAs were profiled using the HTG EdgeSeq platform. We examined associations of plasma miRNAs and NAFLD features using logistic regression after adjusting for age, sex, race, and other key covariates. Ingenuity Pathways Analysis was used to identify biological functions of miRNAs that were associated with multiple histological features of NAFLD.
We identified 16 upregulated plasma miRNAs, including miR-193a-5p and miR-193b-5p, and 22 downregulated plasma miRNAs, including miR-1282 and miR-6734-5p, in adolescents with NAFLD. Moreover, 52, 16, 15, and 9 plasma miRNAs were associated with NASH, fibrosis, ballooning degeneration, and lobular inflammation, respectively. Collectively, 16 miRNAs were associated with two or more histological features of NAFLD. Among those miRNAs, miR-411-5p was downregulated in NASH, ballooning, and fibrosis, while miR-122-5p, miR-1343-5p, miR-193a-5p, miR-193b-5p, and miR-7845-5p were consistently and positively associated with all histological features of NAFLD. Pathway analysis revealed that most common pathways of miRNAs associated with multiple NAFLD features have been associated with tumor progression, while we also identified linkages between miR-122-5p and hepatitis C virus and between miR-199b-5p and chronic hepatitis B.
Plasma miRNAs were associated with NAFLD features in adolescent with severe obesity. Larger studies with more heterogeneous NAFLD phenotypes are needed to evaluate miRNAs as potential biomarkers of NAFLD.
Core Tip: Nonalcoholic fatty liver disease (NAFLD) is the most common chronic liver disease in the world, and its prevalence in adolescents is increasing. Studies suggest plasma microRNAs (miRNAs) are dysregulated in NAFLD, but relevant observational studies are scarce. In this study, we analyzed the expression of plasma miRNA in adolescents diagnosed with NAFLD by liver biopsy. We identified associations between histological features of NAFLD and plasma miRNA expression. Further, we found consistent expression of miRNA across different features of NAFLD. Although these results need further testing and validation, our findings suggest these miRNAs could be diagnostic and prognostic biomarkers of NAFLD.
- Citation: Li YJ, Baumert BO, Stratakis N, Goodrich JA, Wu HT, He JX, Zhao YQ, Aung MT, Wang HX, Eckel SP, Walker DI, Valvi D, La Merrill MA, Ryder JR, Inge TH, Jenkins T, Sisley S, Kohli R, Xanthakos SA, Baccarelli AA, McConnell R, Conti DV, Chatzi L. Circulating microRNA expression and nonalcoholic fatty liver disease in adolescents with severe obesity. World J Gastroenterol 2024; 30(4): 332-345
- URL: https://www.wjgnet.com/1007-9327/full/v30/i4/332.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i4.332
Nonalcoholic fatty liver disease (NAFLD) is defined by excessive fat accumulation in the liver and the presence of steatosis without heavy alcohol use[1,2]. NAFLD is comprised of two conditions: Nonalcoholic fatty liver (NAFL) and nonalcoholic steatohepatitis (NASH). Although both NAFL and NASH include accumulation of fat in hepatocytes, the histopathological abnormality in NASH involves further hepatocellular ballooning, fibrosis, and lobular inflammation[3-6]. In the United States, the overall estimated prevalence of NAFL and NASH are approximately 30% and 5%, res-pectively[7]. The prevalence of NAFLD in adolescents is 18.5%[8] and has more than doubled over the past 20 years to affect approximately one-half of adolescents with obesity[9,10]. Studies suggest that histopathological features of pediatric NAFLD are different from adult NAFLD[11,12], and children with NAFLD may experience increased risk of severe liver disease and higher liver-related mortality in adulthood[13].
Thus, early diagnosis and prevention of NAFLD among adolescents are crucial. Liver biopsy is the gold standard to diagnose NAFLD[14] yet is invasive and costly[15]. Alternately, noninvasive assessments for NAFLD such as blood tests of aspartate aminotransferase and alanine aminotransferase are commonly used[15,16], but are less predictive of more advanced NAFLD features such as NASH and fibrosis[17,18]. Nonetheless, more robust, noninvasive diagnostic biomarkers of the full spectrum of NAFLD disease severity, are needed.
MicroRNAs (miRNAs) are non-coding RNA that regulate gene expression[19]. In addition to intracellular activities, miRNA can be encapsulated in circulating extracellular vesicles that can convey biological information to recipient cells[20]. Hence, miRNAs play crucial roles in various aspects of metabolism and are frequently dysregulated in the context of diseases[21]. Evidence suggests that dysregulation of miRNA is associated with NAFLD pathogenesis[22-26], via multiple pathways, including lipid metabolism, insulin signaling, hepatocyte apoptosis, hepatic inflammation, and liver fibrosis[22,23]. To date, NAFLD–miRNA association studies in humans are scarce and have presented inconsistent results[25-39], and only two studies included adolescents[38,39]. Moreover, only one study measured associations between lobular inflammation and ballooning degeneration with miRNA expression[32], and it focused solely on the expression of miR-34a, miR-122, miR-191, miR-192, and miR-200a. Therefore, it is critical to further investigate the relationship between NAFLD and miRNA expression in adolescents.
The objectives of this study were to: (1) Examine the associations between circulating miRNA levels and histological characteristics of NAFLD in adolescents with obesity; and (2) investigate the pathways of identified NAFLD-related miRNA.
This study was based on data from the Teen-Longitudinal Assessment of Bariatric Surgery (Teen-LABS study, ClinicalTrials.gov NCT00465829), a prospective, multicenter, observational study of adolescents (≤ 19 years of age) with severe obesity who underwent bariatric surgery in 2007-2012 and enrolled at participating clinical centers in the United States: Cincinnati Children’s Hospital Medical Center (Cincinnati, Ohio), Nationwide Children’s Hospital (Columbus, Ohio), University of Pittsburgh Medical Center (Pittsburgh, Pennsylvania), Texas Children’s Hospital (Houston, Texas), and Children’s Hospital of Alabama (Birmingham, Alabama)[10,40-42]. The protocol, assent/consent forms, and monitoring plans for data and safety were approved by the institutional review boards of each institution, the independent data and safety monitoring board prior to study initiation, and the University of Southern California review board[10,40-42]. Detailed cohort information is described in previous studies[10,40-42].
Liver biopsies were obtained by a laparoscopically controlled, transabdominal core needle biopsy technique after induction of anesthesia and before performing the bariatric procedure[10,40-42]. Biopsies were evaluated by an experienced hepatopathologist using the NASH Clinical Research Network scoring system[43]. NAFLD features were categorized as definite NASH, borderline NASH, NAFLD not NASH (NAFL), and no NAFLD[10,44]. Other histological features of NAFLD were also categorized, including ballooning, lobular inflammation, and fibrosis[10].
Plasma samples were collected at baseline, typically within 30 days of bariatric surgery and stored at -70 ℃. Analyses were performed at HTG Molecular Diagnostics, Inc. (Tucson, AZ) for HTG EdgeSeq miRNA sequencing. HTG EdgeSeq uses EdgeSeq miRNA Whole Transcriptome Assay and Illumina HiSeq 4000 to quantify 2083 mature miRNA. For quality control, triplicate internal human brain tissue controls were sequenced. HTG EdgeSeq Parser software (version 5.0.535.3181) was used for alignment to a priori defined target sequences. The following quality control measures were implemented: (1) Percentage of bases with a quality score 30 ≥ 87%; (2) percentage of clusters passing filter ≥ 75%; (3) cluster density of 180-290 k/mm2; and (4) all samples passing HTG-defined criteria, including > 500000 reads, < 14% reads aligned to positive control probes, and > 0.08 relative standard deviation of reads allocated to each probe with each sample. To correct technical batch effects, we used the ComBat_seq function of the sva package in R[45]. Sequencing reads were normalized within and across plates using relative log expression[46]. The reliability of each probe was determined by calculating the coefficient of variation for each miRNA across human brain tissue control samples. Analysis included a total of 843 miRNAs with coefficient of variation ≤ 0.25 in replicate control samples. All final counts were converted to counts per million and log2-transformed prior to analysis.
Standardized methods for Teen-LABS data collection have been described previously[10,40-42]. We included participant characteristics as important confounders, including: Age[47-49], body mass index (BMI)[49-51], sex[49,52,53], weight loss prior to surgery[54,55], and covariates, such as race[56,57], parents’ income[58,59] and clinical site of surgery. Data were collected within 30 days of bariatric surgery at in-person visits with trained study personnel. Detailed descriptions of methods, comorbidities, data definitions, medical record data, and laboratory testing can be found in previous publications[10,40-42].
Due to the low frequency of borderline NASH (n = 22, 16.3%) and definite NASH (n = 8, 5.9%) in the study population, these two categories were combined and referred to as general NASH (n = 30, 22%). Similarly, we grouped the two ballooning degeneration conditions, which were prominent (n = 5, 3.7%) and less characteristics (n = 16, 11.9%), to create a general ballooning group (n = 21, 15.6%). These groupings ensured an adequate sample size for meaningful analysis and interpretation of results. To investigate associations between histological features of NAFLD and miRNA, we used multivariate logistic regression to investigate miRNA expression in participants with NAFLD (NAFL and NASH). Additional comparisons between each histological grouping, including NASH (NASH vs. NAFL), fibrosis, lobular inflammation, and ballooning, were performed using independent logistic regression models for each comparison. Coefficient estimates of miRNA expression change (log odds ratio), standard errors, and P value for each miRNA relationship were calculated. To account for multiple comparisons, we applied the false discovery rate (FDR) approach with a threshold of 0.05 to adjust P values from each regression analysis. All models were adjusted by covariates. All statistical analysis was conducted in RStudio version 1.0.143 (RStudio: Integrated Development for R. RStudio, Inc., Boston, MA, United States, http://www.rstudio.com/).
We investigated pathways of NAFLD-related miRNA in both miRbase and the Kyoto Encyclopedia for Genes and Genomes from Ingenuity Pathway Analysis (IPA) (Qiagen Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis)[60-62]. Given that an individual miRNA can participate in numerous pathways, we used the miRNA Target Filter tool from IPA and selected experimentally observed diseases and functions of NAFLD-related miRNA in humans.
The analysis consisted of 135 study participants with complete data. The mean age was 16.9 years (SD = 1.5), mean BMI was 53.8 kg/m2 (SD = 9.8), and 73.3% were female. Because more than half of study participants were recruited from a single clinical site, we re-categorized study site as a binary variable for subsequent regression models. Study population characteristics are summarized in Table 1.
| Characteristics | Mean (SD)/n (%) |
| Age (yr) | 16.86 (1.53) |
| BMI (kg/m2) | 53.80 (9.81) |
| Weight loss prior to surgery (kg) | 0.69 (8.36) |
| Sex | |
| Female | 99 (73.33) |
| Male | 36 (26.67) |
| Race (binary) | |
| White or Caucasian | 93 (68.89) |
| Others | 42 (31.11) |
| Parents’ income | |
| < $25000 | 53 (39.26) |
| $25000-$74999 | 57 (42.22) |
| ≥ $75000 | 25 (18.52) |
| NAFLD | |
| NAFL | 51 (37.78) |
| Borderline NASH | 22 (16.30) |
| Definite NASH | 8 (5.93) |
| No NAFLD | 54 (40.00) |
| Fibrosis | |
| Presence | 26 (19.26) |
| None | 109 (80.74) |
| Ballooning degeneration | |
| Many, prominent | 5 (3.70) |
| Less characteristics | 16 (11.86) |
| None | 114 (84.44) |
| Lobular inflammation | |
| Presence | 97 (71.85) |
| None | 38 (28.15) |
By histological analysis, 40% of participants did not have NAFLD, while 37.8% were diagnosed with NAFL, 16.3% with borderline NASH, and 5.9% with definite NASH at the time of surgery. Notably, a high proportion of participants from the Teen-LABS cohort exhibited progressive histopathological features associated with NAFLD-19.3% were diagnosed with fibrosis, and 71.9% were diagnosed with lobular inflammation. Furthermore, 3.7% of participants were diagnosed with prominent ballooning degeneration, while 11.9% exhibited ballooning with fewer characteristics. Distribution of NAFLD features is summarized in Table 1.
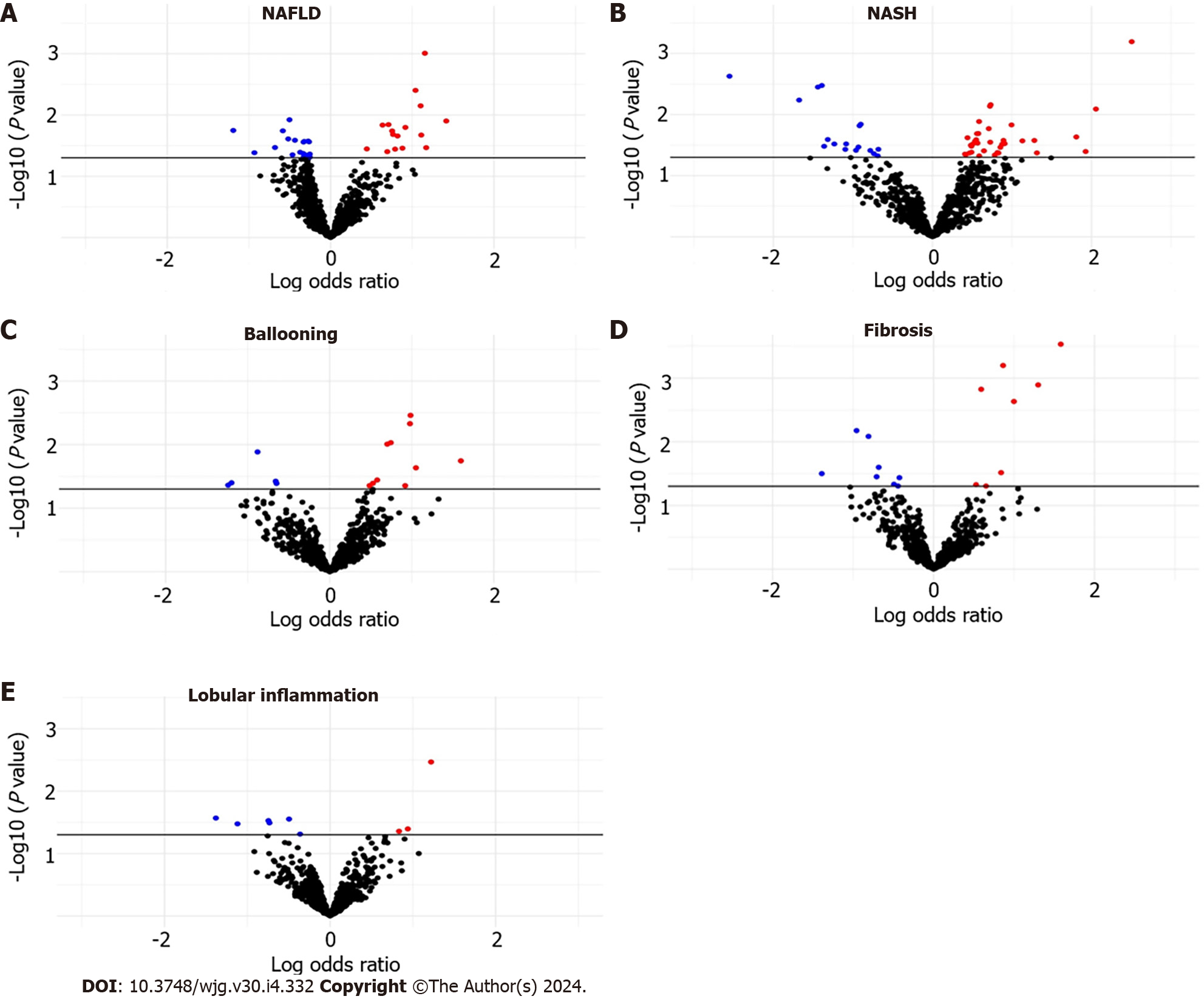
The distribution of NAFLD–miRNA associations is shown in Supplementary Table 1, Figure 1A. We found 38 associations between NAFLD and plasma miRNA expression levels. A subset of 16 miRNA displayed upregulation, while a subset of 22 miRNAs demonstrated downregulation. There was dysregulation of 17 downregulated miRNAs and 35 upregulated miRNAs in patients with NASH relative to those with NAFL (Supplementary Table 2, Figure 1B). However, these findings did not retain significance after applying multiple comparison adjustments (FDR > 0.05).
Within the group of participants with fibrosis (n = 26), we observed downregulation of 8 miRNAs and upregulation of 8 miRNAs compared to participants without fibrosis (n = 109). Additionally, in participants with ballooning (n = 21), we identified 15 altered miRNAs, including downregulation of miR-1224-5p, miR-369-5p, miR-411-5p, and miR-500b-5p. Among participants diagnosed with lobular inflammation (n = 97), we identified downregulation of 6 miRNAs and upregulation of miR-1244, miR-125b-2-3p, and miR-365b-5p compared to individuals without lobular inflammation. Associations among ballooning, fibrosis, and lobular inflammation with plasma miRNA levels are depicted in Supple
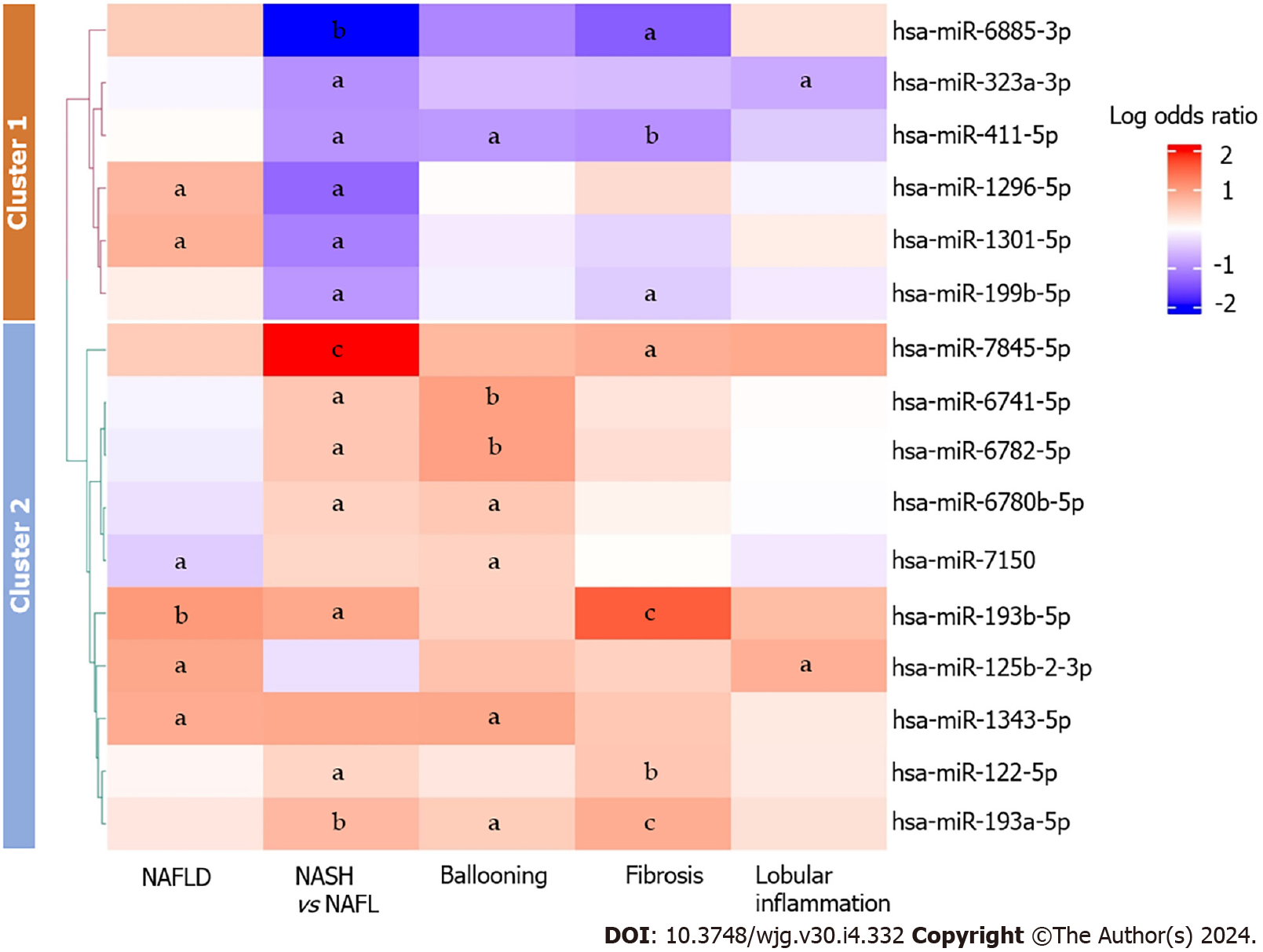
A total of 16 miRNAs exhibited differences in expression across two or more histological features of NAFLD (Figure 2). MiR-193a-5p was consistently upregulated in NASH, ballooning and fibrosis; miR-193b-5p was consistently upregulated in NAFLD, NASH, and fibrosis; expression of miR-411-5p was downregulated in NASH, ballooning, and fibrosis. Additionally, we observed inconsistent expression patterns of miR-1301-5p and miR-1296-5p between NAFLD and NASH-miR-1301-5p and miR-1296-5p were upregulated in NAFLD yet downregulated in NASH. Additionally, we observed downregulation of miR-7150 in NAFLD, and this miRNA was conversely upregulated in individuals with ballooning.
The 16 miRNAs associated with two or more histological features of NAFLD were subsequently subjected to scaling and grouped into two distinct clusters using the k-means clustering algorithm and the elbow method[63-66]. Cluster 1 comprised 6 miRNAs, the majority of which were upregulated in individuals with NAFLD. However, these miRNAs were mostly downregulated in patients with NASH, fibrosis, lobular inflammation, and ballooning. Cluster 2 encompassed 10 miRNAs, most of which were upregulated in NASH, fibrosis, and ballooning. In addition to overall inconsistency between the clusters, we noted consistent upregulation of miR-122-5p, miR-1343-5p, miR-193a-5p, miR-193b-5p, and miR-7845-5p across histological features of NAFLD. Figure 2 shows a graphical representation of associations between multiple histological features of NAFLD and miRNA expression.
We conducted pathway analysis on the 16 miRNAs associated with two or more histological features of NAFLD. Analysis revealed 16 experimentally confirmed pathways predominantly involving 6 overlapping NAFLD-related miRNAs in humans (Table 2). Specifically, miR-122-5p, miR-193b-5p, miR-199b-5p, and miR-323-3p were associated with apoptosis of tumor cell lines, while miR-122-5p, miR-193a-5p, and miR-199b-5p were associated with cell migration. Notably, miR-122-5p and miR-199b-5p were associated with multiple pathways in humans. For example, miR-122-5p was associated with production of hepatitis C virus, RNA decay, metastatic hepatocellular carcinoma, replication of viral replicon, and invasion of hepatoma cell lines. Similarly, miR-199b-5p was associated with congenital adrenal hyperplasia, chronic hepatitis B, early-stage invasive cervical squamous cell carcinoma, and proliferation of myeloma cell lines.
| Disease and functions | miRNA |
| Apoptosis of tumor cell lines | miR-122-5p, miR-193b-5p, miR-199b-5p, miR-323-3p |
| Migration of cells | miR-122-5p, miR-193a-5p, miR-199b-5p |
| Apoptosis of myeloma cell lines | miR-122-5p, miR-193a-5p |
| Dedifferentiated liposarcoma | miR-193a-5p, miR-199b-5p |
| Production of hepatitis C virus | miR-122-5p |
| Decay of RNA | miR-122-5p |
| Metastatic hepatocellular carcinoma | miR-122-5p |
| Replication of viral replicon | miR-122-5p |
| Invasion of hepatoma cell lines | miR-122-5p |
| Chemosensitivity of squamous cell carcinoma cell lines | miR-193a-5p |
| Epithelial-mesenchymal transition of adenocarcinoma cell lines | miR-193a-5p |
| Migration of adenocarcinoma cell lines | miR-193a-5p |
| Congenital adrenal hyperplasia | miR-199b-5p |
| Chronic hepatitis B | miR-199b-5p |
| Early-stage invasive cervical squamous cell carcinoma | miR-199b-5p |
| Proliferation of myeloma cell lines | miR-199b-5p |
Our study is the first to show associations between histological features of NAFLD and expression of plasma miRNA in adolescents with severe obesity. The IPA results revealed that miRNAs associated with multiple NAFLD features were linked to cancer, hepatitis B and hepatitis C. Our findings have several important implications. First, our findings were consistent with previous epidemiological studies[26]. Additionally, we identified novel NAFLD-miRNA associations. Moreover, our findings revealed consistent patterns of miRNA expression across various histological features of NAFLD, diagnosed using gold standard methods. The consistency of miRNA expression trends across different NAFLD features strengthen their potential utility as valuable diagnostic and prognostic markers for NAFLD.
Among the 38 NAFLD-related miRNAs identified in our study, several associations are comparable to published epidemiological studies. Our findings demonstrating positive associations between NAFLD and expression of miR-193a-5p, and miR-7150 align with current epidemiological studies[26]. However, it is noteworthy that Soronen et al[67] reported increased expression of hepatic miR-584-5p in those with NAFLD, while we found a negative association between NAFLD and plasma miR-584-5p levels. However, this differential expression may be attributed to variation in miRNA measurement sites, as contrasting expression patterns between hepatic miR-122[30] and serum miR-122[36] have been reported in those with NASH.
Additionally, NASH-related miRNA findings from our study align with current research findings. For example, we identified increased expression of miR-2861, miR-3940-5p, miR-6727-5p, miR-6771-5p, miR-6780b-5p, miR-6845-5p, and miR-7114-3p in participants with NASH, consistent with reported positive associations between these miRNAs in the setting of NAFLD and/or NASH[26]. However, it is important to note that this previous study also demonstrated downregulation of miR-6741-5p, miR-6782-5p, and miR-7108-5p in individuals with NAFLD and/or NASH[26], which contrasts with our observation of positive associations between these miRNA and NASH. We also observed a negative association between NASH and miR-182-5p, while mouse studies suggest this miRNA attenuates NASH[68]. In addition, Katsura et al[69] reported downregulation of miR-301b in mice with NASH, which is consistent with our findings.
We identified more miRNAs specifically associated with ballooning degeneration, fibrosis, and lobular inflammation, respectively. Among these miRNAs, we identified increased levels of plasma miR-34a-5p in participants with liver fibrosis, while epidemiological studies also show upregulation of miR-34a in NAFLD and NASH patients[29].
We identified several miRNAs associated with two or more NAFLD features. Interestingly, plasma levels of miR-122-5p, miR-1343-5p, miR-193a-5p, miR-193b-5p, and miR-7845-5p exhibited consistent increases across all histological features of NAFLD. Similarly, previous studies reported that miR-122-5p and miR-193a-5p were upregulated in individuals with NAFLD[26,33]. Moreover, Johnson et al[26] found strong associations between increased miR-193a-5p levels and NAFLD activity grade and fibrosis stage. Furthermore, Pirola et al[36] reported increased expression of serum miR-125b in individuals with NAFLD. Similarly, we found upregulated plasma miR-125b-2-3p expression with NAFLD and lobular inflammation.
Additionally, our analysis revealed decreased levels of miR-1296-5p, miR-1301-5p, miR-199b-5p, miR-411-5p, and miR-6885-3p in NASH compared to NAFL, while these levels were elevated in NAFLD compared to individuals without NAFLD. These miRNAs also demonstrated predominantly negative associations with fibrosis, lobular inflammation, and ballooning (Figure 2). Notably, the downregulation of miR-411-5p aligned with a recent study by Wan et al[70], which reported decreased expression of serum miR-411-5p in persons with NASH. Collectively, the distinct expression patterns observed across various NAFLD features suggest that these miRNAs may serve as potential biomarkers for NAFLD progression.
An increasing body of research have investigated the associations between NAFLD and miRNA expression, yet little is known about mechanisms underlying the dysregulation of circulating miRNA in NAFLD patients. We first conducted pathway analysis by IPA, and the results revealed that most overlapping miRNA were associated with tumorigenesis. Analysis also highlighted linkage between miR-122-5p and production of hepatitis C virus and between miR-199b-5p and chronic hepatitis B. Given the high prevalence of NAFLD in those with hepatitis C virus and the reported associations among miR-122, hepatitis C virus, and NAFLD[34], the relationship among miR-122-5p, hepatitis C virus, and NAFLD warrants further attention[71]. Furthermore, meta-analyses suggest an inverse association between hepatitis B virus infection and risk of developing NAFLD[72], offering potential insights into the mechanisms involving miR-199b-5p in NAFLD in the context of hepatitis B virus infection.
Experimental studies provide valuable insights into the molecular mechanisms underlying associations between NAFLD and miRNA expression while minimizing confounding variables intrinsic to human observational studies. Particularly, miR-122, a highly expressed hepatic miRNA in hepatocytes, is associated with NAFLD progression by regulating lipid metabolism[23]. For example, Long et al[73] revealed that miR-122 inhibited liver kinase B1/AMP-activated protein kinase signaling pathway, which further induced hepatic lipogenesis and steatosis in NAFLD. Additionally, the inhibition of miR-122-5p may suppress the inflammation and oxidative stress damage in NAFLD[74]. Given the observed upregulation of circulating miR-122 and downregulation of hepatic miR-122 in NASH patients[30,36], the elevated circulating miR-122-5p across NAFLD features in our study might be released by hepatocytes. Furthermore, we identified downregulation of plasma miR-146a-5p, miR-181a-5p, and miR-22-3p in individuals with NAFLD, which is supported by experimental studies of miRNA. For example, miR-146a targeted complex subunit 1 to attenuate lipid accumulation and alleviate NAFLD progression in mice[75]. Additionally, miR-181a was found to downregulate peroxisome proliferator-activated receptor-α and mediate lipid metabolisms in NAFLD in human liver cells[76]. Moreover, miR-22 is a pivotal regulator of lipid and glucose metabolism, playing a crucial role in mitigating NAFLD progression in mice[77]. For example, miR-22 inhibited sirtuin-1 and regulated gluconeogenesis in NAFLD[78]. We also observed increased expression of miR-125b-2-3p in both NAFLD and lobular inflammation, while studies indicated that miR-125b promoted the nuclear factor kappa-light-chain-enhancer of activated B cells-mediated inflammatory response in NAFLD[79]. Furthermore, we observed positive associations between liver fibrosis and expression of miR-34a-5p and miR-375, while experimental research suggested that both miR-34a-5p and miR-375 could alleviate liver fibrosis[80,81]. Together these experimental data support a plausible biological mechanism of NAFLD-miRNA association (Table 3).
| miRNA | Target | Function |
| miR-122 | SIRT-1[73]; FOXO3[74] | miR-122 downregulates SIRT-1 and induces steatosis and hepatic lipogenesis in NAFLD[73]; miR-122-5p inhibits FOXO3 to attenuate inflammatory response and oxidative stress damage in NAFLD[74] |
| miR-125b | TNFAIP3[79] | miR-125b targets TNFAIP3 and promotes the NF-κB-mediated inflammatory response in NAFLD[79] |
| miR-146a | MED1[75] | miR-146a targets MED1 and improves hepatic lipid and glucose metabolism in NAFLD[75] |
| miR-181a | PPARα[76] | miR-181a inhibits PPARα and aggravates lipid accumulation in hepatocytes[76] |
| miR-22 | SIRT-1[78] | miR-22 targets SIRT-1 and inhibits gluconeogenesis[78] |
| miR-34a | TGF-β1/Smad3[80] | miR-34a-5p targets TGF-β1/Smad3 and inhibits liver fibrosis in hepatic stellate cells[80] |
| miR-375 | RAC1[81] | miR-375 inhibits RAC1 and alleviates liver fibrosis[81] |
Our study has several strengths. Liver biopsies are considered the gold standard in NAFLD assessment, thus ensuring robust and accurate diagnoses of our study. Besides, the consistency of our NAFLD–miRNA associations with epidemiological studies further reinforced the importance of circulating miRNA in NAFLD, supporting their potential use as diagnostic markers. Additionally, our study revealed NAFLD-miRNA associations that have only been previously recognized in experimental research, enhancing the translational value of our findings. By bridging the gap between experimental research and clinical observations, our study helps unravel the complexities of NAFLD and its potential management strategies. Furthermore, our study provides comprehensive characterization of more severe NAFLD features through liver biopsies. Previous studies only profiled specific miRNAs, namely miR-34a, miR-122, miR-191, miR-192, and miR-200a, in patients with ballooning and lobular inflammation[32], while our study conducted an analysis of 843 miRNAs across various histological features of NAFLD. By integrating miRNA expression across these histological features of NAFLD, we uncovered consistent expression patterns of plasma miRNA that hold promise as potential NAFLD biomarkers. Conversely, miRNAs that exhibit differential expression across histological features also warrant further investigation to understand their specific roles and mechanisms in NAFLD pathogenesis.
However, this study was not without limitations. The miRNAs were profiled at baseline, limiting the ability to establish a straightforward causal relationship between NAFLD and plasma miRNA expression. Investigations incorporating longitudinal cohort study designs could better elucidate the temporal relationship and causal associations between NAFLD and plasma miRNA expression. Given our specific focus on adolescents with obesity, who are at high risk of developing NAFLD[10,50,51], and the limitations arising from our small sample size, a validation for generalizability is imperative. The limitation in sample size is an inherent consequence of our methodological choice to employ liver biopsy for NAFLD diagnosis. While liver biopsy ensures accurate and definitive diagnosis, its invasiveness and cost present challenges in expanding the participant pool[82]. Studies with larger and more diverse populations would facilitate more robust and conclusive findings regarding NAFLD–miRNA associations.
Our study provides valuable insights into differential miRNA expression in adolescents with NAFLD. In addition to the previously reported miR-122-5p, miR-193a-5p, and miR-34a, our findings reveal the presence of novel NAFLD-associated miRNAs, namely miR-125b-2-3p and miR-193b-5p. Furthermore, our research underscores similar expression trend of specific miRNAs, such as miR-122-5p, miR-1343-5p, miR-193a-5p, miR-193b-5p, and miR-7845-5p, across all histological features of NAFLD, highlighting their potential roles in pathogenesis and promise as diagnostic and prognostic biomarkers for NAFLD. Plasma miRNAs hold potential to distinguish different stages and phenotypes of NAFLD, allowing for more precise clinical disease classification and targeted management strategies.
Nonalcoholic fatty liver disease (NAFLD) is the most prevalent chronic liver diseases in the world, impacting approximately 25% of the population. The gold standard for NAFLD diagnosis is liver biopsy, yet it is invasive and expensive. Therefore, it is essential to provide alternative methods for NAFLD diagnosis. Recent studies propose that plasma microRNAs (miRNAs) are potential biomarkers for NAFLD, though research in this area remains limited.
This study is motivated by the current gaps of concerning associations between plasma miRNAs and NAFLD. This study aims to identify potential biomarkers for NAFLD diagnosis and NAFLD progression.
The objective of this study is to investigate associations between histological features of NAFLD and plasma miRNAs in adolescents with severe obesity.
A total of 135 participants from the Teen Longitudinal Assessment of Bariatric Surgery (Teen-LABS) study were included in this study. Within Teen-LABS, the histological features of NAFLD, including NAFLD, nonalcoholic steatohepatitis (NASH), ballooning degeneration, fibrosis, and lobular inflammation, are characterized based on liver biopsy. Multivariate logistic regression was employed to investigates associations between NAFLD features and 843 plasma miRNAs. Pathway analysis was performed for identified NAFLD-associated miRNA by Ingenuity Pathway Analysis (IPA).
In the present study, we identified 38, 52, 16, 15, and 9 plasma miRNAs associated with NAFLD, NASH, fibrosis, ballooning, and lobular inflammation, respectively. Among these miRNA, miR-122-5p, miR-1343-5p, miR-193a-5p, miR-193b-5p, and miR-7845-5p were consistently upregulated across NAFLD features. In contrast, miR-1296-5p, miR-1301-5p, miR-199b-5p, miR-411-5p, and miR-6885-3p were positively associated with NAFLD, yet displayed predominant decreasing in NASH, fibrosis, ballooning, and lobular inflammation. IPA results suggested that most of NAFLD-associated miRNAs were related to cancer.
Positive and consistent associations were observed between miR-122-5p, miR-1343-5p, miR-193a-5p, miR-193b-5p, and miR-7845-5p and NAFLD features, indicating their potential as biomarkers for NAFLD diagnosis. Additionally, miR-1296-5p, miR-1301-5p, miR-199b-5p, miR-411-5p, and miR-6885-3p showed different patterns of expression in response to NAFLD severity, indicating they had potential for characterizing NAFLD progression.
Conducting studies with larger and more diverse populations would contribute to more conclusive findings regarding NAFLD–miRNA associations. Furthermore, experimental research is imperative to understand the underlying molecular mechanisms of NAFLD-miRNA associations.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gao YT, China; Niu CY, China; Zhou B, China S-Editor: Qu XL L-Editor: A P-Editor: Yu HG
| 1. | Abd El-Kader SM, El-Den Ashmawy EM. Non-alcoholic fatty liver disease: The diagnosis and management. World J Hepatol. 2015;7:846-858. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 233] [Cited by in RCA: 267] [Article Influence: 26.7] [Reference Citation Analysis (5)] |
| 2. | Anstee QM, Targher G, Day CP. Progression of NAFLD to diabetes mellitus, cardiovascular disease or cirrhosis. Nat Rev Gastroenterol Hepatol. 2013;10:330-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1066] [Cited by in RCA: 1317] [Article Influence: 109.8] [Reference Citation Analysis (0)] |
| 3. | Machado MV, Diehl AM. Pathogenesis of Nonalcoholic Steatohepatitis. Gastroenterology. 2016;150:1769-1777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 263] [Cited by in RCA: 392] [Article Influence: 43.6] [Reference Citation Analysis (0)] |
| 4. | Takahashi Y, Fukusato T. Histopathology of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. World J Gastroenterol. 2014;20:15539-15548. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 255] [Cited by in RCA: 321] [Article Influence: 29.2] [Reference Citation Analysis (3)] |
| 5. | Pais R, Pascale A, Fedchuck L, Charlotte F, Poynard T, Ratziu V. Progression from isolated steatosis to steatohepatitis and fibrosis in nonalcoholic fatty liver disease. Clin Res Hepatol Gastroenterol. 2011;35:23-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 46] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 6. | Ong JP, Younossi ZM. Epidemiology and natural history of NAFLD and NASH. Clin Liver Dis. 2007;11:1-16, vii. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 339] [Cited by in RCA: 323] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 7. | Cotter TG, Rinella M. Nonalcoholic Fatty Liver Disease 2020: The State of the Disease. Gastroenterology. 2020;158:1851-1864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 411] [Cited by in RCA: 838] [Article Influence: 167.6] [Reference Citation Analysis (2)] |
| 8. | Arshad T, Paik JM, Biswas R, Alqahtani SA, Henry L, Younossi ZM. Nonalcoholic Fatty Liver Disease Prevalence Trends Among Adolescents and Young Adults in the United States, 2007-2016. Hepatol Commun. 2021;5:1676-1688. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 54] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 9. | Welsh JA, Karpen S, Vos MB. Increasing prevalence of nonalcoholic fatty liver disease among United States adolescents, 1988-1994 to 2007-2010. J Pediatr. 2013;162:496-500.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 337] [Cited by in RCA: 376] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 10. | Xanthakos SA, Jenkins TM, Kleiner DE, Boyce TW, Mourya R, Karns R, Brandt ML, Harmon CM, Helmrath MA, Michalsky MP, Courcoulas AP, Zeller MH, Inge TH; Teen-LABS Consortium. High Prevalence of Nonalcoholic Fatty Liver Disease in Adolescents Undergoing Bariatric Surgery. Gastroenterology. 2015;149:623-34.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 106] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 11. | Takahashi Y, Inui A, Fujisawa T, Takikawa H, Fukusato T. Histopathological characteristics of non-alcoholic fatty liver disease in children: Comparison with adult cases. Hepatol Res. 2011;41:1066-1074. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 29] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 12. | Nobili V, Alisi A, Newton KP, Schwimmer JB. Comparison of the Phenotype and Approach to Pediatric vs Adult Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology. 2016;150:1798-1810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 125] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 13. | Feldstein AE, Charatcharoenwitthaya P, Treeprasertsuk S, Benson JT, Enders FB, Angulo P. The natural history of non-alcoholic fatty liver disease in children: a follow-up study for up to 20 years. Gut. 2009;58:1538-1544. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 572] [Cited by in RCA: 554] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
| 14. | Perumpail BJ, Khan MA, Yoo ER, Cholankeril G, Kim D, Ahmed A. Clinical epidemiology and disease burden of nonalcoholic fatty liver disease. World J Gastroenterol. 2017;23:8263-8276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 543] [Cited by in RCA: 514] [Article Influence: 64.3] [Reference Citation Analysis (6)] |
| 15. | Vilar-Gomez E, Chalasani N. Non-invasive assessment of non-alcoholic fatty liver disease: Clinical prediction rules and blood-based biomarkers. J Hepatol. 2018;68:305-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 331] [Cited by in RCA: 446] [Article Influence: 63.7] [Reference Citation Analysis (0)] |
| 16. | Ferraioli G, Wong VW, Castera L, Berzigotti A, Sporea I, Dietrich CF, Choi BI, Wilson SR, Kudo M, Barr RG. Liver Ultrasound Elastography: An Update to the World Federation for Ultrasound in Medicine and Biology Guidelines and Recommendations. Ultrasound Med Biol. 2018;44:2419-2440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 417] [Cited by in RCA: 370] [Article Influence: 52.9] [Reference Citation Analysis (0)] |
| 17. | Amernia B, Moosavy SH, Banookh F, Zoghi G. FIB-4, APRI, and AST/ALT ratio compared to FibroScan for the assessment of hepatic fibrosis in patients with non-alcoholic fatty liver disease in Bandar Abbas, Iran. BMC Gastroenterol. 2021;21:453. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 62] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 18. | Nallagangula KS, Nagaraj SK, Venkataswamy L, Chandrappa M. Liver fibrosis: a compilation on the biomarkers status and their significance during disease progression. Future Sci OA. 2018;4:FSO250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 101] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 19. | Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3368] [Cited by in RCA: 4113] [Article Influence: 373.9] [Reference Citation Analysis (1)] |
| 20. | Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110:483-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 722] [Cited by in RCA: 789] [Article Influence: 60.7] [Reference Citation Analysis (0)] |
| 21. | Cui M, Wang H, Yao X, Zhang D, Xie Y, Cui R, Zhang X. Circulating MicroRNAs in Cancer: Potential and Challenge. Front Genet. 2019;10:626. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 181] [Cited by in RCA: 318] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 22. | Gjorgjieva M, Sobolewski C, Dolicka D, Correia de Sousa M, Foti M. miRNAs and NAFLD: from pathophysiology to therapy. Gut. 2019;68:2065-2079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 175] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 23. | Fang Z, Dou G, Wang L. MicroRNAs in the Pathogenesis of Nonalcoholic Fatty Liver Disease. Int J Biol Sci. 2021;17:1851-1863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 56] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 24. | Pek SL, Tavintharan S, Woon K, Lin L, Ong CN, Lim SC, Sum CF. MicroRNAs as biomarkers of hepatotoxicity in a randomized placebo-controlled study of simvastatin and ubiquinol supplementation. Exp Biol Med (Maywood). 2016;241:317-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 25. | Auguet T, Aragonès G, Berlanga A, Guiu-Jurado E, Martí A, Martínez S, Sabench F, Hernández M, Aguilar C, Sirvent JJ, Del Castillo D, Richart C. miR33a/miR33b* and miR122 as Possible Contributors to Hepatic Lipid Metabolism in Obese Women with Nonalcoholic Fatty Liver Disease. Int J Mol Sci. 2016;17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 26. | Johnson K, Leary PJ, Govaere O, Barter MJ, Charlton SH, Cockell SJ, Tiniakos D, Zatorska M, Bedossa P, Brosnan MJ, Cobbold JF, Ekstedt M, Aithal GP, Clément K, Schattenberg JM, Boursier J, Ratziu V, Bugianesi E, Anstee QM, Daly AK; LITMUS Consortium Investigators§; LITMUS Consortium Investigators. Increased serum miR-193a-5p during non-alcoholic fatty liver disease progression: Diagnostic and mechanistic relevance. JHEP Rep. 2022;4:100409. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 29] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 27. | Becker PP, Rau M, Schmitt J, Malsch C, Hammer C, Bantel H, Müllhaupt B, Geier A. Performance of Serum microRNAs -122, -192 and -21 as Biomarkers in Patients with Non-Alcoholic Steatohepatitis. PLoS One. 2015;10:e0142661. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 120] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 28. | Chai C, Rivkin M, Berkovits L, Simerzin A, Zorde-Khvalevsky E, Rosenberg N, Klein S, Yaish D, Durst R, Shpitzen S, Udi S, Tam J, Heeren J, Worthmann A, Schramm C, Kluwe J, Ravid R, Hornstein E, Giladi H, Galun E. Metabolic Circuit Involving Free Fatty Acids, microRNA 122, and Triglyceride Synthesis in Liver and Muscle Tissues. Gastroenterology. 2017;153:1404-1415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 81] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 29. | Liu XL, Pan Q, Zhang RN, Shen F, Yan SY, Sun C, Xu ZJ, Chen YW, Fan JG. Disease-specific miR-34a as diagnostic marker of non-alcoholic steatohepatitis in a Chinese population. World J Gastroenterol. 2016;22:9844-9852. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 81] [Cited by in RCA: 80] [Article Influence: 8.9] [Reference Citation Analysis (2)] |
| 30. | Cheung O, Puri P, Eicken C, Contos MJ, Mirshahi F, Maher JW, Kellum JM, Min H, Luketic VA, Sanyal AJ. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. 2008;48:1810-1820. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 583] [Cited by in RCA: 551] [Article Influence: 32.4] [Reference Citation Analysis (0)] |
| 31. | Kim TH, Lee Y, Lee YS, Gim JA, Ko E, Yim SY, Jung YK, Kang S, Kim MY, Kim H, Kim BH, Kim JH, Seo YS, Yim HJ, Yeon JE, Um SH, Byun KS. Circulating miRNA is a useful diagnostic biomarker for nonalcoholic steatohepatitis in nonalcoholic fatty liver disease. Sci Rep. 2021;11:14639. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 43] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 32. | Ezaz G, Trivedi HD, Connelly MA, Filozof C, Howard K, L Parrish M, Kim M, Herman MA, Nasser I, Afdhal NH, Jiang ZG, Lai M. Differential Associations of Circulating MicroRNAs With Pathogenic Factors in NAFLD. Hepatol Commun. 2020;4:670-680. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 33. | Tan Y, Ge G, Pan T, Wen D, Gan J. A pilot study of serum microRNAs panel as potential biomarkers for diagnosis of nonalcoholic fatty liver disease. PLoS One. 2014;9:e105192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 141] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 34. | Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One. 2011;6:e23937. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 423] [Cited by in RCA: 466] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 35. | Celikbilek M, Baskol M, Taheri S, Deniz K, Dogan S, Zararsiz G, Gursoy S, Guven K, Ozbakır O, Dundar M, Yucesoy M. Circulating microRNAs in patients with non-alcoholic fatty liver disease. World J Hepatol. 2014;6:613-620. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 67] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 36. | Pirola CJ, Fernández Gianotti T, Castaño GO, Mallardi P, San Martino J, Mora Gonzalez Lopez Ledesma M, Flichman D, Mirshahi F, Sanyal AJ, Sookoian S. Circulating microRNA signature in non-alcoholic fatty liver disease: from serum non-coding RNAs to liver histology and disease pathogenesis. Gut. 2015;64:800-812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 373] [Cited by in RCA: 444] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 37. | Yamada H, Suzuki K, Ichino N, Ando Y, Sawada A, Osakabe K, Sugimoto K, Ohashi K, Teradaira R, Inoue T, Hamajima N, Hashimoto S. Associations between circulating microRNAs (miR-21, miR-34a, miR-122 and miR-451) and non-alcoholic fatty liver. Clin Chim Acta. 2013;424:99-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 257] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 38. | Lin H, Mercer KE, Ou X, Mansfield K, Buchmann R, Børsheim E, Tas E. Circulating microRNAs Are Associated With Metabolic Markers in Adolescents With Hepatosteatosis. Front Endocrinol (Lausanne). 2022;13:856973. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 39. | Zhou X, Huang K, Jia J, Ni Y, Yuan J, Liang X, Lin H, Peng W, Wu W, Dong G, Fu J. Exosomal miRNAs Profile in Children's Nonalcoholic Fatty Liver Disease and the Correlation with Transaminase and Uric Acid. Ann Nutr Metab. 2020;76:44-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 40. | Inge TH, Zeller M, Harmon C, Helmrath M, Bean J, Modi A, Horlick M, Kalra M, Xanthakos S, Miller R, Akers R, Courcoulas A. Teen-Longitudinal Assessment of Bariatric Surgery: methodological features of the first prospective multicenter study of adolescent bariatric surgery. J Pediatr Surg. 2007;42:1969-1971. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 98] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 41. | Inge TH, Zeller MH, Jenkins TM, Helmrath M, Brandt ML, Michalsky MP, Harmon CM, Courcoulas A, Horlick M, Xanthakos SA, Dolan L, Mitsnefes M, Barnett SJ, Buncher R; Teen-LABS Consortium. Perioperative outcomes of adolescents undergoing bariatric surgery: the Teen-Longitudinal Assessment of Bariatric Surgery (Teen-LABS) study. JAMA Pediatr. 2014;168:47-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 209] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 42. | Inge TH, Courcoulas AP, Jenkins TM, Michalsky MP, Helmrath MA, Brandt ML, Harmon CM, Zeller MH, Chen MK, Xanthakos SA, Horlick M, Buncher CR; Teen-LABS Consortium. Weight Loss and Health Status 3 Years after Bariatric Surgery in Adolescents. N Engl J Med. 2016;374:113-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 455] [Cited by in RCA: 509] [Article Influence: 56.6] [Reference Citation Analysis (0)] |
| 43. | Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, Ferrell LD, Liu YC, Torbenson MS, Unalp-Arida A, Yeh M, McCullough AJ, Sanyal AJ; Nonalcoholic Steatohepatitis Clinical Research Network. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313-1321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6807] [Cited by in RCA: 8224] [Article Influence: 411.2] [Reference Citation Analysis (5)] |
| 44. | Brunt EM, Kleiner DE, Wilson LA, Belt P, Neuschwander-Tetri BA; NASH Clinical Research Network (CRN). Nonalcoholic fatty liver disease (NAFLD) activity score and the histopathologic diagnosis in NAFLD: distinct clinicopathologic meanings. Hepatology. 2011;53:810-820. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 997] [Cited by in RCA: 950] [Article Influence: 67.9] [Reference Citation Analysis (0)] |
| 45. | Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882-883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2446] [Cited by in RCA: 3973] [Article Influence: 305.6] [Reference Citation Analysis (0)] |
| 46. | Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12015] [Cited by in RCA: 11481] [Article Influence: 765.4] [Reference Citation Analysis (0)] |
| 47. | McPherson S, Hardy T, Dufour JF, Petta S, Romero-Gomez M, Allison M, Oliveira CP, Francque S, Van Gaal L, Schattenberg JM, Tiniakos D, Burt A, Bugianesi E, Ratziu V, Day CP, Anstee QM. Age as a Confounding Factor for the Accurate Non-Invasive Diagnosis of Advanced NAFLD Fibrosis. Am J Gastroenterol. 2017;112:740-751. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 644] [Cited by in RCA: 644] [Article Influence: 80.5] [Reference Citation Analysis (0)] |
| 48. | Lin Y, Feng X, Cao X, Miao R, Sun Y, Li R, Ye J, Zhong B. Age patterns of nonalcoholic fatty liver disease incidence: heterogeneous associations with metabolic changes. Diabetol Metab Syndr. 2022;14:181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 49. | Ameling S, Kacprowski T, Chilukoti RK, Malsch C, Liebscher V, Suhre K, Pietzner M, Friedrich N, Homuth G, Hammer E, Völker U. Associations of circulating plasma microRNAs with age, body mass index and sex in a population-based study. BMC Med Genomics. 2015;8:61. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 140] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 50. | Marchesini G, Bugianesi E, Forlani G, Cerrelli F, Lenzi M, Manini R, Natale S, Vanni E, Villanova N, Melchionda N, Rizzetto M. Nonalcoholic fatty liver, steatohepatitis, and the metabolic syndrome. Hepatology. 2003;37:917-923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1907] [Cited by in RCA: 1917] [Article Influence: 87.1] [Reference Citation Analysis (0)] |
| 51. | Loomis AK, Kabadi S, Preiss D, Hyde C, Bonato V, St Louis M, Desai J, Gill JM, Welsh P, Waterworth D, Sattar N. Body Mass Index and Risk of Nonalcoholic Fatty Liver Disease: Two Electronic Health Record Prospective Studies. J Clin Endocrinol Metab. 2016;101:945-952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 173] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 52. | Lonardo A, Suzuki A. Sexual Dimorphism of NAFLD in Adults. Focus on Clinical Aspects and Implications for Practice and Translational Research. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 95] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 53. | Lonardo A, Nascimbeni F, Ballestri S, Fairweather D, Win S, Than TA, Abdelmalek MF, Suzuki A. Sex Differences in Nonalcoholic Fatty Liver Disease: State of the Art and Identification of Research Gaps. Hepatology. 2019;70:1457-1469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 689] [Cited by in RCA: 692] [Article Influence: 115.3] [Reference Citation Analysis (0)] |
| 54. | Koutoukidis DA, Astbury NM, Tudor KE, Morris E, Henry JA, Noreik M, Jebb SA, Aveyard P. Association of Weight Loss Interventions With Changes in Biomarkers of Nonalcoholic Fatty Liver Disease: A Systematic Review and Meta-analysis. JAMA Intern Med. 2019;179:1262-1271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 176] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 55. | Brunner KT, Henneberg CJ, Wilechansky RM, Long MT. Nonalcoholic Fatty Liver Disease and Obesity Treatment. Curr Obes Rep. 2019;8:220-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 88] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 56. | Bonacini M, Kassamali F, Kari S, Lopez Barrera N, Kohla M. Racial differences in prevalence and severity of non-alcoholic fatty liver disease. World J Hepatol. 2021;13:763-773. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (2)] |
| 57. | Riazi K, Swain MG, Congly SE, Kaplan GG, Shaheen AA. Race and Ethnicity in Non-Alcoholic Fatty Liver Disease (NAFLD): A Narrative Review. Nutrients. 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 46] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 58. | Tang M, Liu M, Zhang Y, Xie R. Association of family income to poverty ratio and vibration-controlled transient elastography quantified degree of hepatic steatosis in U.S. adolescents. Front Endocrinol (Lausanne). 2023;14:1160625. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 31] [Reference Citation Analysis (0)] |
| 59. | Tan SY, Georgousopoulou EN, Cardoso BR, Daly RM, George ES. Associations between nut intake, cognitive function and non-alcoholic fatty liver disease (NAFLD) in older adults in the United States: NHANES 2011-14. BMC Geriatr. 2021;21:313. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 60. | Krämer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523-530. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2902] [Cited by in RCA: 4189] [Article Influence: 349.1] [Reference Citation Analysis (0)] |
| 61. | Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140-D144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3282] [Cited by in RCA: 3540] [Article Influence: 186.3] [Reference Citation Analysis (0)] |
| 62. | Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18868] [Cited by in RCA: 24599] [Article Influence: 984.0] [Reference Citation Analysis (0)] |
| 64. | Marutho DHH, Wijaya S, Muljono E. The Determination of Cluster Number at k-Mean Using Elbow Method and Purity Evaluation on Headline News. IEEE. 2018;. [DOI] [Full Text] |
| 65. | Jain AK. Data clustering: 50 years beyond K-means. Pattern Recognit Lett. 2010;31:651-666. [RCA] [DOI] [Full Text] [Cited by in Crossref: 5232] [Cited by in RCA: 1798] [Article Influence: 119.9] [Reference Citation Analysis (0)] |
| 66. | Thorndike RL. Who belongs in the family? Psychometrika. 1953;18:267-276. [DOI] [Full Text] |
| 67. | Soronen J, Yki-Järvinen H, Zhou Y, Sädevirta S, Sarin AP, Leivonen M, Sevastianova K, Perttilä J, Laurila PP, Sigruener A, Schmitz G, Olkkonen VM. Novel hepatic microRNAs upregulated in human nonalcoholic fatty liver disease. Physiol Rep. 2016;4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 43] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 68. | Liang Q, Chen H, Xu X, Jiang W. miR-182-5p Attenuates High-Fat -Diet-Induced Nonalcoholic Steatohepatitis in Mice. Ann Hepatol. 2019;18:116-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 69. | Katsura A, Morishita A, Iwama H, Tani J, Sakamoto T, Tatsuta M, Toyota Y, Fujita K, Kato K, Maeda E, Nomura T, Miyoshi H, Yoneyama H, Himoto T, Fujiwara S, Kobara H, Mori H, Niki T, Ono M, Hirashima M, Masaki T. MicroRNA profiles following metformin treatment in a mouse model of non-alcoholic steatohepatitis. Int J Mol Med. 2015;35:877-884. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 70. | Wan Z, Yang X, Liu X, Sun Y, Yu P, Xu F, Deng H. M2 macrophage-derived exosomal microRNA-411-5p impedes the activation of hepatic stellate cells by targeting CAMSAP1 in NASH model. iScience. 2022;25:104597. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 36] [Reference Citation Analysis (0)] |
| 71. | Adinolfi LE, Rinaldi L, Guerrera B, Restivo L, Marrone A, Giordano M, Zampino R. NAFLD and NASH in HCV Infection: Prevalence and Significance in Hepatic and Extrahepatic Manifestations. Int J Mol Sci. 2016;17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 85] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 72. | Iacob DG, Rosca A, Ruta SM. Circulating microRNAs as non-invasive biomarkers for hepatitis B virus liver fibrosis. World J Gastroenterol. 2020;26:1113-1127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 27] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (2)] |
| 73. | Long JK, Dai W, Zheng YW, Zhao SP. miR-122 promotes hepatic lipogenesis via inhibiting the LKB1/AMPK pathway by targeting Sirt1 in non-alcoholic fatty liver disease. Mol Med. 2019;25:26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 188] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 74. | Hu Y, Peng X, Du G, Zhang Z, Zhai Y, Xiong X, Luo X. MicroRNA-122-5p Inhibition Improves Inflammation and Oxidative Stress Damage in Dietary-Induced Non-alcoholic Fatty Liver Disease Through Targeting FOXO3. Front Physiol. 2022;13:803445. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 27] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 75. | Li K, Zhao B, Wei D, Wang W, Cui Y, Qian L, Liu G. miR146a improves hepatic lipid and glucose metabolism by targeting MED1. Int J Mol Med. 2020;45:543-555. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 76. | Huang R, Duan X, Liu X, Cao H, Wang Y, Fan J, Wang B. Upregulation of miR-181a impairs lipid metabolism by targeting PPARα expression in nonalcoholic fatty liver disease. Biochem Biophys Res Commun. 2019;508:1252-1258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 77. | Gjorgjieva M, Sobolewski C, Ay AS, Abegg D, Correia de Sousa M, Portius D, Berthou F, Fournier M, Maeder C, Rantakari P, Zhang FP, Poutanen M, Picard D, Montet X, Nef S, Adibekian A, Foti M. Genetic Ablation of MiR-22 Fosters Diet-Induced Obesity and NAFLD Development. J Pers Med. 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 78. | Yadav AK, Sata TN, Verma D, Sah AK, Mishra AK, Mrinalini, Hossain MM, Pant K, Venugopal SK. Free fatty acid-induced miR-22 inhibits gluconeogenesis via SIRT-1-mediated PGC-1α expression in nonalcoholic fatty liver disease. iLIVER. 2023;2:1-9. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 3] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 79. | Zhang Q, Yu K, Cao Y, Luo Y, Liu Y, Zhao C. miR-125b promotes the NF-κB-mediated inflammatory response in NAFLD via directly targeting TNFAIP3. Life Sci. 2021;270:119071. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 80. | Feili X, Wu S, Ye W, Tu J, Lou L. MicroRNA-34a-5p inhibits liver fibrosis by regulating TGF-β1/Smad3 pathway in hepatic stellate cells. Cell Biol Int. 2018;42:1370-1376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 81. | Liang Z, Li J, Zhao L, Deng Y. miR‑375 affects the hedgehog signaling pathway by downregulating RAC1 to inhibit hepatic stellate cell viability and epithelial‑mesenchymal transition. Mol Med Rep. 2021;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 82. | Spengler EK, Loomba R. Recommendations for Diagnosis, Referral for Liver Biopsy, and Treatment of Nonalcoholic Fatty Liver Disease and Nonalcoholic Steatohepatitis. Mayo Clin Proc. 2015;90:1233-1246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 207] [Article Influence: 20.7] [Reference Citation Analysis (0)] |