Published online Mar 21, 2021. doi: 10.3748/wjg.v27.i11.1064
Peer-review started: December 24, 2020
First decision: January 10, 2021
Revised: January 21, 2021
Accepted: March 1, 2021
Article in press: March 1, 2021
Published online: March 21, 2021
Processing time: 82 Days and 8.6 Hours
Hepatitis C virus (HCV) infection is responsible for a chronic liver inflammation, which may cause end-stage liver disease and hepatocellular carcinoma. Apolipoprotein E (protein: ApoE, gene: APOE), a key player in cholesterol metabolism, is mainly synthesized in the liver and APOE polymorphisms may influence HCV-induced liver damage.
To determine whether APOE alleles affect outcomes in HCV-infected patients with liver cirrhosis following orthotopic liver transplantation (OLT).
This was a cohort study in which 179 patients, both genders and aged 34-70 years, were included before or after (up to 10 years follow-up) OLT. Liver injury severity was assessed using different criteria, including METAVIR and models for end-stage liver disease. APOE polymorphisms were analyzed by quantitative real-time polymerase chain reaction.
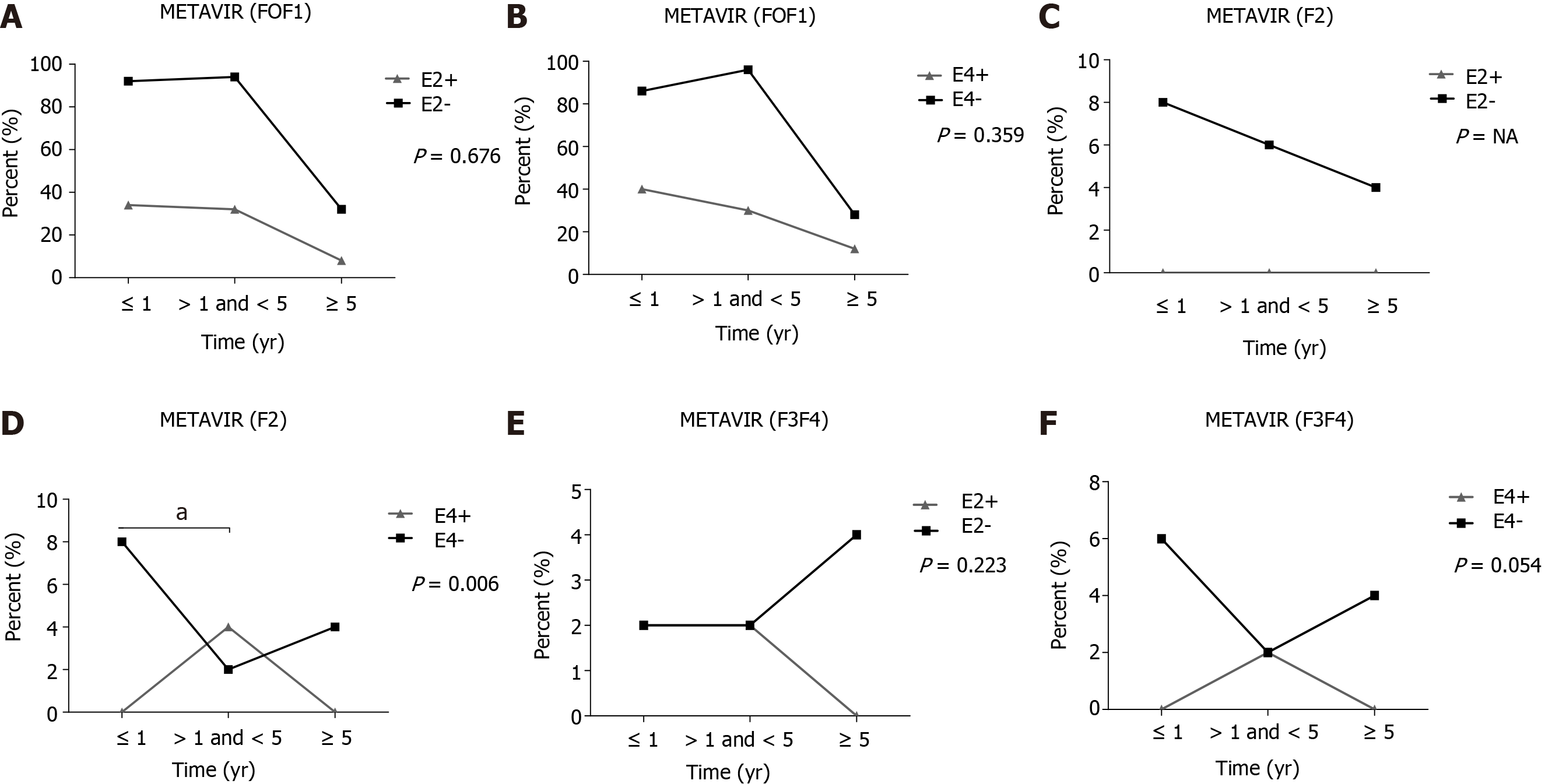
The APOE3 allele was the most common (67.3%). In inflammation severity of biopsies from 89 OLT explants and 2 patients in pre-transplant, the degree of severe inflammation (A3F4, 0.0%) was significantly less frequent than in patients with minimal and moderate degree of inflammation (≤ A2F4, 16.2%) P = 0.048, in patients carrying the APOE4 allele when compared to non-APOE4. In addition, a significant difference was also found (≤ A2F4, 64.4% vs A3F4, 0.0%; P = 0.043) and (A1F4, 57.4% vs A3F4, 0.0%; P = 0.024) in APOE4 patients when compared to APOE3 carriers. The fibrosis degree of the liver graft in 8 of 91 patients and the lack of the E4 allele was associated with more moderate fibrosis (F2) (P = 0.006).
Our results suggest that the E4 allele protects against progression of liver fibrosis and degree of inflammation in HCV-infected patients.
Core Tip: Hepatitis C virus (HCV) infection is responsible for a chronic liver inflammation, which may cause end-stage liver disease. Apolipoprotein E (protein: ApoE, gene: APOE) is key for lipid metabolism. In this study, the APOE4 allele, which has been associated with increased risk of acquiring late-onset Alzheimer’s disease, was found to have a protective effect against the progression of inflammation and/or fibrosis in liver damage induced by HCV pre- and post-liver transplantation. Further studies are needed to unravel the possible contribution of ApoE from the donor liver to this protection.
- Citation: Nascimento JCR, Pereira LC, Rêgo JMC, Dias RP, Silva PGB, Sobrinho SAC, Coelho GR, Brasil IRC, Oliveira-Filho EF, Owen JS, Toniutto P, Oriá RB. Apolipoprotein E polymorphism influences orthotopic liver transplantation outcomes in patients with hepatitis C virus-induced liver cirrhosis. World J Gastroenterol 2021; 27(11): 1064-1075
- URL: https://www.wjgnet.com/1007-9327/full/v27/i11/1064.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i11.1064
The World Health Organization reported in 2015, that around 71 million people had chronic hepatitis C virus (HCV) infection worldwide, with a global prevalence of 1% and that 399,000 died of hepatocellular carcinoma (HCC) or cirrhosis[1].
HCV infection causes a chronic liver inflammatory condition leading to chronic hepatitis[2,3]. The evolution from acute to chronic hepatitis C occurs in up to 80% of cases when HCV infection lasts for more than six months. Without characteristic symptoms, HCV evolves slowly, for years, with approximately 20% of those chronically infected progressing to cirrhosis, and between 1% to 5% to HCC [4,5].
Human apolipoprotein E (protein: ApoE, gene: APOE) is a 34-KDa glycoprotein of 299 amino acids. ApoE is an important protein constituent of very-low-density lipoproteins (LDL), high-density lipoproteins, and chylomicrons in plasma, and a ligand for the LDL receptor[6]. ApoE is synthesized mainly in the liver (90%), but also in spleen, kidney, lungs, gonads, monocyte-macrophages, and in the nervous system[7,8].
The human APOE gene is polymorphic with three common APOE alleles on chromosome 19 (19q13), named E2, E3 and E4, giving six possible genotypes: E2/E2, E2/E3, E2/E4, E3/E3, E3/E4 and E4/E4[9]. These alleles form different ApoE isoforms due to amino acid substitutions at positions 112 and 158: E2 = Cys-112/Cys-158; E3 = Cys-112/Arg-158; and E4 = Arg-112/Arg-158[10].
Several studies have documented the association between ApoE isoforms and chronic illnesses such as Alzheimer's disease[11], atherosclerosis[12], herpes simplex virus infection[13] and early childhood diarrhea[12]. The ApoE4 allele has been recently associated with a high risk for severe coronavirus disease 2019 infection, independent of preexisting cardiovascular disease, type-2 diabetes and dementia[14]. In addition, the APOE3/3 genotype is implicated in fibrosis progression in HCV-infected patients, likely through mechanisms of competition for viral entry and replication[15].
Although some studies report that APOE4 exerts a protective effect in HCV-induced liver damage, no studies have investigated the role of APOE genotypes in modifying the natural history of HCV-induced liver injury in liver transplanted patients in a Brazilian setting. Our aim, therefore, was to establish whether APOE4 genotype recipients were associated with more benign HCV-related liver injuries compared to patients with other APOE genotypes.
This is a cohort study conducted at the University Hospital Walter Cantídio and the Fortaleza General Hospital (HGF). A total of 179 consecutive patients were enrolled from May 2017 to July 2019 for the collection of buccal cells and medical record data. Medical records from liver transplant recipients before May 2017 were also collected retrospectively and prospectively each year. Patients in the orthotopic liver transplantation (OLT) queue, or liver transplanted from May 2017 onwards, had data collected prospectively and annually during the study period.
The study included 179 patients of both genders and aged 34-70 years, with HCV-related end-stage liver disease, 105 of them complicated with HCC and 74 without HCC, in pre- and/or post-OLT. At enrolment, 143 known HCV-infected patients were on antiviral treatment, 126 with direct-acting antivirals and 17 treated with the combination of interferon & ribavirin or pegylated interferon & ribavirin; 36 patients had not received any treatment. The HCV status of all patients was confirmed by identification of serum HCV-RNA and HCV genotypes. Exclusion criteria were: patient refusal, coinfection with HBV or HIV, and HCC associated with metastases.
The study protocol was approved by the Research Ethics Committee of the Federal University of Ceará, Protocol No 2.018.768 and the HGF, Protocol No 2.062.278. The research team explained the study protocol to all patients and clarified that failure to participate in the study would not cause discontinuation of care or medical treatment. Patients then read and signed the informed consent form. All protocols of this study were in accordance with the Helsinki Declaration.
Of 179 patients, oral cells for DNA extraction were obtained at single time points from 56 (31.3%) pre-OLT patients, and from 123 (68.7%) who underwent liver transplantation. Oral cell DNA was extracted using the Gentra Puregene system (Qiagen, MD, United States)[12].
APOE genotyping was detected by quantitative real-time polymerase chain reaction (qRT-PCR), through enhancement of a fluorescent signal (SYBR® Green) interspersed in the double strand of the amplified DNA[16]. The primers were combined in three PCR amplification mixtures according to Calero et al[16] in a Light Cycler® Nano (Roche) with a 32-well RT-PCR system.
Demographic, clinical and laboratory data were collected after thorough reviews of medical records during the preoperative period (179 patients) and after OLT with follow-up of the 144 transplanted patients over 10 years (Supplementary Figure 1). Data included serological markers such as anti-HCV antibodies to define the agent and quantitative HCV-RNA; severity markers of liver cirrhosis [models for end-stage liver disease (MELD)]; liver imaging (computed tomography, nuclear magnetic resonance, and ultrasonography) for identification of liver tumor and classification according to the Milan criteria[17]. Out of 144 patients who underwent OLT based on Milan classification, 24 (16.7%) were subjected to one or more sessions of chemoembolization to reduce tumor size before Milan criteria were reached.
Liver damage severity was categorized into different criteria. Less severe cases were identified by Milan scores (single nodule < 5 cm, or up to 3 nodules < 3 cm), METAVIR (≤ A2 and ≤ F2), and MELD < 25. Patients with more severe liver injury were scored according to the Milan expanded criteria of the University of San Francisco[18] (1 nodule ≤ 6.5 cm; ≤ 3 nodules, each ≤ 4.5 cm with total diameter ≤ 8 cm), METAVIR score (A3 and ≥ F3), and MELD > 25. METAVIR scoring was categorized to assess liver inflammation/fibrosis severity (Supplementary Table 1).
Liver fibrosis was assessed by using aminotransferase-platelet ratio index (APRI) and fibrosis-4 (FIB4) scores. APRI = [AST (U/L)/35 (ULN, the upper limit of normal AST is estimated at 35)]/platelets count (109/L) × 100 and FIB4 = AST (U/L) × age (years)/[platelets count (109/L) × ALT1/2 (U/L)]. The cut-off points of severity are: for significant liver fibrosis (APRI ≥ 1.5, METAVIR F3-F4, and FIB4 ≥ 3.25) and for low-degree fibrosis (APRI ≤ 0.5, METAVIR F0-F1, and FIB4 ≤ 1.45)[19].
Liver biopsy data were obtained from liver explants patients in pre-OLT and of post-OLT liver grafts. Liver biopsies were performed post-transplant in 91 of the 144 transplant recipients, who presented positive HCV viral load, or high risk of viral recurrence or rejection. All recipients had a minimum 1-year follow-up period post-OLT, and donated at least one liver biopsy more than 1 year after their OLT.
Data were analyzed with the SPSS statistical 22.0 software. For normality, D’Agostino and Kolmogorov–Smirnov tests were performed. The absolute and relative frequencies were calculated for the categorical variables and mean ± SD for the numerical variables. Fisher’s exact test or the Mann–Whitney test was used to compare frequencies or means, respectively, when appropriate. Multilinear regression and correlation analyses were performed to avoid other potential confounders. Either one-way or two-way ANOVA test followed by Bonferroni's or Kruskal-Wallis, and Dunn's test were used for multiple comparisons. P < 0.05 was considered significant.
A total of 179 patients (145 males, 34 females) were enrolled in this study with a median age of 61 (range = 34-70). All patients were diagnosed with HCV-induced chronic liver cirrhosis; 105 of them (58.6%) complicated with HCC.
Demographic and clinical data were collected from all patients only in pre-OLT. Their APOE allele stratification is reported in Table 1; no statistical difference was found in any comparisons.
| Variables | APOE2 | APOE3 | APOE4 | P value |
| Age (yr) (mean ± SD) | 60.8 ± 7.26 | 60.3 ± 7.19 | 60.3 ± 7.44 | 0.956 |
| BMI (mean ± SD) | 27.6 ± 4.36 | 26.4 ± 5.01 | 27.0 ± 4.81 | 0.167 |
| Gender (n1 = 179; n2 = 358), n2 (%) | ||||
| Male (n1 = 145; n2 = 290) | 52 (17.9) | 193 (66.6) | 45 (15.5) | 0.650 |
| Female (n1 = 34; n2 = 68) | 9 (13.2) | 48 (70.6) | 11 (16.2) | |
| BMI (n1 = 179; n2 = 358), n2 (%) | ||||
| Non-obese (n1 = 137; n2 = 274) | 49 (17.9) | 185 (67.5) | 40 (14.6) | 0.519 |
| Obese (n1 = 42; n2 = 84) | 12 (14.3) | 56 (66.7) | 16 (19.0) | |
| Etiology (n1 = 179; n2 = 358), n2 (%) | ||||
| HCV (n1 = 74; n2 = 148) | 24 (16.2) | 96 (64.9) | 28 (18.9) | 0.357 |
| HCV + HCC (n1 = 105; n2= 210) | 37 (17.6) | 145 (69.0) | 28 (13.3) | |
| HCC Milan (n1 = 105; n2= 210), n2 (%) | ||||
| Within the criteria (n1 = 83; n2 = 166) | 29 (17.5) | 115 (69.3) | 22 (13.2) | 0.885 |
| Outside the criteria (n1 = 22; n2 = 44) | 8 (18.2) | 29 (65.9) | 7 (15.9) |
The APOE allele frequency according to group, HCV viral load and liver inflammation by the METAVIR score are depicted in Table 2. The APOE allele frequencies were 67.3%, 17.1% and 15.6% for E3, E2 and E4, respectively. The most frequent genotype was E3/E3 (51.4%).
| APOE genotype | Group | HCV-RNA | METAVIR | METAVIR | |||||
| All | HCV | HCV + HCC | Positive | Negative | ≤ A2F4 | A3F4 | A1F4 | A3F4 | |
| n1 (179) | n1 (74) | n1 (105) | n1 (33) | n1 (146) | n1 (80) | n1 (11) | n1 (27) | n1 (11) | |
| E2/E2 | 15 (8.4) | 6 (8.1) | 9 (8.6) | 7 (21.2) | 8 (5.5) | 9 (11.2) | 1 (9.1) | 4 (14.8) | 1 (9.1) |
| E2/E3 | 25 (14.0) | 9 (12.2) | 16 (15.2) | 3 (9.1) | 22 (15.1) | 11 (13.8) | 2 (18.2) | 4 (14.8) | 2 (18.2) |
| E2/E4 | 6 (3.3) | 3 (4.1) | 3 (2.9) | 0 (0.0) | 6 (4.1) | 2 (2.5) | 0 (0.0) | 1 (3.7) | 0 (0.0) |
| E3/E3 | 92 (51.4) | 36 (48.6) | 56 (53.3) | 16 (48.5) | 76 (52.0) | 39 (48.8) | 8 (72.7) | 12 (44.5) | 8 (72.7) |
| E3/E4 | 32 (17.9) | 15 (20.3) | 17 (16.2) | 5 (15.1) | 27 (18.5) | 14 (17.5) | 0 (0.0) | 3 (11.1) | 0 (0.0) |
| E4/E4 | 9 (5.0) | 5 (6.7) | 4 (3.8) | 2 (6.1) | 7 (4.8) | 5 (6.2) | 0 (0.0) | 3 (11.1) | 0 (0.0) |
| APOE alleles | n2 (%) | n2 (%) | n2 (%) | n2 (%) | n2 (%) | n2 (%) | n2 (%) | n2 (%) | n2 (%) |
| E2 | 61 (17.1) | 24 (16.2) | 37 (17.6) | 17 (25.8) | 44 (15.1) | 31 (19.4) | 4 (18.2) | 13 (24.1) | 4 (18.2) |
| E3 | 241 (67.3) | 96 (64.9) | 145 (69.1) | 40 (60.6) | 201 (68.8) | 103 (64.4) | 18 (81.8) | 31 (57.4) | 18 (81.8) |
| E4 | 56 (15.6) | 28 (18.9) | 28 (13.3) | 9 (13.6) | 47 (16.1) | 26 (16.2) | 0(0.0) | 10 (18.5) | 0 (0.0) |
| All | 358 | 148 | 210 | 66 | 292 | 160 | 22 | 54 | 22 |
APOE allele frequencies were associated with liver inflammation based on METAVIR score, assessed in biopsies of liver explants from 89 patients and from 2 patients in pre-OLT. The degree of severe inflammation (A3F4, 0.0%) was significantly less frequent than in patients with minimal and moderate degree of inflammation (≤ A2F4, 16.2%) P = 0.048, in patients carrying the APOE4 allele when compared to non-APOE4. In addition, a significant difference was also found regarding METAVIR score (≤ A2F4, 64.4% vs A3F4, 0.0%; P = 0.043) and (A1F4, 57.4% vs A3F4, 0.0%; P = 0.024) in APOE4 patients compared to APOE3 carriers (Table 3). All patients with advanced liver inflammation (A3) were treated with antivirals only in the post-OLT period.
| METAVIR score | APOE2 | APOE3 | APOE4 | ||||||
| Yes | No | Yes | No | Yes | No | ||||
| n2 (%) | n2 (%) | P value | n2 (%) | n2 (%) | P value | n2 (%) | n2 (%) | P value | |
| METAVIR (n1 = 38; n2 = 76) | |||||||||
| A1F4 (n1 = 27; n2 = 54) | 13 (24.1) | 41 (75.9) | 0.764 | 31 (57.4)3 | 23 (42.6) | 0.064 | 10 (18.5) | 44 (81.5) | 0.055 |
| A3F4 (n1 = 11; n2 = 22) | 4 (18.2) | 18 (81.8) | 18 (81.8) | 4 (18.2) | 0 (0.0)1 | 22 (100.0) | |||
| METAVIR (n1 = 91; n2 = 182) | |||||||||
| ≤ A2F4 (n1 = 80; n2 = 160) | 31 (19.4) | 129 (80.6) | 1.000 | 103 (64.4)4 | 57 (35.6) | 0.148 | 26 (16.2) | 134 (83.8) | 0.048a |
| A3F4 (n1 = 11; n2 = 22) | 4 (18.2) | 18 (81.8) | 18 (81.8) | 4 (18.2) | 0 (0.0)2 | 22 (100.0) | |||
Among patients with less severe liver disease (MELD ≤ 25), the degree of severe inflammation (A3F4, 0.0%) was significantly less frequent in APOE4 carriers when compared to non-APOE4 patients with minimal and moderate degree of inflammation (≤ A2F4, 15.7%, P = 0.046), and with minimal degree of inflammation (A1F4, 18.2%, P = 0.044). These results were also significant in APOE4 patients when compared to APOE3, as categorized by their METAVIR scores [≤ A2F4, 65.7% vs A3F4, 0.0% (P = 0.042) and A1F4, 61.4% vs A3F4, 0.0% (P = 0.040)] (Table 4).
| Variables | MELD≤ 25 | MELD> 25 | MELD≤ 25 | MELD> 25 | ||||||||||
| APOE2 | APOE3 | APOE4 | APOE2 | APOE3 | APOE4 | APOE4 | APOE4 | |||||||
| (Yes) | (No) | (Yes) | (No) | |||||||||||
| n2 (%) | n2 (%) | n2 (%) | P value | n2 (%) | n2 (%) | n2 (%) | P value | n2 (%) | n2 (%) | P value | n2 (%) | n2 (%) | P value | |
| METAVIR (n1 = 38; n2 = 76) | ||||||||||||||
| A1F4 (n1 = 27; n2 = 54) | 9 (20.4) | 27 (61.4)3 | 8 (18.2) | 0.085 | 4 (40.0) | 4 (40.0) | 2 (20.0) | NA | 8 (18.2) | 36 (81.8) | 0.044a | 2 (20.0) | 4 (80.0) | NA |
| A3F4 (n1 = 11; n2 = 22) | 4 (18.2) | 18 (81.8) | 0 (0.0)3 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 22 (100.0) | 0 (0.0) | 0 (0.0) | ||||
| METAVIR (n1 = 91; n2 = 182) | ||||||||||||||
| ≤ A2F4 (n1 = 80; n2 = 160) | 26 (18.6) | 92 (65.7)4 | 22 (15.7) | 0.123 | 5 (25.0) | 11 (55.0) | 4 (20.0) | NA | 22 (15.7) | 118 (84.3) | 0.046a | 4 (20.0) | 16 (80.0) | NA |
| A3F4 (n1 = 11; n2 = 22) | 4 (18.2) | 18 (81.8) | 0 (0.0)4 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 22 (100.0) | 0 (0.0) | 0 (0.0) | ||||
Logistic regression model predicting moderate and severe (A2A3) vs minimal (A1) degree of liver inflammation included as significant predictors male-gender (P = 0.032), and mean-BMI (P = 0.017). In the other analyses, there was no significance without adjustment or with adjustment for potential confounders (MELD, age and BMI) (Table 5).
With respect to the fibrosis degree, using METAVIR scores, of liver grafts in 91 non-APOE4 patients undergoing OLT, the frequency of patients with a moderate degree of fibrosis (F2) was significantly higher in up to 1 year when compared to those between 1 and 5 years (P = 0.006) (Figure 1D). No other significant differences were found (Figure 1). Of note, patients who progressed to moderate (F2) fibrosis in the post-OLT follow-up were treated with antiviral therapy only in the post-OLT period.
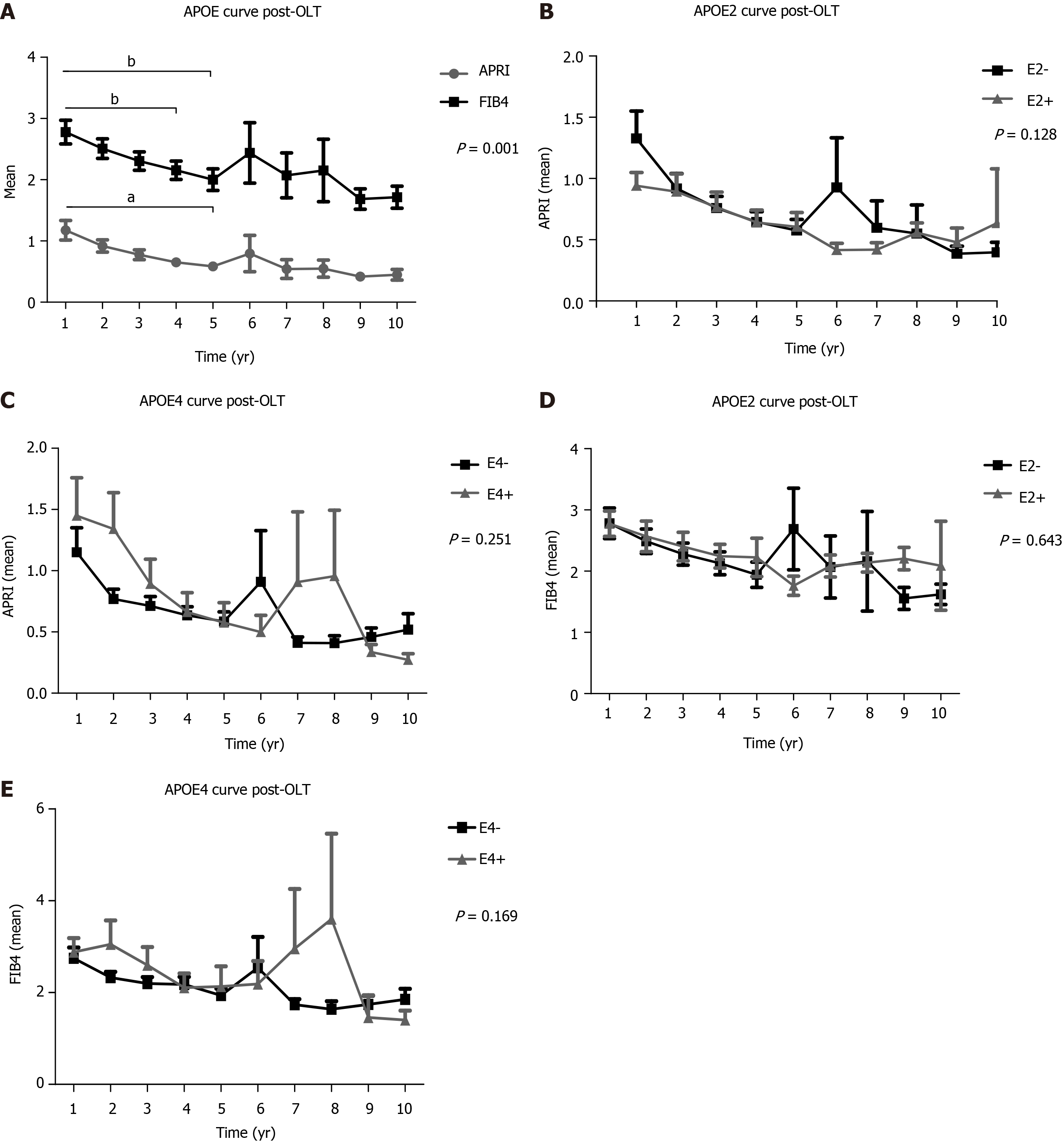
In a 10-year follow-up post-OLT, based on non-invasive tests (APRI and FIB4) of the total transplanted population, a significant higher mean of the 1st year APRI score was found when compared to the 4th and 5th years (P < 0.001), as well as between the 1st year FIB4 and the 5th year scores (P < 0.001) (Figure 2A). APRI and FIB4 scores over the follow-up time did not significantly change regardless of the E2 and E4 alleles (Figure 2).
Increasing evidence associates APOE polymorphisms with progression of chronic liver disease[20,21]. Here, we evaluate the impact of ApoE genetic background in patients with HCV-induced liver cirrhosis, with or without HCC, transplanted or non-transplanted, and with positive or negative viral loads; in particular, in a Brazilian population and with a focus on the influence of E2, E3, and E4 alleles. We related E2, E3, and E4 carriers with the degree of liver inflammation, fibrosis and severity of the disease assessed by MELD scores, using liver biopsy and/or non-invasive indices, such as APRI and FIB4, and the METAVIR score. In addition, we also associated APOE alleles with co-morbidities and lipid blood levels.
As expected, we found E3 the most common APOE allele (67.3%), though slightly lower than the expected 70%-80% seen in the general Brazilian population[22] and in other countries[15,23] The E2 allele frequency was 17.1%, higher than in the Brazilian general population[22] and even greater than the E4 allele (15.6%). This E2 frequency in our study population was also higher than that reported by Wozniak et al[24] (7.7%).
Our liver biopsies from 89 liver explants and from 2 pre-OLT patients showed that cirrhotic E4 carriers were less likely to present with severe inflammation. These results were also evident in patients with MELD score ≤ 25. All these patients with severe hepatic inflammation were treated with antivirals only post-OLT. In agreement, others identified a protective role of APOE4 against severe HCV-related liver damage, when comparing to patients with mild liver disease[24], while another study found that APOE4 allele was under-represented in 996 patients chronically infected with HCV[23]. Other researchers have also noted a higher frequency of the E4 allele among patients with chronic non-cirrhotic hepatitis C, suggesting that the E4 allele is protective against severe HCV infection[25] However, somewhat inconsistent with these findings a 2003 report by Mueller et al[26] was unable to associate the E4 allele in chronic HCV-infected patients with a strong antiviral treatment response, although a later study by Price et al[27] found an association of the E2 and E4 alleles with reduced likelihood of chronic infection in HCV patients.
In 2012, Ahn et al[20] suggested that high ApoE serum levels in patients with liver cirrhosis may be due to liver inflammation. ApoE is known to modulate immune function by inhibiting CD4 and CD8 lymphocyte proliferation, reducing lymphocyte-derived production of IL-2, a key cytokine in regulating lymphocyte differentiation[28]. We speculate that a reduction in ApoE plasma levels, which is recognized for APOE4 carriers[6], could be protective to support OLT and to reduce over-inflammation and fibrosis caused by chronic HCV infection.
In the follow-up of our 144 liver transplanted patients, we identified the E4 allele as protective against the progression of liver fibrosis in 91 (63.2%) recipients. This protection of APOE4 against severe HCV-related liver fibrosis agrees with an early report[24]. Other investigators also showed a benefit of the APOE4 allele against fibrosis progression in liver transplanted patients diagnosed with HCV recurrence. Additionally, it is reported that liver transplanted patients carrying at least one E4 allele may present with reduced graft fibrosis progression during HCV recurrence. Indeed, ApoE polymorphism can be an important tool to monitor fibrosis progression in patients with hepatitis C and normal values of alanine aminotransferase, as there may be competition mechanisms for viral entry and replication in cells[15].
ApoE is a component of several lipoprotein classes and important for lipid transport. ApoE isoforms have several effects on lipoprotein entry into cells, and this mechanism might explain our results, supporting previous investigations in which ApoE4 protects against HCV infection[29,30].
In the follow-up of liver fibrosis progression evaluated between 1 to 10 years post-OLT of our transplanted population, the average score of APRI and FIB4, tended to decrease significantly over the years, implying liver graft survival without progression to fibrosis. Thus, non-invasive methods are now widely used in clinical practice to stage the degree of fibrosis[31,32].
Our findings suggest that APOE4 can be important tool in the medical management of patients following inflammation and liver fibrosis, since the carriage of APOE4 may select patients with a more benign clinical course of liver disease. Of note, patients with a degree of severe inflammation and moderate degree of fibrosis (F2) were cured for HCV only in the post-OLT period.
This study has some limitations: Data from liver transplanted patients were obtained retrospectively from medical records; no data from liver graft donors, including APOE genotypes, were collected. The sample size, may have been insufficient to draw strong, robust conclusions. Isoform studies have previously shown that transplanted donor livers supply > 90% of plasma ApoE[10]. The remainder is synthesized by circulating macrophages and immune cells, or by tissues such as kidney, adipose and muscle, and hence retains the phenotype of the recipient. However, to date, there are no reports of how each source, hepatic ApoE or circulating non-liver ApoE, particularly that of macrophages, might affect the inflammation and fibrosis status of the transplanted liver.
Our results indicate that APOE4 genotype may protect against HCV-induced severe hepatic inflammation and fibrosis in pre- and post-OLT patients. Additionally, the APOE2 allele was over-represented in these patients, suggesting that E2 carriers have increased risk and worse outcomes following HCV infection. Further studies are needed to better understand how ApoE levels via liver and extrahepatic derived sources, and biochemical activities, are affected by donor and recipient genetic backgrounds after liver transplantation.
Hepatitis C virus (HCV) can cause chronic liver inflammation, end-stage liver disease and hepatocellular carcinoma (HCC). Apolipoprotein E (protein: ApoE, gene: APOE) is mainly liver synthesized and APOE polymorphisms may affect HCV-induced liver damage after orthotopic liver transplantation (OLT).
Although APOE4 may protect against HCV-induced liver damage, the role of APOE genotypes in modifying HCV-induced liver injury in post-OLT has not been reported.
To establish if APOE4 genotype OLT recipients have more benign HCV-related liver injuries compared to patients with APOE2 or APOE4 genotypes.
Patients with HCV-related end-stage liver disease, 105 of 179 complicated with HCC, were assessed pre-OLT (179) and post-OLT (144; with a 1-year follow-up for 132 patients). Liver injury analyses included METAVIR and models for end-stage liver disease scores, while APOE genotype was determined by qRT-PCR.
HCV positive recipients with severe hepatic inflammation had low APOE4 genotype frequency, compared to those with minimal and moderate inflammation. In addition, liver fibrosis was lower in patients carrying APOE4 genotype compared to those carrying the most common APOE3 genotype.
We found that carriage of APOE4 genotype protects pre-OLT patients against HCV-induced severe hepatic inflammation, and against fibrosis progression post-OLT.
We propose that carriage of APOE4 genotype protects against progression of inflammation and liver fibrosis in recurrent HCV hepatitis after OLT, but additional studies are needed to assess whether donor-derived ApoE4 protein directly affects these processes.
The authors deeply thank patients and family members for agreeing to participate in this study and also staff and technicians from study hospitals for their support.
Manuscript source: Invited manuscript
Specialty type: Transplantation
Country/Territory of origin: Brazil
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Hilmi IA, Zhang L, Zheng H S-Editor: Zhang H L-Editor: A P-Editor: Wang LL
| 1. | World Health Organization. Global hepatitis report. 2017. [cited 19 December 2020]. Available from: https://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/. |
| 2. | Fauteux-Daniel S, Larouche A, Calderon V, Boulais J, Béland C, Ransy DG, Boucher M, Lamarre V, Lapointe N, Boucoiran I, Le Campion A, Soudeyns H. Vertical Transmission of Hepatitis C Virus: Variable Transmission Bottleneck and Evidence of Midgestation In Utero Infection. J Virol. 2017;91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 3. | Westbrook RH, Dusheiko G. Natural history of hepatitis C. J Hepatol. 2014;61:S58-S68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 552] [Cited by in RCA: 616] [Article Influence: 56.0] [Reference Citation Analysis (0)] |
| 4. | Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57:1333-1342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1770] [Cited by in RCA: 1847] [Article Influence: 153.9] [Reference Citation Analysis (3)] |
| 5. | Toshikuni N, Arisawa T, Tsutsumi M. Hepatitis C-related liver cirrhosis - strategies for the prevention of hepatic decompensation, hepatocarcinogenesis, and mortality. World J Gastroenterol. 2014;20:2876-2887. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 22] [Cited by in RCA: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 6. | Mahley RW. Apolipoprotein E: from cardiovascular disease to neurodegenerative disorders. J Mol Med (Berl). 2016;94:739-746. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 311] [Cited by in RCA: 354] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 7. | Tudorache IF, Trusca VG, Gafencu AV. Apolipoprotein E - A Multifunctional Protein with Implications in Various Pathologies as a Result of Its Structural Features. Comput Struct Biotechnol J. 2017;15:359-365. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 127] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 8. | Mahley RW, Rall SC Jr. Apolipoprotein E: far more than a lipid transport protein. Annu Rev Genomics Hum Genet. 2000;1:507-537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1164] [Cited by in RCA: 1270] [Article Influence: 55.2] [Reference Citation Analysis (0)] |
| 9. | Huebbe P, Rimbach G. Evolution of human apolipoprotein E (APOE) isoforms: Gene structure, protein function and interaction with dietary factors. Ageing Res Rev. 2017;37:146-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 122] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 10. | Nascimento JCR, Matos GA, Pereira LC, Mourão AECCB, Sampaio AM, Oriá RB, Toniutto P. Impact of apolipoprotein E genetic polymorphisms on liver disease: An essential review. Ann Hepatol. 2020;19:24-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 11. | Saunders AM, Trowers MK, Shimkets RA, Blakemore S, Crowther DJ, Mansfield TA, Wallace DM, Strittmatter WJ, Roses AD. The role of apolipoprotein E in Alzheimer's disease: pharmacogenomic target selection. Biochim Biophys Acta. 2000;1502:85-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 52] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 12. | Pereira LC, Nascimento JCR, Rêgo JMC, Canuto KM, Crespo-Lopez ME, Alvarez-Leite JI, Baysan A, Oriá RB. Apolipoprotein E, periodontal disease and the risk for atherosclerosis: a review. Arch Oral Biol. 2019;98:204-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 13. | Linard M, Letenneur L, Garrigue I, Doize A, Dartigues JF, Helmer C. Interaction between APOE4 and herpes simplex virus type 1 in Alzheimer's disease. Alzheimers Dement. 2020;16:200-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 97] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 14. | Kuo CL, Pilling LC, Atkins JL, Masoli JAH, Delgado J, Kuchel GA, Melzer D. APOE e4 Genotype Predicts Severe COVID-19 in the UK Biobank Community Cohort. J Gerontol A Biol Sci Med Sci. 2020;75:2231-2232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 249] [Cited by in RCA: 266] [Article Influence: 53.2] [Reference Citation Analysis (0)] |
| 15. | Fabris C, Vandelli C, Toniutto P, Minisini R, Colletta C, Falleti E, Smirne C, Pirisi M. Apolipoprotein E genotypes modulate fibrosis progression in patients with chronic hepatitis C and persistently normal transaminases. J Gastroenterol Hepatol. 2011;26:328-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 16. | Calero O, Hortigüela R, Bullido MJ, Calero M. Apolipoprotein E genotyping method by real time PCR, a fast and cost-effective alternative to the TaqMan and FRET assays. J Neurosci Methods. 2009;183:238-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 75] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 17. | Mazzaferro V, Regalia E, Doci R, Andreola S, Pulvirenti A, Bozzetti F, Montalto F, Ammatuna M, Morabito A, Gennari L. Liver transplantation for the treatment of small hepatocellular carcinomas in patients with cirrhosis. N Engl J Med. 1996;334:693-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5110] [Cited by in RCA: 5312] [Article Influence: 183.2] [Reference Citation Analysis (0)] |
| 18. | Yao FY, Ferrell L, Bass NM, Watson JJ, Bacchetti P, Venook A, Ascher NL, Roberts JP. Liver transplantation for hepatocellular carcinoma: expansion of the tumor size limits does not adversely impact survival. Hepatology. 2001;33:1394-1403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1594] [Cited by in RCA: 1695] [Article Influence: 70.6] [Reference Citation Analysis (0)] |
| 19. | Yen YH, Kuo FY, Kee KM, Chang KC, Tsai MC, Hu TH, Lu SN, Wang JH, Hung CH, Chen CH. APRI and FIB-4 in the evaluation of liver fibrosis in chronic hepatitis C patients stratified by AST level. PLoS One. 2018;13:e0199760. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 48] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 20. | Ahn SJ, Kim DK, Kim SS, Bae CB, Cho HJ, Kim HG, Kim YJ, Lee JH, Lee HJ, Lee MY, Kim KB, Cho JH, Cho SW, Cheong JY. Association between apolipoprotein E genotype, chronic liver disease, and hepatitis B virus. Clin Mol Hepatol. 2012;18:295-301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 21. | Shen Y, Li M, Ye X, Bi Q. Association of apolipoprotein E with the progression of hepatitis B virus-related liver disease. Int J Clin Exp Pathol. 2015;8:14749-14756. [PubMed] |
| 22. | Fuzikawa AK, Peixoto SV, Taufer M, Moriguchi EH, Lima-Costa MF. Apolipoprotein E polymorphism distribution in an elderly Brazilian population: the Bambuí Health and Aging Study. Braz J Med Biol Res. 2007;40:1429-1434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 23. | Mueller T, Fischer J, Gessner R, Rosendahl J, Böhm S, van Bömmel F, Knop V, Sarrazin C, Witt H, Marques AM, Kovacs P, Schleinitz D, Stumvoll M, Blüher M, Bugert P, Schott E, Berg T. Apolipoprotein E allele frequencies in chronic and self-limited hepatitis C suggest a protective effect of APOE4 in the course of hepatitis C virus infection. Liver Int. 2016;36:1267-1274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 24. | Wozniak MA, Itzhaki RF, Faragher EB, James MW, Ryder SD, Irving WL; Trent HCV Study Group. Apolipoprotein E-epsilon 4 protects against severe liver disease caused by hepatitis C virus. Hepatology. 2002;36:456-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 132] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 25. | Teama SHH, Agwa S, Makhlouf M, Nashaat E, Sayed M, Yousry W, Mansour A, Ibrahim W, Elshafie A. Apolipoprotein-E gene polymorphism and possible role of ApoE e4 allele with a lower probability of progression to HCV-related liver cirrhosis in Egyptian patients. Merit Res J Med Med Sci. 2016;4:440-447. |
| 26. | Mueller T, Gessner R, Sarrazin C, Graf C, Halangk J, Witt H, Köttgen E, Wiedenmann B, Berg T. Apolipoprotein E4 allele is associated with poor treatment response in hepatitis C virus (HCV) genotype 1. Hepatology. 2003;38:1592; author reply 1592-1592; author reply 1593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 26] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 27. | Price DA, Bassendine MF, Norris SM, Golding C, Toms GL, Schmid ML, Morris CM, Burt AD, Donaldson PT. Apolipoprotein epsilon3 allele is associated with persistent hepatitis C virus infection. Gut. 2006;55:715-718. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 73] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 28. | Kelly ME, Clay MA, Mistry MJ, Hsieh-Li HM, Harmony JA. Apolipoprotein E inhibition of proliferation of mitogen-activated T lymphocytes: production of interleukin 2 with reduced biological activity. Cell Immunol. 1994;159:124-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 99] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 29. | Wozniak MA, Lugo Iparraguirre LM, Dirks M, Deb-Chatterji M, Pflugrad H, Goldbecker A, Tryc AB, Worthmann H, Gess M, Crossey MM, Forton DM, Taylor-Robinson SD, Itzhaki RF, Weissenborn K. Apolipoprotein E-ε4 deficiency and cognitive function in hepatitis C virus-infected patients. J Viral Hepat. 2016;23:39-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 30. | Weller R, Hueging K, Brown RJP, Todt D, Joecks S, Vondran FWR, Pietschmann T. Hepatitis C Virus Strain-Dependent Usage of Apolipoprotein E Modulates Assembly Efficiency and Specific Infectivity of Secreted Virions. J Virol. 2017;91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 31. | McGoogan KE, Smith PB, Choi SS, Berman W, Jhaveri R. Performance of the AST-to-platelet ratio index as a noninvasive marker of fibrosis in pediatric patients with chronic viral hepatitis. J Pediatr Gastroenterol Nutr. 2010;50:344-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 52] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 32. | Karic U, Pesic-Pavlovic I, Stevanovic G, Korac M, Nikolic N, Radovanovic-Spurnic A, Barac A, Mitrovic N, Markovic A, Markovic M, Petkovic A, Ostojic I, Perunicic S, Kekic N, Glidzic M, Djonin-Nenezic M, Brmbolic B, Milosevic I. FIB-4 and APRI scores for predicting severe fibrosis in chronic hepatitis C - a developing country's perspective in DAA era. J Infect Dev Ctries. 2018;12:178-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |