Published online Apr 28, 2021. doi: 10.3748/wjg.v27.i16.1785
Peer-review started: December 22, 2020
First decision: January 23, 2021
Revised: February 4, 2021
Accepted: March 11, 2021
Article in press: March 11, 2021
Published online: April 28, 2021
Processing time: 119 Days and 18 Hours
Gastric carcinoma (GC) is a digestive system disease with high morbidity and mortality. However, early clinical detection is difficult, and the therapeutic effect for advanced disease is not satisfactory. Thus, finding new tumor markers and therapeutic targets conducive to the treatment of GC is imperative. MRPL35 is a member of the large subunit family of mitochondrial ribosomal protein. MRPL35 shows the characteristic of oncogene in colorectal cancer and esophageal cancer, which promotes the exploration of the correlation between MRPL35 and GC. We proposed that the expression of MRPL35 might be critical in GC.
To study the effect of MRPL35 knockdown on GC cell proliferation.
The expression of MRPL35 in GC was evaluated based on data from the public tumor database UALCAN (http://www.ualcan.path.uab.edu). The effect of the expression of MRPL35 on the prognosis was evaluated with KMplot (http://www.kmplot.com). The expression of MRPL35 was assessed on the tissue microarray by immunohistochemistry and the level of MRPL35 mRNA in 25 pairs of clinical GC tissues and matched adjacent tissues was detected by quantitative reverse transcription-polymerase chain reaction. Celigo cell count assay, colony formation assay, and flow cytometry were used to assess the role of MRPL35 in GC cell proliferation and apoptosis in vitro. Additionally, tumor formation experiment in BALB/c nude mice was utilized to determine the effect of MRPL35 on GC cell proliferation. After knockdown of MRPL35, related proteins were identified by isobaric tags for relative and absolute quantification analysis, and the expression of related proteins was detected by Western blot.
The expression of MRPL35 was up-regulated in GC (P = 1.77 × 10-4). The Kaplan-Meier plots of the overall survival indicated that high expression of MRPL35 was associated with a poor survival in GC. Compared with adjacent tissues, the expression of MRPL35 in GC tissues was increased, which was related to age (P = 0.03), lymph node metastasis (P = 0.007), and pathological tumor-node-metastasis stage (P = 0.024). Knockdown of MRPL35 inhibited GC cell proliferation and colony formation and induced apoptosis. Animal experiment results showed that knockdown of MRPL35 inhibited tumor formation in BALB/c nude mice. Western blotting analysis showed that after knockdown of MRPL35, the expression of PICK1 and BCL-XL proteins decreased, and that of AGR2 protein increased.
Collectively, our findings demonstrate that knockdown of MRPL35 inhibits GC cell proliferation through related proteins including PICK1, BCL-XL, and AGR2.
Core Tip: MRPL35 is a member of the large subunit family of mitochondrial ribosomal protein. Our results showed that compared with adjacent tissues, the expression of MRPL35 in gastric carcinoma (GC) tissues was increased significantly, which was related to age, lymph node metastasis, and pathological tumor-node-metastasis stage of GC patients. Knockdown of MRPL35 inhibited GC cell proliferation, clone formation, and tumor formation in BALB/c nude mice, induced apoptosis, reduced the expression of PICK1 and BCL-XL proteins, and increased that of AGR2 protein. These data indicate that MRPL35 might be an oncogene and be used as a new therapeutic target for GC.
- Citation: Yuan L, Li JX, Yang Y, Chen Y, Ma TT, Liang S, Bu Y, Yu L, Nan Y. Depletion of MRPL35 inhibits gastric carcinoma cell proliferation by regulating downstream signaling proteins. World J Gastroenterol 2021; 27(16): 1785-1804
- URL: https://www.wjgnet.com/1007-9327/full/v27/i16/1785.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i16.1785
Gastric carcinoma (GC) is a common digestive system disease, the fourth most common cancer, and the second leading cause of cancer-related mortality worldwide. About 1033701 new cases of GC and 782685 GC-related deaths occurred worldwide in 2018, accounting for 8.2% of all cancer deaths in this year[1,2]. In China, the incidence of GC has always been high, and the mortality rate of the disease has also increased gradually, which is threatening human life and health.
At present, the treatment for GC mainly includes surgery, chemotherapy, radiotherapy, and drug treatment[3]. Most patients with early and advanced GC can be treated by surgery. However, the metastasis of GC is a major problem faced by surgery. The surgical treatment cannot cope with the distant metastasis of GC. Chemotherapy and radiotherapy reduce the recurrence and metastasis of GC, treat residual lesions, protect surrounding normal tissues and important organs, and improve survival[4]. During chemotherapy and radiotherapy, patients have to bear a specific degree of discomfort and pain, resulting in lower patient compliance, which in turn, affects the treatment effect. Therefore, finding effective drug treatment target is a major issue in the treatment of GC.
MRPL35, a 39S large subunit, encoded by a nuclear gene, is a member of the large subunit family of mitochondrial ribosomal protein. It is a specific component of mitochondrial ribosomes and plays a key role in the assembly of cytochrome C oxidase, involved in the translation of mitochondrial proteins[5,6]. The MRPL35 gene is localized on chromosome 2p11.2, and the protein contains 188 amino acids and has a molecular weight of about 25 ku. Survival analysis revealed that high expression of MRPL35 indicated a prolonged survival in patients with glioblastoma[7]. The expression of MRPL35 in colorectal cancer tissues is higher than that of the matched adjacent tissues[8]. Furthermore, down-regulation of MRPL35 leads to increased production of reactive oxygen species and DNA damage and can inhibit cell proliferation, block the G2/M phase cell cycle, reduce the mitochondrial membrane potential, and induce apoptosis and autophagy in vitro. After the transfection of esophageal cancer TE-1 cells with lentivirus, compared with shCtrl (negative control virus) group, the expression of MRPL35 was reduced in the shMRPL35 (lentiviral particles of MRPL35) group, and MRPL35 silencing suppresses the proliferation of esophageal cancer TE-1 cells and promotes apoptosis[9]. Other studies found that MRPL35 was negatively correlated with the survival time of patients after surgery[10]. MRPL35 knockdown in colorectal cancer HCT116 cell and SW480 cell inhibited cell proliferation, promoted apoptosis, and blocked the G2/M phase of the cell cycle.
The MRP family regulates cells by participating in the transcriptional translation function of ribosomal proteins, which is closely related to tumor genesis and development[11]. Previous studies have confirmed that MRPS18-2 is an oncogene[12]; MRPL44 is up-regulated in the tissues of cervical lymph node metastasis of thyroid cancer[13]; MRPS22, which is found in large cell lung cancer, is highly expressed in squamous cell carcinoma and adenocarcinoma, and is low in other types of cancer[14]; MRPL41 is down-regulated in colorectal cancer, pancreatic cancer, and other cancers[15]; MRPS23, down-regulated in breast cancer, can activate P21 WAF1/CIP1 and P53, inhibit cell proliferation, and promote apoptosis[16]; and MRPL35 is associated with the occurrence and development of glioblastoma, esophageal cancer, and colorectal cancer. However, the expression and functional role of MRPL35 in GC remain largely unknown. Therefore, we analyzed the correlation between MRPL35 and GC by tissue microarray, cell function experiments, nude mouse tumor formation experiments, and proteomic analysis. The expression of MRPL35 might be critical in GC according to the experimental results, and it could be a potential therapeutic target for GC.
GC tissues and matched adjacent tissues were obtained from Department of Hepatobiliary Surgery, General Hospital of Ningxia Medical University. Following surgical removal, the tissue samples were immediately stored in liquid nitrogen before total RNA extraction. All tissues were histopathologically identified as GC or adjacent tissues. None of the patients had received preoperative adjuvant therapy, and all patients provided informed consent in accordance with the protocol approved by the Ethics Committee of Ningxia Medical University.
Immunohistochemistry (IHC) staining was performed on the tissue microarray (TMA)[17,18]. The method of staining is immunohistochemical streptavidin-peroxidase (SP) method. The section contained 64 pairs of GC tissues and matched adjacent tissues. The section was deparaffinized in xylene and washed in 700 mL/L ethanol. The antigen was recovered using citrate buffer (pH 6.0) under high pressure for 10 min. Endogenous peroxidase was blocked with 30 mL/L H2O2 (MERCK, Germany) in methanol for 10 min. Then, the section was incubated overnight with primary antibody (mouse polyclonal anti-MRPL35 antibody, HPA026631; Sigma-Aldrich, St. Louis, MO, United States) at 4 °C and secondary antibody successively, and developed with diaminobenzidine (DAB) reagent. The section was washed in TBST three times for 5 min, rinsed with tap water for 5 min, and then counterstained with hematoxylin. A negative control was run by omission of primary antibody (phosphate buffered saline instead of primary antibody) on another slide that was for testing the proper antibody concentration. Subsequently, the TMA was analyzed independently by two pathologists without knowing the patients’ clinical information. The areas of the brown-yellow particles are the signs of positively stained tumor cells which represent the MRPL35 protein in the tissues. The staining intensity was scored from 0-3 as negative (0), weak (1), medium (2), or strong (3). The degree of staining was scored by the area of positively stained tumor cells relative to the entire tumor area: 0 (0%), 1 (1%-25%), 2 (26%-50%), 3 (51%-75%), and 4 (76%-100%). The overall protein expression score (the overall score range was 0-12) was calculated by multiplying the intensity and the positive score.
Human GC cell lines (AGS, NCI-N87, MKN-45, and HGC-27) were purchased from the Cell Bank of the Chinese Academy of Sciences. The cell lines were cultured in a cell incubator at 37 °C and 50 mL/L CO2 in RPMI-1640 medium (Gibco, New York, United States) containing 100 mL/L fetal bovine serum (FBS, Gibco) and penicillin and streptomycin (Invitrogen, Carlsbad, United States). shMRPL35 and shCtrl were designed and produced by GeneChem (Shanghai, China). Lentiviral transfection and the specific steps were performed according to the manufacturer’s instructions.
Total RNA of the cultured cells was extracted using TRIzol reagent (PuFei, Shanghai, China). PrimeScript RT Master Mix Perfect Real-Time (TaKaRa, Shiga, Japan) was used to synthesize first-strand cDNA from total RNA. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed using SYBR Master Mixture (TaKaRa) according to the manufacturer’s instructions. The primers used are as follows: MRPL35: Forward, 5’-TTGGCATCTTCAACCTACCGC-3’ and reverse, 5’-GGAGGAAACAACTGGTGTCTGA-3’; GAPDH: Forward, 5’-TGACTTCAACAGCGACACCCA-3’ and reverse, 5’-CACCCTGTTGCTGTAGCCAAA-3’. Data are presented as the mean ± SD for duplicate runs. The relative quantification of MRPL35 expression was evaluated using the 2-∆∆CT method. All experiments were repeated three times.
The infected cells were seeded in a six-well plate at a density of 2000 cells/well and cultured at 37 °C and 50 mL/L CO2. On the following day, the Celigo assay was performed daily for 5 consecutive days according to the manufacturer’s instructions. All experiments were repeated three times.
The infected cells were inoculated in a six-well plate at a density of 1000 cells/well. The medium was changed every 3 d during the interval. After 2 wk, the colonies were fixed with 4% paraformaldehyde for 40 min and stained with 0.1% crystal violet (Sigma-Aldrich) for 4 min. The visible colonies were counted manually. Triplicate wells were assessed for each treatment group. All experiments were repeated three times.
Apoptotic and necrotic cells were evaluated with Annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) (KeyGEN, Jiangsu, China). All the samples were analyzed in triplicate by flow cytometry (BD Bioscience, Bedford, MA, United States). All experiments were repeated three times.
Four-week-old male BABL/c nude mice of specific pathogen-free (SPF) grade were purchased from the Animal Experiment Center of Ningxia Medical University. All animals were housed in polypropylene cages and fed standard laboratory chow and water ad libitum, and maintained at a temperature of 22 ± 1 °C, with 50% ± 5% relative humidity and 12:12 h light-dark cycles. First, MKN-45 cells were infected with shMRPL35 and shCtrl (n = 10 per group). A microsyringe was used to extract 200 µL of the infected and uninfected MKN-45 cell suspension at a concentration of 1.25 × 107/mL, and injected into the skin of nude mice. The tumor volume was measured daily using the formula V = W2× L × 0.5 (V, volume; W, width; L, length). The animal protocols (IACUC-NYLAC-2019-083) were approved by the Institutional Animal Care and Use Committee of Ningxia Medical University.
SDT lysis method was used for GC cell sample preparation, and the bicinchoninic acid (BCA) method (Beyotime, Shanghai, China) was used for protein quantification. After protein samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), Coomassie brilliant blue staining, and filter aided sample preparation enzymolysis, the resulting peptides were quantified using NanoDrop (Thermo Scientific ND2000, Shanghai, China), and the sample peptides were labeled according to the instructions of the isobaric tags for relative and absolute quantification (iTRAQ) labeling kit (AB SCIEX, Framingham, MA, United States). The labeled peptides of each group were mixed and graded using Agilent 1260 infinity II HPLC system, and Easy nLC chromatographic system (Thermo Scientific) analysis and mass spectrometry were performed. The high-resolution mass spectrometer Q Exactive Plus (Thermo Scientific) and Proteome Discoverer 2.1 (Thermo Scientific) software were used for iTRAQ analysis, and the database search was performed on the MASCOT2.5 (Matrix Science, Boston, MA, United States) server. The Protein Database used was Uniprot_HomoSapiens_159615_20170811.fasta (http:// www.uniprot.org). A homology search was first performed for the identified sequences with a localized sequence comparison tool NCBI BLAST+ against the NCBI database, and top 10 aligned sequences were selected according to the E value from the lowest to the highest for subsequent analysis. Then, Blast2GO Command Line was utilized to perform GO (Gene Ontology) annotation on the identified proteins. The enrichment analysis of GO annotation or KEGG pathway annotation was performed on the identified proteins through Fisher's exact test. The expression of identified proteins was analyzed with Cluster 3.0 software, and hierarchical clustering heat map was generated with Java Trewview software.
Total protein was extracted using the Whole Cell Lysis Assay (KeyGEN, Jiangsu, China), and the concentration was determined by BCA Protein Quantitation Assay (KeyGEN, Jiangsu, China). The protein was separated by SDS-PAGE (60-100 mL/L gel) with a sample amount of 50 µg and transferred to polyvinylidene difluoride (PVDF) membranes that were probed with specific primary antibodies overnight at 4 °C. Then, the membranes were incubated with secondary antibodies for 1 h. The immune-reactive bands were visualized using ECL Reagent (Affinity Biosciences, Jiangsu, China) and images were analyzed by Bio-Rad Laboratories (Inc., Hercules, United States). All the primary antibodies and secondary antibodies were from Abcam (MA, United States).
The statistical methods of this study were reviewed by Sun J from Department of Occupational and Environmental Health and Department of Medical Statistics, Institute of Public Health and Management, Ningxia Medical University. All analyses were performed using GraphPad Prism 7.0. All the data are expressed as the mean ± SE. Statistical significance was analyzed using one-way ANOVA. P < 0.05 was considered statistically significant.
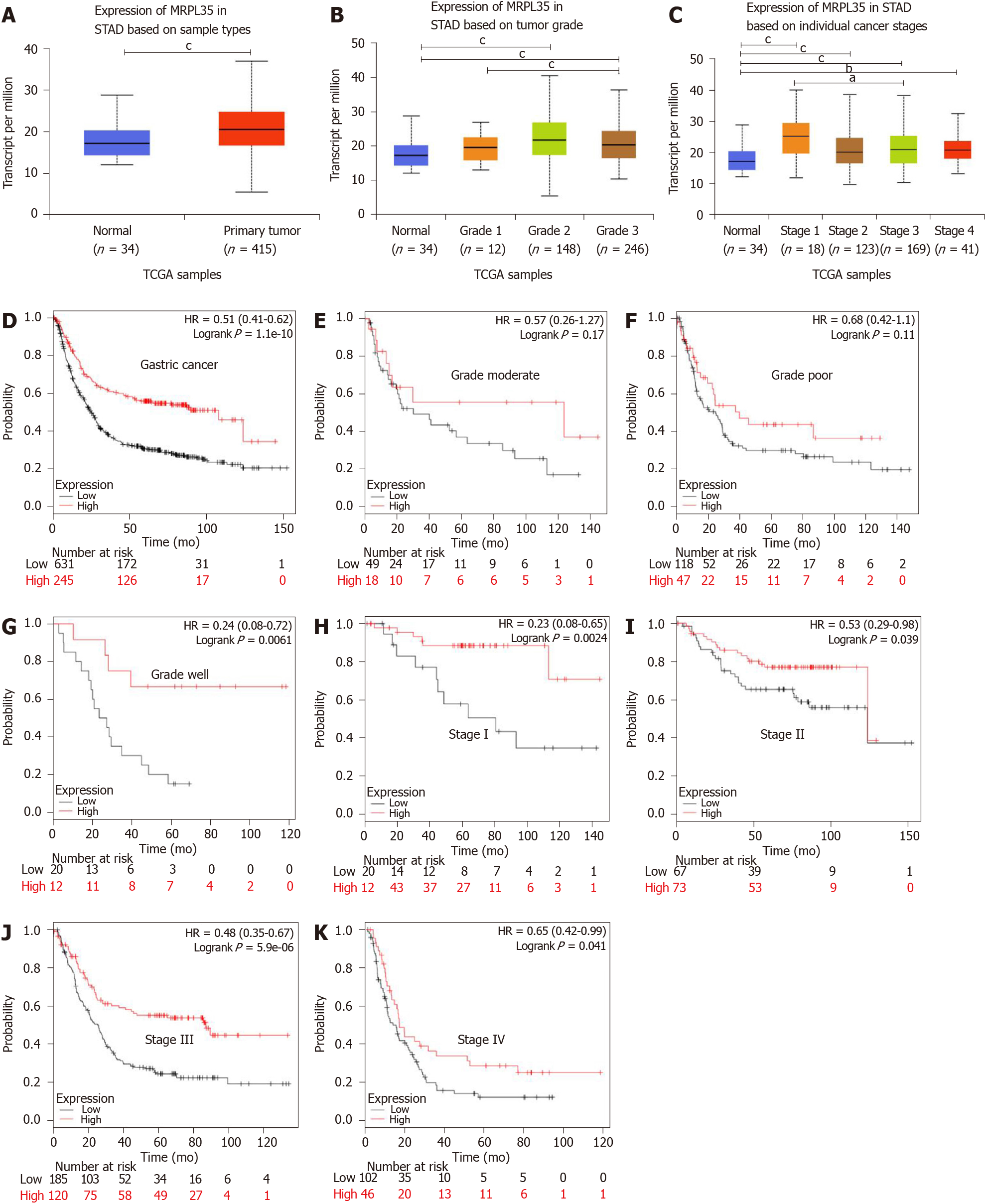
The expression of MRPL35 between GC tissues and adjacent tissues was compared through the public tumor database UALCAN (www.ualcan.path.uab.edu). The results showed that the expression of MRPL35 was increased in GC tissues in comparison with matched adjacent tissues (P = 1.77 × 10-4; 10-4; Figure 1A). In different pathological grades, the expression of MRPL35 in moderate and well grades was higher than that in matched adjacent tissues (P = 8.25 × 10-5; P = 4.19 × 10-5; 10-5; Figure 1B). In different pathological stages, the expression of MRPL35 in stages I, II, III, and IV was higher than that in matched adjacent tissues (P = 0.0012, P < 6.42 × 10-4, P = 4.36 × 10-4, and P = 0.0021, respectively; Figure 1C). Next, the effect of MRPL35 expression on the prognosis was evaluated with KMplot (http://www.kmplot.com). The Kaplan-Meier plots of the overall survival indicated that high expression of MRPL35 was associated with a poor survival in GC (Figure 1D). When stratified according to different histological types and disease stages, patients with high MRPL35 expression had a poor overall survival in well grade and in stage I, stage II, stage III, and stage IV (Figure 1G-K), but not in poor and moderate grades (Figure 1E and F).
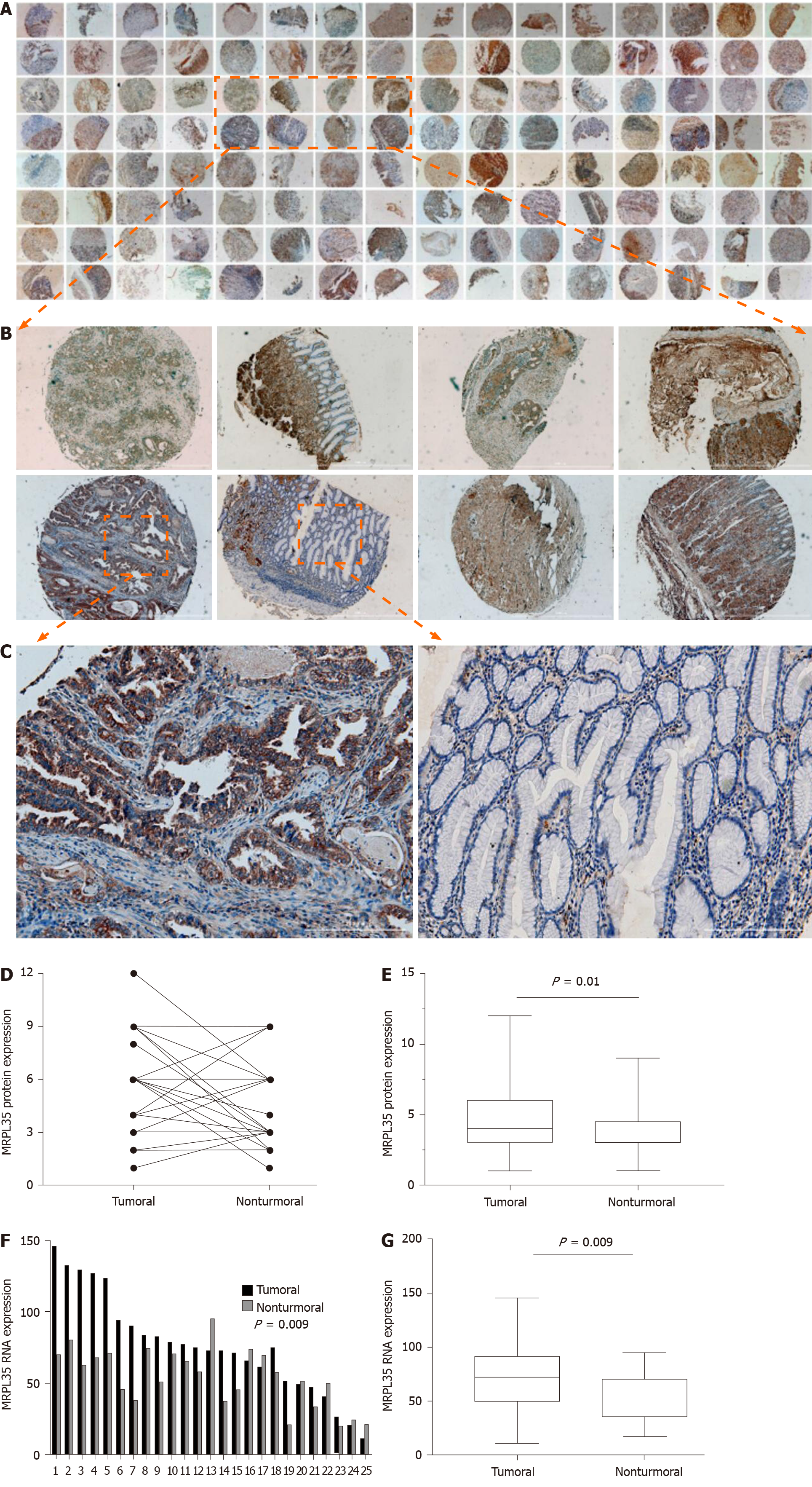
Furthermore, the expression of MRPL35 in clinical samples was detected by IHC method in 64 pairs of GC tissues and matched adjacent tissues. The immunostaining of MRPL35 protein was mainly located in the cell (Figure 2A-C). The negative control pictures are shown in the supplementary materials (Supplementary Figure 1). According to the staining site, the MRPL35 protein expression score was calculated by multiplying the intensity and the positive score (total score range 0-12). The expression of MRPL35 protein in GC tissues was higher than that in matched adjacent tissues (P < 0.05) (Figure 2D and E). These data indicated that up-regulation of MRPL35 might play an important role in the development of GC. In addition, the expression of MRPL35 mRNA in 25 pairs of clinical GC tissues and matched adjacent tissues was detected by qRT-PCR, and the expression of MRPL35 mRNA was up-regulated in GC tissues (P = 0.009; Figure 2F and G).
The correlation between the high expression of MRPL35 and clinicopathological factors in patients with GC proved the importance of high expression of MRPL35 in GC patients (Table 1). According to the average expression (staining score), 68 patients were divided into two groups; for the expression of MRPL35, < 6 was defined as low expression and ≥ 6 defined as high expression. The result of statistical analysis showed that high expression of MRPL35 was correlated with age (P = 0.03), lymph node metastasis (P = 0.007), and pathological tumor-node-metastasis stage (p-TNM stage) (P = 0.024). On the other hand, the correlation between the expression of MRPL35 and other clinicopathological features was not significant.
| Pathological feature | n | Expression of MRPL35 | ||
| High | Low | P value | ||
| 68 | 28 | 40 | ||
| Gender | ||||
| Male | 48 | 19 | 29 | 0.685 |
| Female | 20 | 9 | 11 | |
| Age (yr) | ||||
| < 65 | 33 | 18 | 15 | 0.03a |
| ≥ 65 | 35 | 10 | 25 | |
| p-TNM stage | ||||
| I/II | 33 | 9 | 24 | 0.024a |
| III/IV | 35 | 19 | 16 | |
| Size of tumour | ||||
| ≤ 5 cm | 29 | 9 | 20 | 0.1 |
| > 5 cm | 39 | 19 | 20 | |
| Pathological T grade | ||||
| T1/T2 | 11 | 3 | 8 | 0.313 |
| T3/T4 | 57 | 25 | 32 | |
| Lymph node metastasis (pN) | ||||
| N0/N1 | 30 | 7 | 23 | 0.007b |
| N2/N3 | 38 | 21 | 17 | |
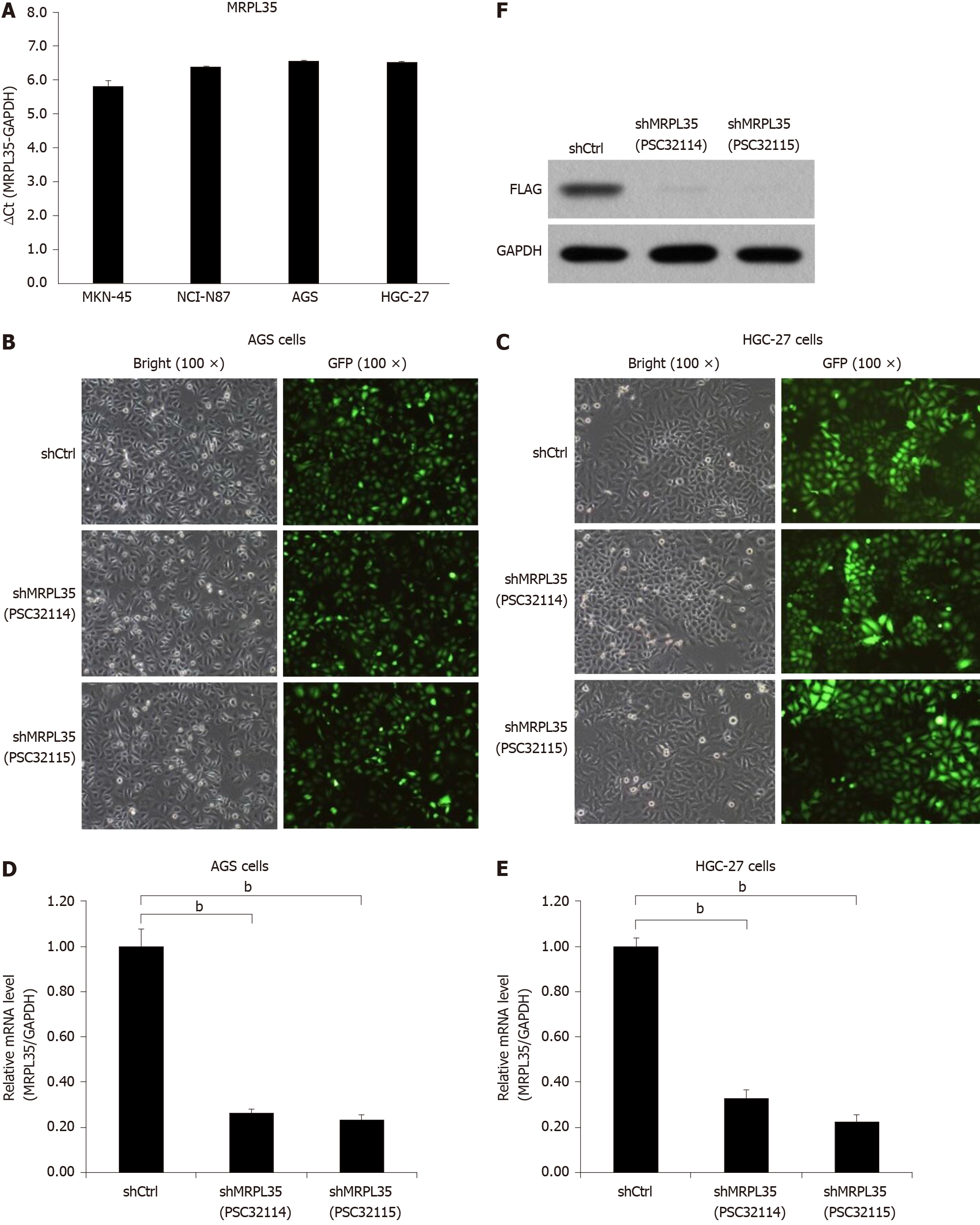
Next, qRT-PCR analysis was performed to determine the expression of MRPL35 in four human GC cell lines. The result showed that the expression of MRPL35 was elevated in AGS and HGC-27 cell lines, but was lower in MKN-45 and NCI-N87 cell lines (Figure 3A). Thus, lentiviral vectors were used to knock down MRPL35 in both cell lines. At 72 h post-transfection, bright light and fluorescence observation under a microscope showed that the cell transfection rate was > 80% (Figure 3B and C). qRT-PCR data revealed that the expression of MRPL35 mRNA (PSC32114 and PSC32115) in AGS and HGC-27 cells was inhibited after shRNA lentivirus transfection (P < 0.05), and the knockdown efficiency reached 73.7% and 76.5% in AGS cells, and 67% and 77.5% in HGC-27 cells (Figure 3D and E). Subsequently, the expression of MRPL35 protein after MRPL35 knockdown in AGS cells was assessed by Western blot; in contrast with the shCtrl group, the expression of MRPL35 (PSC32114 and PSC32115) was reduced in AGS cells (P < 0.05) (Figure 3F).
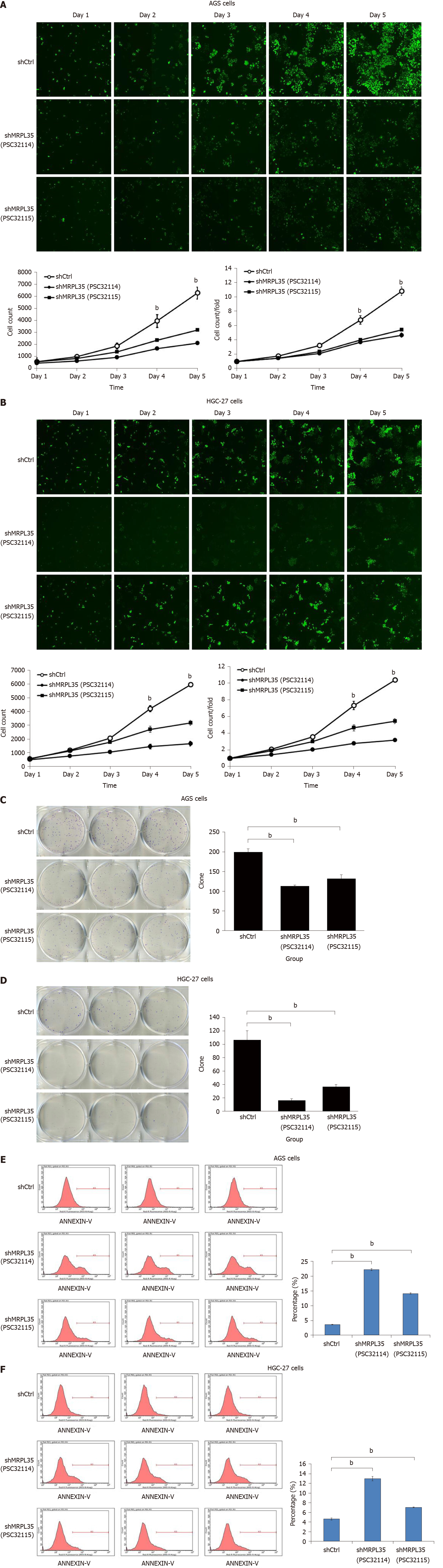
In contrast with the shCtrl group, the shMRPL35 (PSC32114 and PSC32115) group had inhibited proliferation of the two cell lines as monitored by Celigo cell count assay (P < 0.01), indicating that knockdown of MRPL35 had an inhibitory effect on cell proliferation (Figure 4A and B). In addition, knockdown of MRPL35 greatly reduced the colony-forming ability of AGS and HGC-27 cells (Figure 4C and D). Finally, the apoptosis was analyzed using flow cytometry. The numbers of apoptosis of AGS cells and HGC-27 cells in the shMRPL35 (PSC32114 and PSC32115) group increased compared with the shCtrl group (P < 0.01) (Figure 4E and F). These result indicated that MRPL35 might be essential for the proliferation of GC cells.
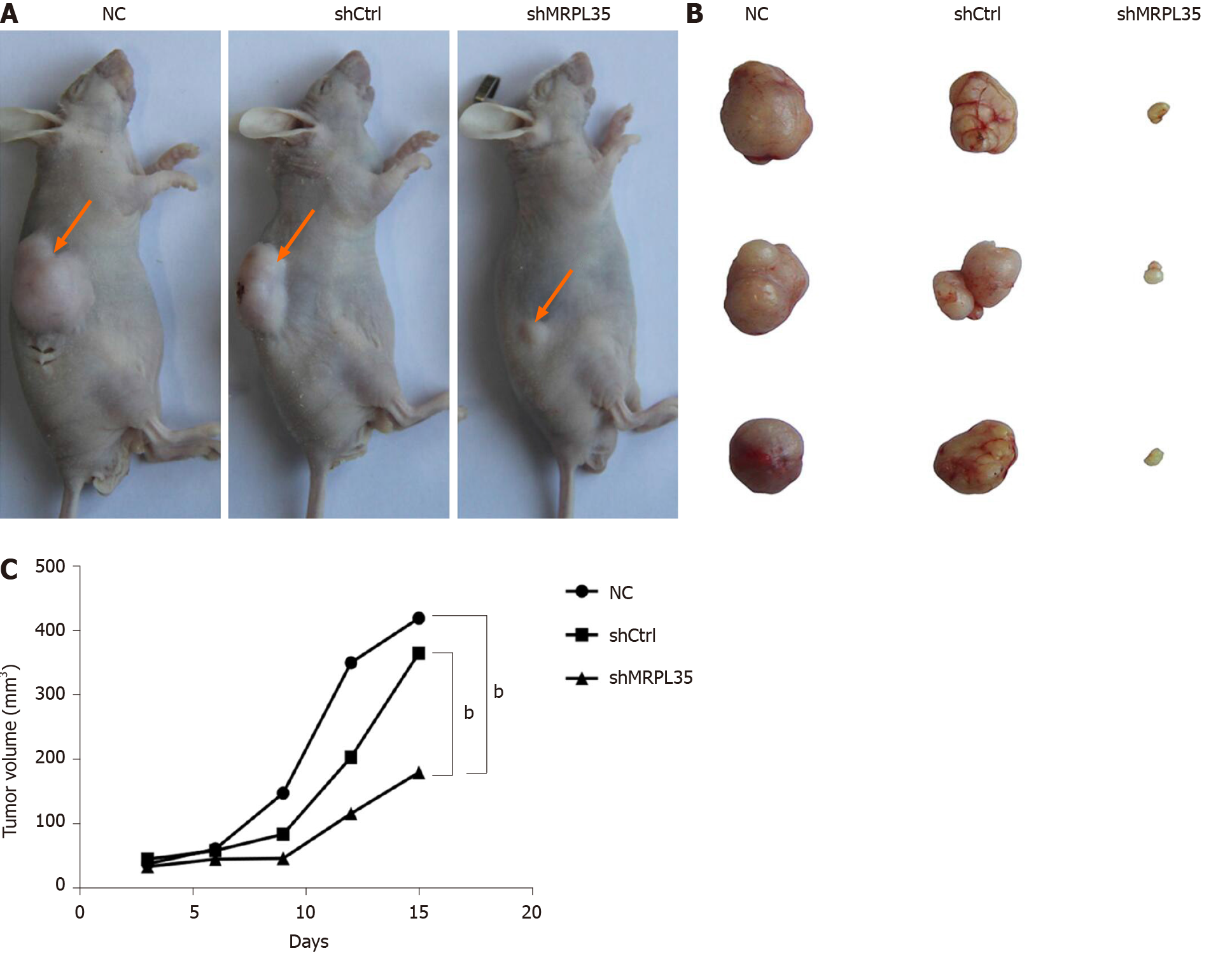
The tumor volume of the shMRPL35 group was smaller than that of the shCtrl group (Figure 5A and B). Within 3 wk of tumor formation, the tumor growth curve was drawn. Compared with the shCtrl and the NC groups, the tumor growth rate of the shMRPL35 group was lower (Figure 5C) (P < 0.01).
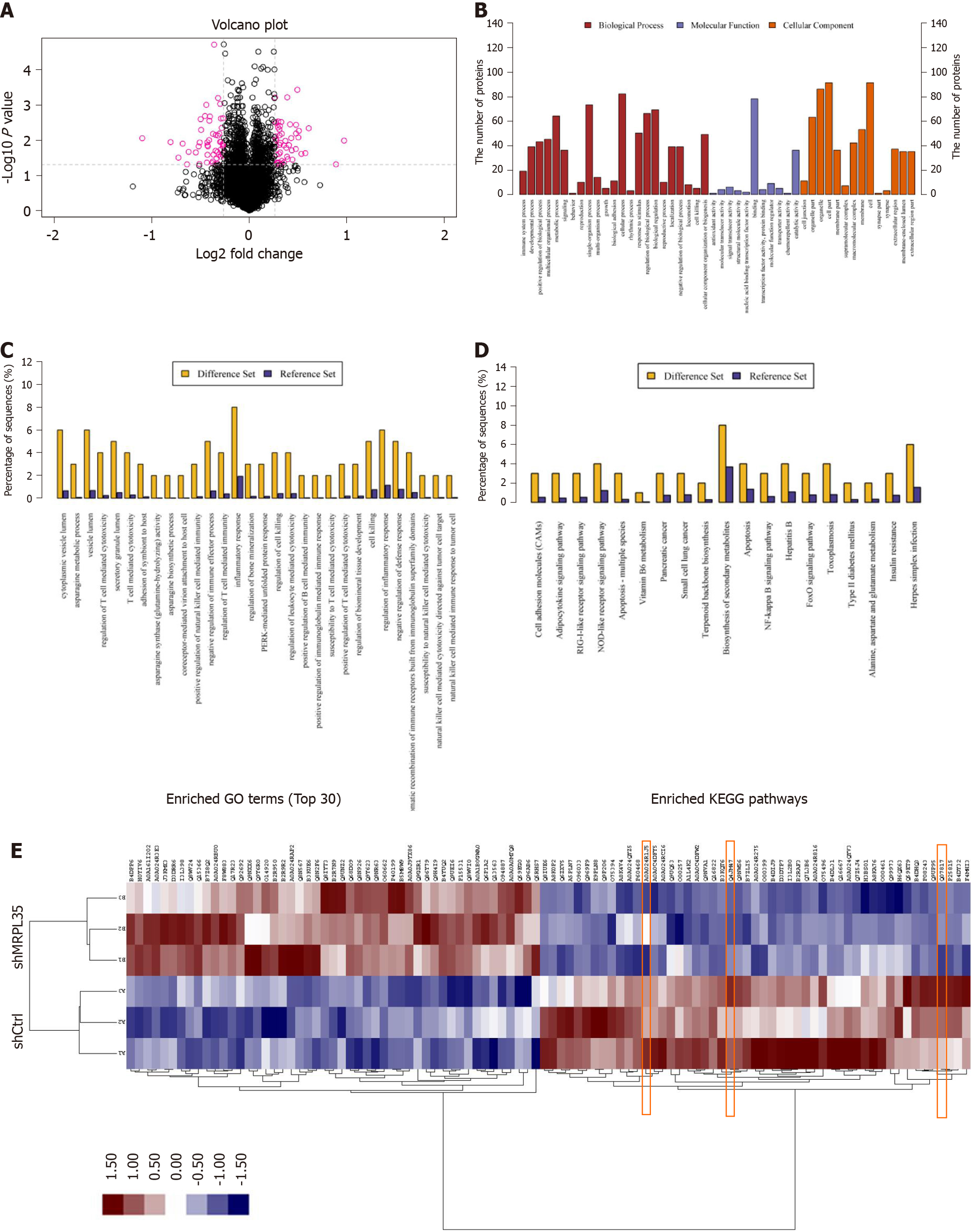
Among 5993 proteins, 100 differentially expressed proteins (DEPs) were identified (Figure 6A). In comparison with the shCtrl group, 49 DEPs were up-regulated and 51 were down-regulated in the shMRPL35 group. The DEPs were screened based on the multiple of expression difference > 1.2 times and P value (t-test) < 0.05. The enrichment analysis was performed through GO functional annotation, describing the differential proteins from a large variety of biological processes, including cellular process, single-organism process, biological regulation, regulation of the biological process, and metabolic process (Figure 6B and C). The significance level of protein enrichment in each pathway was obtained by analyzing DEPs based on the KEGG database pathway annotation analysis (Figure 6D). The result of cluster analysis showed that the expression of all DEPs was different in the groups of shMRPL35 and shCtrl (Figure 6E).
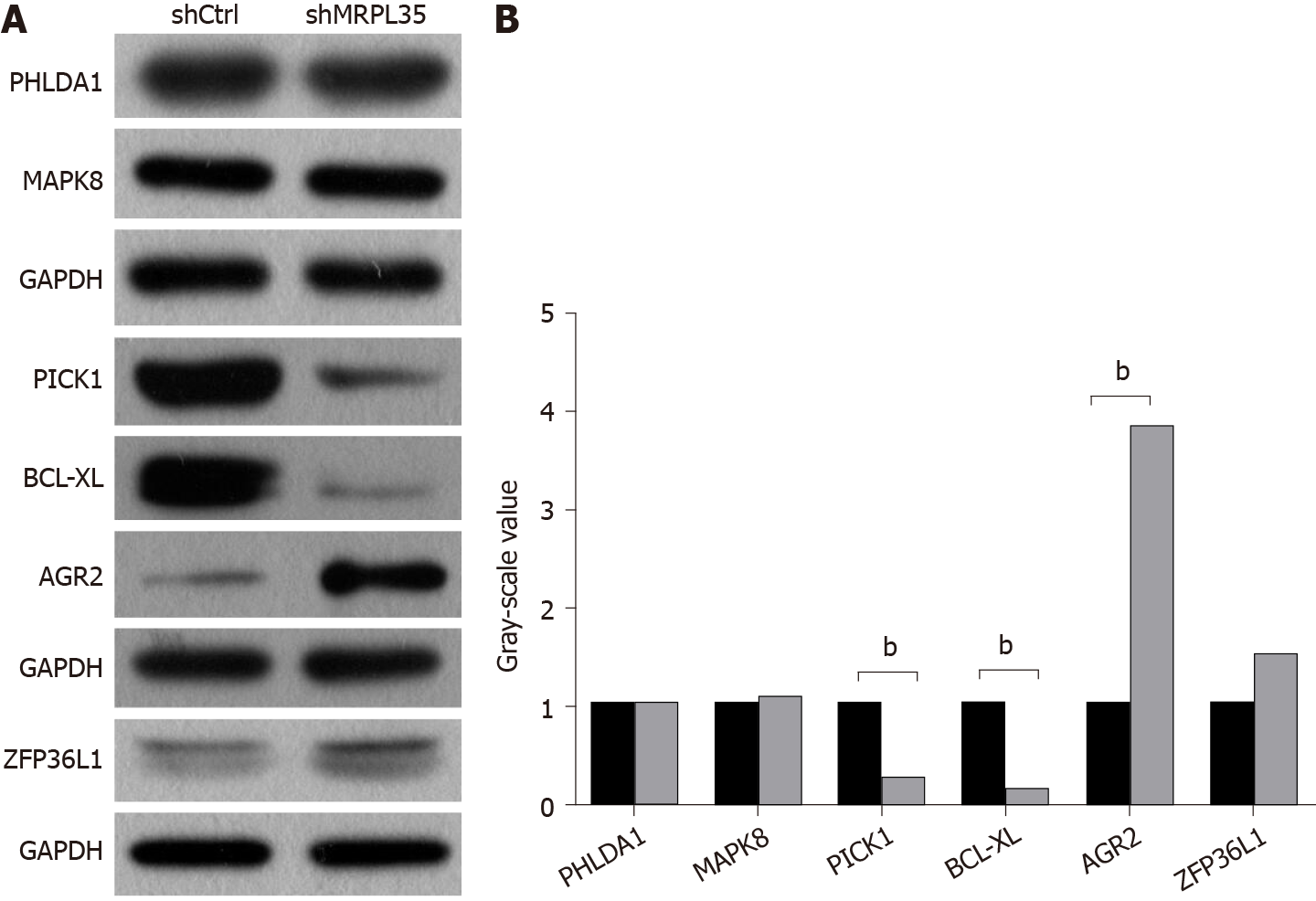
PHLDA1, MAPK8, PICK1, BCL-XL, AGR2, and ZFP36L1 proteins were selected for Western blot verification, and these proteins ranked the top 30 in biological process, molecular function, and cellular component. The expression of PHLDA1, MAPK8, PICK1, BCL-XL, AGR2, and ZFP36L1 proteins in AGS cell was detected by Western blot, and these proteins were contained in DEPs. Compared with the shCtrl group, the expression of PICK1 and BCL-XL proteins was decreased, and that of AGR2 was increased in the shMRPL35 group (P < 0.01) (Figure 7A and B). Based on the above result, knockdown of MRPL35 may inhibit the proliferation of GC cells through PICK1, BCL-XL, and AGR2 proteins.
A major finding of this study is that knockdown of MRPL35 could inhibit the proliferation of human GC cells and promote apoptosis through nude mouse tumor formation experiment in vivo, together with cell function experiment in vitro. In addition, proteomic analysis found that related downstream proteins changed after knockdown of MRPL35. And then, the six proteins were assessed by Western blot, which showed that the expression of PICK1 and BCL-XL proteins decreased, while that of AGR2 protein increased. In addition, the expression of MRPL35 was up-regulated in GC tissues, and high expression of MRPL35 was associated with a low survival rate in GC. Sixty-four pairs of clinical samples of GC tissues and matched adjacent tissues were examined for the expression of MRPL35 by IHC. The expression of MRPL35 in GC tissues was increased compared with matched adjacent tissues, and high expression of MRPL35 was correlated with age, lymph node metastasis, and p-TNM stage. Moreover, the results of other studies on MRPL35 in other cancers were similar to those of the current study, suggested that MRPL35 has the potential to serve as a new therapeutic target for GC. It is necessary to explain the number of tissues on the TMA. Originally, there were 75 pairs of GC tissues and matched adjacent tissues on the section, but a few of them were worn out during the IHC process. Finally, the tissues that could present on the slide and be used for analysis included 68 GC tissues and 64 matched adjacent tissues, that is to say, there were 64 GC tissues and 64 matched adjacent tissues, as well as the extra four GC tissues of which the matched adjacent tissues were worn out. We performed clinicopathological feature analysis on the remaining 68 GC tissues, and then obtained the data in Table 1. However, we carried out the IHC analysis on the remaining 64 pairs of GC tissues and matched adjacent tissues, because it should make a comparative analysis between GC tissues and the matched adjacent tissues. So only 64 pairs of tissues are shown in Figure 2A, for there were no matched adjacent tissues to be compared with the extra four GC tissues. The figures of the extra four GC tissues are shown in the supplementary materials (Supplementary Figure 2).
From the previous studies, we noticed that the expression of MRPL35 was decreased and cell proliferation inhibited, while apoptosis was promoted after knockdown of MRPL35 in esophageal cancer TE-1 cells[9]. This phenomenon was similar to the results of the current study. The expression of MRPL35 in cancer tissues was higher than that in matched adjacent tissues in colorectal cancer, and MRPL35 knockdown in vitro can inhibit cell proliferation and promote apoptosis[8]. The expression of MRPL35 was decreased in the nude mouse xenograft model of colorectal cancer[19]. Indeed, the tumor growth rate in the experimental group was slow, and the tumor was smaller than that in the control group, which was the same as our experimental results. The gastrointestinal tract contains the mouth, esophagus, stomach, small intestine, and large intestine. The cells contained in these tissues were developed from the endoderm of the early embryo. Perhaps because the esophagus, stomach, and large intestine all come from the endoderm, the inhibitory effects of MRPL35 in GC, esophageal cancer, and colorectal cancer are similar.
Furthermore, the PICK1 protein contains a PDZ domain, which binds to a variety of membrane proteins and organizes the subcellular localization of adaptor protein[20]. It may also participate in receptor aggregation by acting on the level of receptor internalization. Decreased expression of MRPL35 regulates the expression of MRPL27, reducing the expression of PICK1. The protein encoded by BCL-XL belongs to the BCL-2 protein family[21]. It can be used as an anti- or pro-apoptotic regulator involved in various cell activities and regulation of the opening of the mitochondrial outer membrane channel (VDAC). MRPL35 regulates the expression of BCL-XL by regulating the family member BAX. AGR2 encodes a member of the disulfide isomerase family of endoplasmic reticulum protein and plays a role in cell migration, cell differentiation, and growth, and is associated with cancer progression[22]. Consequently, the expression of MRPL35 and MRPL2 is altered, which in turn, affects that of AGR2. These results fully indicated that MRPL35 might be an oncogene, and the correlation between MRPL35 and GC is worthy of in-depth exploration.
In short, the current study suggested that MRPL35 has a causal correlation with GC, and related downstream proteins of MRPL35 have been identified. Nevertheless, the current study has some deficiencies. At present, only a few studies have investigated MRPL35, which is not sufficient to clarify its role as a therapeutic target for GC. On the premise that there are no relevant studies on MRPL35 and GC, follow-up research would be divided into three stages. In the first stage, the correlation between PICK1, BCL-XL, and AGR2 genotype and GC phenotype would be explored. In the second stage, the correlation between MRPL35 and its downstream proteins PICK1, BCL-XL, and AGR2 would be examined. In the third stage, the mechanism underlying the interaction between MRPL35 and PICK1, BCL-XL, and AGR2 would be assessed, and two modes of action were considered: Direct and through tool protein. Also, covalent and non-covalent interactions occurred between the proteins. Anyway, our study aimed to provide a scientific basis for the treatment of GC and find effective drug target for the early prevention of GC.
MRPL35 is related to esophageal cancer and colorectal cancer, and has not been reported in GC. Our experimental data combined with the data in the public tumor database show that MRPL35 is of extraordinary importance in GC tissues and cells, which has also been verified in nude mice. The expression of MRPL35 in GC tissues is higher than that of the matched adjacent tissues, and knockdown of MRPL35 can inhibit the proliferation of GC cells and induce apoptosis. This suggests that MRPL35 is a potential target for the treatment of GC, and it is necessary to further study the effect of MRPL35 on GC.
Gastric carcinoma (GC) is one of the most common cancers, and the existing treatment methods cannot meet the treatment needs for GC. At the same time, MRPL35, a member of the large subunit family of mitochondrial ribosomal protein, shows the characteristics of oncogene in certain cancers.
The existing treatment methods for GC mainly include drugs, chemotherapy, and surgery, all of which have certain defects. Finding new therapeutic targets will be of great benefit to patients with GC.
The present study aimed to explore the correlation between MRPL35 and GC, and the effect of knockdown of MRPL35 on GC cells.
The expression of MRPL35 in GC and the effect of MRPL35 on the prognosis of GC were evaluated based on data from public databases. Immunohistochemistry staining and pathological factors analysis were performed on 64 pairs of GC tissues and matched adjacent tissues. The effect of MRPL35 on the proliferation and apoptosis of GC cells was determined by Celigo cell count assay, flow cytometry, and tumor formation experiment in BABL/c nude mice. The related proteins that changed after knockdown of MRPL35 was identified by proteomic analysis and tested by Western blot.
The expression of MRPL35 was up-regulated in GC and high expression of MRPL35 was associated with a poor survival in GC. The expression of MRPL35 in GC tissues was increased in comparison with matched adjacent tissues, which was related to age, lymph node metastasis, and pathological tumor-node-metastasis stage. Knockdown of MRPL35 inhibited GC cell proliferation and colony formation and induced apoptosis. After knockdown of MRPL35, the expression of PICK1 and BCL-XL protein decreased, and that of AGR2 protein increased.
MRPL35 is up-regulated in GC, and knockdown of MRPL35 could inhibit the proliferation of GC cells and induce apoptosis.
MRPL35 can be used for targeted therapy of GC, and can also be used as a new biomarker for GC.
The authors would like to acknowledge Sun J for statistical analysis assistance.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cheng TH, Exbrayat JM S-Editor: Gao CC L-Editor: Wang TQ P-Editor: Liu JH
| 1. | Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, Znaor A, Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941-1953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3585] [Cited by in RCA: 4890] [Article Influence: 698.6] [Reference Citation Analysis (1)] |
| 2. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55772] [Article Influence: 7967.4] [Reference Citation Analysis (132)] |
| 3. | Song Z, Wu Y, Yang J, Yang D, Fang X. Progress in the treatment of advanced gastric cancer. Tumour Biol. 2017;39:1010428317714626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 396] [Cited by in RCA: 643] [Article Influence: 80.4] [Reference Citation Analysis (0)] |
| 4. | Slagter AE, Vollebergh MA, Jansen EPM, van Sandick JW, Cats A, van Grieken NCT, Verheij M. Towards Personalization in the Curative Treatment of Gastric Cancer. Front Oncol. 2020;10:614907. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 5. | Brown A, Amunts A, Bai XC, Sugimoto Y, Edwards PC, Murshudov G, Scheres SHW, Ramakrishnan V. Structure of the large ribosomal subunit from human mitochondria. Science. 2014;346:718-722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 248] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 6. | Box JM, Kaur J, Stuart RA. MrpL35, a mitospecific component of mitoribosomes, plays a key role in cytochrome c oxidase assembly. Mol Biol Cell. 2017;28:3489-3499. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 7. | Alshabi AM, Vastrad B, Shaikh IA, Vastrad C. Identification of Crucial Candidate Genes and Pathways in Glioblastoma Multiform by Bioinformatics Analysis. Biomolecules. 2019;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 8. | Zhang L, Lu P, Yan L, Yang L, Wang Y, Chen J, Dai J, Li Y, Kang Z, Bai T, Xi Y, Xu J, Sun G, Yang T. MRPL35 Is Up-Regulated in Colorectal Cancer and Regulates Colorectal Cancer Cell Growth and Apoptosis. Am J Pathol. 2019;189:1105-1120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 9. | Wang AF, Zhang Q, Zhang DM, Xiu YT, Ding YM, Liu LL. Effect of silencing mitochondrial ribosomal protein L35 gene on the growth of human esophageal carcinoma TE-1 cells. Jilin Daxue Xuebao. 2019;45:28-32. |
| 10. | Wang YT. Preliminary study on the role of MRPL35 protein in the development of colorectal cancer. M.Sc. Thesis, Shanxi Medical University. 2017. Available from: https://kns.cnki.net/KXReader/Detail?TIMESTAMP=637472036637783203&DBCODE=CJFQ&TABLEName=CJFDLAST2019&FileName=BQEB201901006&RESULT=1&SIGN=hPOiLoUIMEDlnsKQkf7paQxOVMs%3d. |
| 11. | Gopisetty G, Thangarajan R. Mammalian mitochondrial ribosomal small subunit (MRPS) genes: A putative role in human disease. Gene. 2016;589:27-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 12. | Buchynska LG, Iurchenko NP, Kashuba EV, Brieieva OV, Glushchenko NM, Mints M, Lukianova NY, Chekhun VF. Overexpression of the mitochondrial ribosomal protein S18-2 in the invasive breast carcinomas. Exp Oncol. 2018;40:303-308. [PubMed] |
| 13. | Lee J, Seol MY, Jeong S, Lee CR, Ku CR, Kang SW, Jeong JJ, Shin DY, Nam KH, Lee EJ, Chung WY, Jo YS. A metabolic phenotype based on mitochondrial ribosomal protein expression as a predictor of lymph node metastasis in papillary thyroid carcinoma. Medicine (Baltimore). 2015;94:e380. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 14. | Lazar V, Suo C, Orear C, van den Oord J, Balogh Z, Guegan J, Job B, Meurice G, Ripoche H, Calza S, Hasmats J, Lundeberg J, Lacroix L, Vielh P, Dufour F, Lehtiö J, Napieralski R, Eggermont A, Schmitt M, Cadranel J, Besse B, Girard P, Blackhall F, Validire P, Soria JC, Dessen P, Hansson J, Pawitan Y. Integrated molecular portrait of non-small cell lung cancers. BMC Med Genomics. 2013;6:53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 15. | Kim MJ, Yoo YA, Kim HJ, Kang S, Kim YG, Kim JS, Yoo YD. Mitochondrial ribosomal protein L41 mediates serum starvation-induced cell-cycle arrest through an increase of p21(WAF1/CIP1). Biochem Biophys Res Commun. 2005;338:1179-1184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 16. | Gao Y, Li F, Zhou H, Yang Y, Wu R, Chen Y, Li W, Li Y, Xu X, Ke C, Pei Z. Down-regulation of MRPS23 inhibits rat breast cancer proliferation and metastasis. Oncotarget. 2017;8:71772-71781. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 17. | Ong JR, Bamodu OA, Khang NV, Lin YK, Yeh CT, Lee WH, Cherng YG. SUMO-Activating Enzyme Subunit 1 (SAE1) Is a Promising Diagnostic Cancer Metabolism Biomarker of Hepatocellular Carcinoma. Cells. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 18. | Sun Z, Zhu Y, Aminbuhe, Fan Q, Peng J, Zhang N. Differential expression of APE1 in hepatocellular carcinoma and the effects on proliferation and apoptosis of cancer cells. Biosci Trends. 2018;12:456-462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 19. | Yan LH. The effect of down-regulation of MRPL35 and RASGRF2 on tumor growth and metastasis of colorectal carcinoma xenograft model in nude mice. M.Sc. Thesis, Shanxi Medical University. 2018. Available from: https://kns.cnki.net/KXReader/Detail?TIMESTAMP=637472036637783203&DBCODE=CJFQ&TABLEName=CJFDLAST2019&FileName=BQEB201901006&RESULT=1&SIGN=hPOiLoUIMEDlnsKQkf7paQxOVMs%3d. |
| 20. | Lei B, Wang D, Zhang M, Deng Y, Jiang H, Li Y. miR-615-3p promotes the epithelial-mesenchymal transition and metastasis of breast cancer by targeting PICK1/TGFBRI axis. J Exp Clin Cancer Res. 2020;39:71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 21. | Qi G, Yu N, Xu K, Xie X, Mao Y, Chen X, Ran X, Lin G, Hu C. Grass carp (Ctenopharyngodon idella) Bcl-xl: transcriptional regulation and anti-apoptosis analysis. Fish Physiol Biochem. 2020;46:483-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 22. | Li J, Hu J, Luo Z, Zhou C, Huang L, Zhang H, Chi J, Chen Z, Li Q, Deng M, Chen J, Tao K, Wang G, Wang L, Wang Z. AGR2 is controlled by DNMT3a-centered signaling module and mediates tumor resistance to 5-Aza in colorectal cancer. Exp Cell Res. 2019;385:111644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |