Published online May 14, 2020. doi: 10.3748/wjg.v26.i18.2203
Peer-review started: December 31, 2019
First decision: January 19, 2020
Revised: March 27, 2020
Accepted: April 17, 2020
Article in press: April 17, 2020
Published online: May 14, 2020
Processing time: 134 Days and 16.4 Hours
Folic acid has been shown to improve non-alcoholic steatohepatitis (NASH), but its roles in hepatic lipid metabolism, hepatic one-carbon metabolism, and gut microbiota are still unknown.
To demonstrate the role of folic acid in lipid metabolism and gut microbiota in NASH.
Twenty-four Sprague-Dawley rats were assigned into three groups: Chow diet, high-fat diet (HFD), and HFD with folic acid administration. At the end of 16 wk, the liver histology, the expression of hepatic genes related to lipid metabolism, one-carbon metabolism, and gut microbiota structure analysis of fecal samples based on 16S rRNA sequencing were measured to evaluate the effect of folic acid. Palmitic acid-exposed Huh7 cell line was used to evaluate the role of folic acid in hepatic lipid metabolism.
Folic acid treatment attenuated steatosis, lobular inflammation, and hepatocellular ballooning in rats with HFD-induced steatohepatitis. Genes related to lipid de novo lipogenesis, β-oxidation, and lipid uptake were improved in HFD-fed folic acid-treated rats. Furthermore, peroxisome proliferator-activated receptor alpha (PPARα) and silence information regulation factor 1 (SIRT1) were restored by folic acid in HFD-fed rats and palmitic acid-exposed Huh7 cell line. The restoration of PPARα by folic acid was blocked after transfection with SIRT1 siRNA in the Huh7 cell line. Additionally, folic acid administration ameliorated depleted hepatic one-carbon metabolism and restored the diversity of the gut microbiota in rats with HFD-induced steatohepatitis.
Folic acid improves hepatic lipid metabolism by upregulating PPARα levels via a SIRT1-dependent mechanism and restores hepatic one-carbon metabolism and diversity of gut microbiota, thereby attenuating HFD-induced NASH in rats.
Core tip: The roles of folic acid in hepatic lipid metabolism, hepatic one-carbon metabolism, and gut microbiota in high-fat diet (HFD)-induced steatohepatitis are still unknown. This study confirmed that folic acid ameliorated HFD-induced steatohepatitis by restoring PPARα levels via a SIRT1 dependent mechanism. Moreover, folic acid restored depleted hepatic one-carbon metabolism and the diversity of gut microbiota. All these findings further clarified the improvement effect of folic acid on HFD-induced steatohepatitis and suggested that folic acid may become a therapeutic drug to treat non-alcoholic fatty liver disease in the future.
- Citation: Xin FZ, Zhao ZH, Zhang RN, Pan Q, Gong ZZ, Sun C, Fan JG. Folic acid attenuates high-fat diet-induced steatohepatitis via deacetylase SIRT1-dependent restoration of PPARα. World J Gastroenterol 2020; 26(18): 2203-2220
- URL: https://www.wjgnet.com/1007-9327/full/v26/i18/2203.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i18.2203
Non-alcoholic fatty liver disease (NAFLD) has become one of the main causes of chronic liver disease worldwide[1]. The prevalence of NAFLD in China has increased from 18% to 29% in the past ten years[2,3]. Similar trends have been observed in other parts of world. Non-alcoholic steatohepatitis (NASH), which is a subtype of NAFLD, increases the risk of cirrhosis, hepatocellular carcinoma, and liver-related death[4]. However, there are still no drugs approved for treatment of NASH[5]. Therefore, NAFLD has become a serious global health burden and it is critical to find new drug targets for treatment of NASH.
Folic acid is an important substrate for the synthesis of methyl donors as an essential water-soluble vitamin metabolized by the intestinal flora and the human body[6]. Dietary folic acid could be absorbed and metabolized through the small intestine and liver. Finally, 5-methyltetrahydrofolic acid (5-MTHF) is the active form in blood circulation[7]. Folic acid deficiency could induce hyperhomocysteinemia and NAFLD. Dietary folic acid is essential for whole body folate homeostasis[8]. Additional folic acid supplementation could attenuate liver injury under high-fat diet (HFD)-fed or binge drinking conditions[9,10]. Dietary folic acid has been shown to ameliorate liver lipid accumulation[11-13]. All present data indicates that folic acid may become a potential drug target for treatment of NASH. However, further molecular mechanisms of folic acid on hepatic lipid and one-carbon unit metabolism are still unclear. The effect of folic acid on gut microbiota in NASH is also unknown. Taken together, it is necessary to further access the effect of folic acid on NASH and its possible mechanism.
To address the problems mentioned above, we conducted this research in HFD-induced NASH rats and palmitic acid (PA)-treated Huh7 cell line. Liver histology, hepatic one-carbon metabolism, and gut microbiota were evaluated in vivo to investigate the effect of folic acid in NASH. Genes related to lipid metabolism were evaluated both in vivo and vitro to illustrate the role of folic acid in hepatic lipid metabolism in NASH.
The animal experiments were performed in a way that discomfort for animals was minimized. A total of 24 six-week-old specific-pathogen-free (SPF) male Sprague-Dawley rats (Sippurbec Laboratory Animal Co., Ltd., Shanghai, China) were fed in a controlled environment (24 ± 1 °C, 50% ± 5% humidity, 12-h light-dark cycle, free access to water and standard chow diet). After 1 wk of adaptive feeding, the rats were fed a chow diet or HFD (88% standard diet, 10% lard, and 2% cholesterol) for 8 wk. Then, rats fed an HFD were randomly divided into two groups and fed folic acid (15 mg/kg·d) or saline by gavage once daily for 8 wk. All rats were fasted overnight and then euthanized with pentobarbital sodium at the end of 16 wk.
All animal experiments followed the National Research Council’s Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee of SHRM (SHRM-IACUC-015).
Fecal samples from rats were collected immediately upon defecation and then stored at -80 °C after being snap frozen in liquid nitrogen. Total fecal DNA was extracted using a TIANamp DNA Kit (Tiangen, Beijing, China) according to the manufacturer’s protocol. The quality and quantity of DNA were verified with a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, United States) and agarose gel. Extracted DNA was diluted to a concentration of 1 ng/µl and stored at -20 °C until further processing. The V4-V5 variable regions of 16S rRNA genes were amplified with universal primers 515F and 907R for bacterial diversity analysis. Amplicons were purified with the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, United States) and quantified using QuantiFluor™-ST (Promega, Wisconsin, United States) according to the manufacturer’s instructions. Equal amounts of purified amplicon were pooled for subsequent sequencing. Raw sequencing data were given in FASTQ format. Paired-end reads were preprocessed using Trimmomatic software. Clean reads were subjected to primer sequence removal and clustering to generate operational taxonomic units using Vsearch software with a 97% similarity cutoff. All representative reads were annotated and blasted against the Silva database using the Ribosomal Database Project classifier (confidence threshold was 70%).
The body weight and liver mass were recorded after the rats were euthanized. Approximately 1.0 cm × 1.0 cm × 1.5 cm liver tissues were fixed in 4% paraformaldehyde for hematoxylin-eosin (HE), Masson, and Sirius red staining. Approximately 1.0 cm × 1.0 cm × 1.0 cm liver tissues were snap frozen in liquid nitrogen and then frozen at -80 °C for oil red O staining. The other liver tissues were stored at -80 °C for further analyses. Steatosis (S), activity (A), and fibrosis (F) (SAF) score was used for analyzing hepatic histological alterations[14]. Approximately 0.5 cm-long sections of the terminal ileum were gently rinsed with phosphate-buffered saline and then fixed in 4% paraformaldehyde for HE and immunohistochemical staining.
Serum was obtained by centrifugation of whole blood at 3000 r/min at 4 °C. Serum folic acid, alanine aminotransferase (ALT), aspartate aminotransferase (AST), fasting blood glucose (FBG), triglycerides (TG), total cholesterol (TC), high-density lipoprotein (HDL), low-density lipoprotein (LDL), total bile acid (TBA), and homocysteine (Hcy) were measured with an automated analyzer (Sysmex CHEMIX-180, Japan). The liver TG and cholesterol levels were measured with assay kits (Applygen Technologies Inc., Beijing, China). Samples and the standard curve were measured according to the manufacturer’s instructions.
Total RNA was extracted from liver tissue using TRIzol (D9108B, Takara, Dalian, China). The concentration and purity of RNA samples were assessed on a NanoDrop 2000 spectrophotometer (Nanodrop Technologies). Total RNA (1000 ng) was converted to cDNA with RT master mix (RR036A, Takara, Dalian, China). Real-time quantitative polymerase chain reaction (qRT-PCR) was performed with the Applied Biosystems Vii7 with SYBR® Green Master Mix (Low Rox Plus) (11202ES08, YEASEN, Shanghai, China). The primer sequences are shown in Table 1. The specificity of the primers was determined by dissociation curves using Vii7 system SDS software. RPS18 (B661201-0001, Sangon Biotech) was used as the internal control. The 2-ΔΔCT method was used to analyze relative gene expression.
| Gene name | Forward sequence | Reverse sequence |
| TNF-α | TGCCTCAGCCTCTTCTCATT | GAGCCCATTTGGGAACTTCT |
| IL-6 | AGTTGCCTTCTTGGGACTGA | CCTCCGACTTGTGAAGTGGT |
| IL-1β | GAAGTCAAGACCAAAGTGG | TGAAGTCAACTATGTCCCG |
| CCR2 | CACCGTATGACTATGATGATG | CAGGAGAGCAGGTCAGAGAT |
| p47phox | GCCCAAAGATGGCAAGAATA | ATGACCTCAATGGCTTCACC |
| p67phox | AGCAGAAGAGCAGTTAGCATTGG | TGCTTTCCATGGCCTTGTC |
| p22phox | GTAGATGCCGCTCGCAATGGCCAG | ATGGGGCAGATCGAGTGGGCCATGT |
| gp91phox | CTGAGCGAATTGTACGTG | CTTATCACAGCCACAAGC |
| αSMA | TGTGCTATGTCGCTCTGGAC | CCAATGAAAGATGGCTG GAA |
| TGFβ1 | ATTCCTGGCGTTACCTTGG | AGCCCTGTATTCCGTCTCCT |
| Col1a1 | TGTTCAGCTTTGTGGACCT | CAGCTGACTTCAGGGATGT |
| Col2a1 | ACCTCAGGGTGTTCAAGGTG | CGGATTCCAATAGGACCAGA |
| Col3a1 | GGTGGCTTTCAGTTCAGCTATG | GTCTTGCTCCATTCACCAGTGT |
| MAT1A | CAATGTGCTCGTGGCTCTGGAG | TCCTCTGTCTCGTCAGTGGCATAG |
| ALDH1L1 | GCACGGCTCCATCATCTACCATC | GTCATCTGGAAGCACCTCACACTC |
| SREBP1c | CCAGCCTTTGAGGATAACCA | TGCAGGTCAGACACAGGAAG |
| SCD | AGCTGGTGATGTTCCAGAGG | CAAGAAGGTGCTGACGAACA |
| ACACA | GAATATCCAGATGGCCGAGA | CCTTCTGCTCTGGCAAGTTC |
| FASN | GCCTAACACCTCTGTGCAGT | GGCAATACCCGTTCCCTGAA |
| PPARγ | ACAAGAGCTGACCCAATGGT | GGCTCTTCATGTGGCCTGTT |
| ACADL | ACTCCGCCTCCGCTTCCATG | TACCACCGTAGATCGGCTGAACTC |
| FABP1 | GTCTGCCTGAGGACCTCATCCAG | TCATGGTCTCCAGTTCGCACTCC |
| CPT1α | CCACGAAGCCCTCAAACAGA | CACACCCACCACCACGATAA |
| FATP2 | CACGACAGAGTTGGAGACACCTTC | CCGATGCGACCTTCATGACCTG |
Protein levels of methionine adenosyltransferase 1A (MAT1A), silence information regulation factor 1 (SIRT1), peroxisome proliferator-activated receptor alpha (PPARα), carnitine palmitoyltransferase 1A (CPT1α), and fatty acid binding protein 1 (FABP1) in rat liver and SIRT1 and PPARα in the Huh7 cell line were determined by Western blot analysis. Briefly, liver proteins (45 µg) and cell proteins (15 µg) were separated by 8%, 10%, or 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and then proteins were transferred from the gel to a polyvinylidene fluoride (PVDF) membrane under constant current, cold conditions. The membranes were blocked with Quick-block buffer (P0252, Beyotime, Shanghai, China) for 25 min at room temperature and were then incubated with primary antibody overnight at 4 °C. Primary antibodies include anti-MAT1A polyclonal antibody (AB217005, Abscitech, United States), anti-SIRT1 monoclonal antibody (189494, Abcam, Cambridge, United Kingdom), anti-PPARα polyclonal antibody (A6697, Abclonal, Wuhan, China), anti-CPT1α polyclonal antibody (128568, Abcam), anti-FABP1 polyclonal antibody (A5311, Abclonal), and anti-GAPDH monoclonal antibody (#5147, CST, United States). Horseradish peroxidase (HRP)-conjugated anti-rabbit or anti-mouse IgG (Beyotime) were used as secondary antibodies, and the membranes were incubated at room temperature for 1 h. Protein bands were detected using a Western chemiluminescent HRP substrate (Millipore Corporation, Billerica, MA, United States).
The Huh7 cell line was obtained from American Type Culture Collection (ATCC; Manassas, VA, United States) and was cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM; HyClone) supplemented with 10% fetal bovine serum (Gibco, CA, United States). PA powder (Sigma, St. Louis, United States) was dissolved in 1% fatty acid-free BSA (Sigma, St. Louis, United States) Milli-Q water at 70 °C and filtrated with a 0.22 μm filter. The concentration of the stock solution was 5 mmol/L. The concentration of working PA solution was 0.3 mmol/L. The intervention included 0.3 mmol/L PA and 1 or 10 µg/mL 5-MTHF (Sigma-Aldrich, United States). Briefly, after 12 h of serum-free treatment, cells with 5-MTHF or the same amount of phosphate-buffered saline were incubated as pretreatment for 12 h, and then cells were incubated in PA with or without 5-MTHF for another 12 h. The proteins were isolated according to the manufacturer’s instructions.
The Huh7 cell line was transfected with 50 nmol/L SIRT1 siRNA (Genomeditech, Shanghai, China) or its negative control (NC; Genomeditech) with Lipofectamine 3000 (Invitrogen, Carlsbad, United States) in Opti-MEM medium (Gibco, CA, United States). After 18 h, the medium was replaced by high-glucose DMEM without fetal bovine serum. Pretreatment and intervention were performed 24 h after transfection.
All the data are expressed as the mean ± SE. The results were analyzed using two-tailed Student’s t-test between two groups and one-way ANOVA followed by Dunnett’s test among multiple groups. Nonparametric tests were used for discontinuous data. P < 0.05 was considered statistically significant. All the statistical methods mentioned above were reviewed by Guang-Yu Chen from the Clinical Epidemiology Center, Shanghai Jiao Tong University.
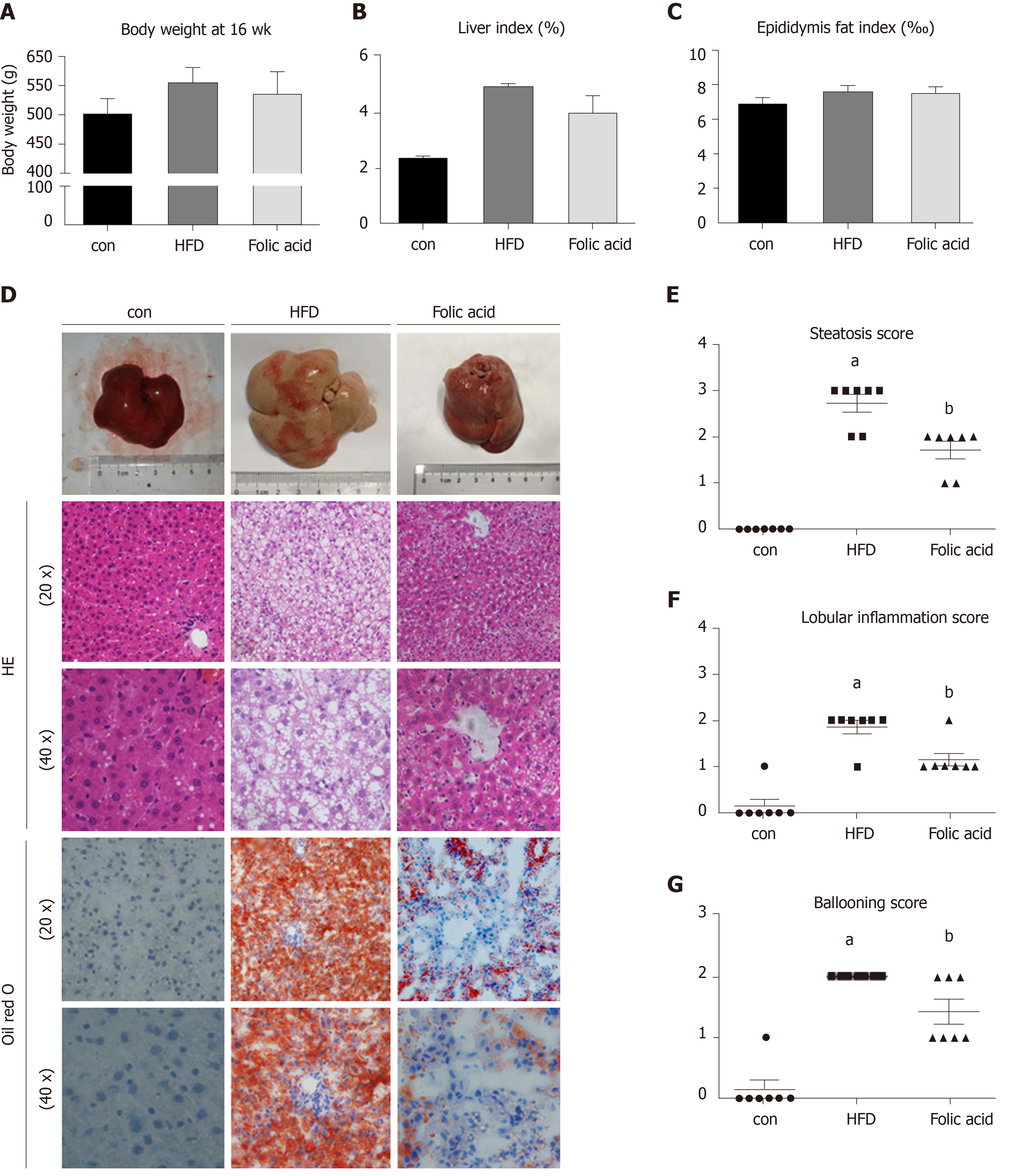
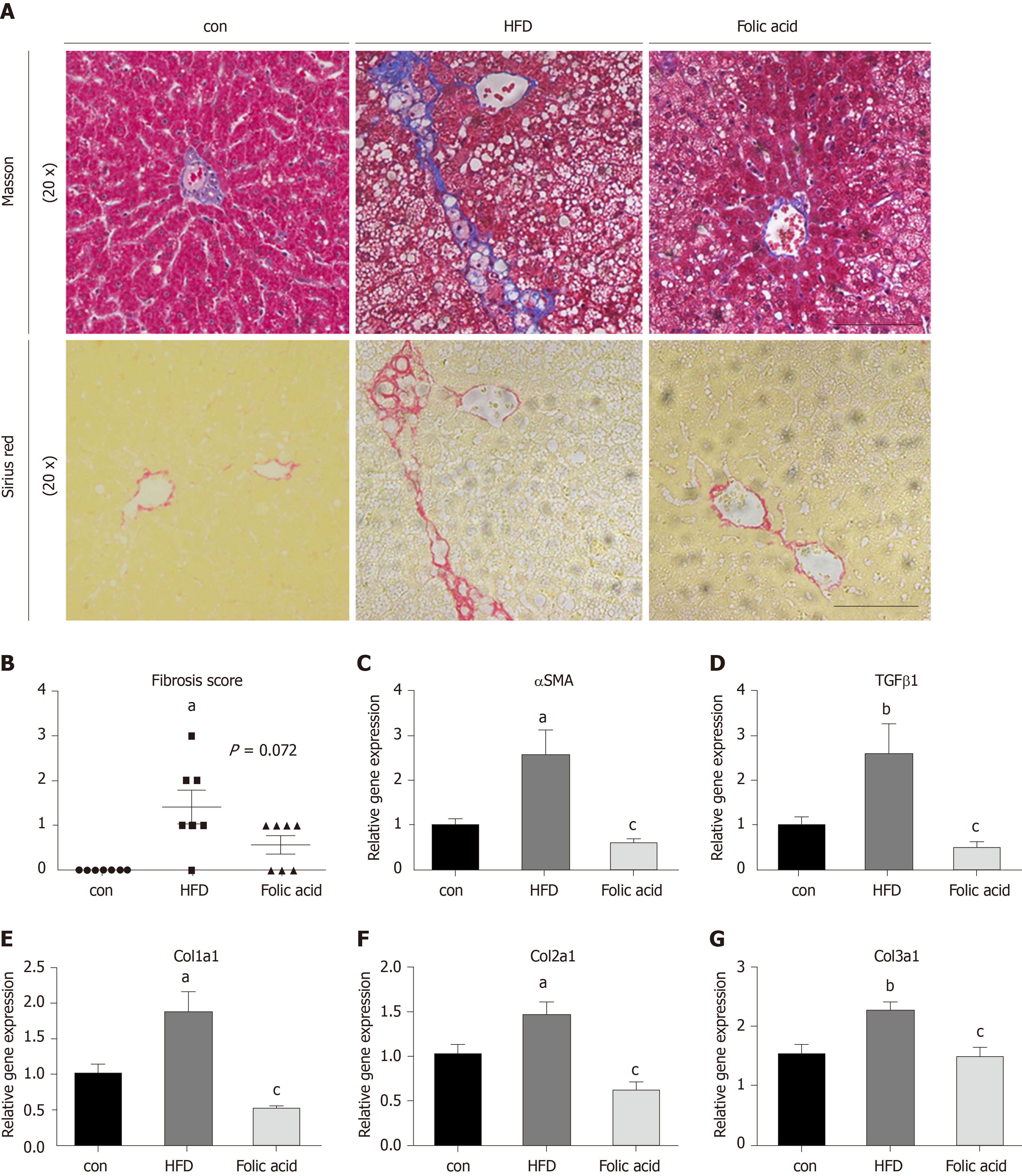
After a 16-wk experimental period, all rats in the HFD group developed typical NASH characteristics. Body weight (Figure 1A) and liver index (Figure 1B) were significantly elevated in the HFD group compared with the control group. Administration of folic acid had no effect on body weight or epididymis fat of rats (Figure 1A and C). But it ameliorated HFD-induced NASH hepatic lesions in rats. As shown in Figure 1B, liver index showed a certain reduction in the folic acid group compared with the HFD group. Additionally, folic acid improved the liver imaging results to a certain extent and ameliorated hepatic lipid deposition, ballooning degeneration, and inflammatory infiltration (Figure 1D). Moreover, steatosis score (Figure 1E), lobular inflammation score (Figure 1F), and ballooning score (Figure 1G) were much lower after folic acid intervention. The rats fed an HFD for 16 wk showed bridging fibrosis through Masson and Sirius red staining (Figure 2A). Treatment with folic acid resulted in less severe fibrosis based on the pathological sections. Furthermore, folic acid downregulated the expression levels of α-smooth muscle actin (Figure 2C), transforming growth factor beta 1 (Figure 2D), collagen type I alpha 1 (Figure 2E), collagen type II alpha 1 (Figure 2F), and collagen type III alpha 1 (Figure 2G). Although folic acid could reduce the fibrosis score, the difference did not reach statistical significance (P = 0.072, Figure 2B).
Rats in the HFD group showed significant dyslipidemia. Serum ALT (P < 0.01), AST (P < 0.01), FBG (P < 0. 01), TG (P < 0.01), TC (P < 0.01), and LDL (P < 0.01) levels were significantly elevated compared with those in the control group, accompanied by lower HDL (P < 0.01) levels (Table 2). The folic acid group showed a significant reduction in FBG (P < 0.01), TG (P < 0.01), TC (P < 0.01), and LDL (P < 0.01) levels. However, there was no significant difference in HDL levels between the HFD and folic acid groups. Abnormal bile acid metabolism and Hcy metabolism were detected in the HFD group. HFD rats had higher TBA (P < 0.01) and Hcy (P < 0.01) levels than the control group. Folic acid significantly reduced serum TBA (P < 0.05) and Hcy (P < 0.01) levels compared with those in the HFD group (Table 2). The results above suggested that folic acid ameliorates HFD-induced hepatic lipid accumulation, inflammation, and fibrosis.
| Control | HFD | Folic acid | |
| ALT (U/L) | 38.50 ± 1.58 | 134.0 ± 8.02b | 82.13 ± 7.19d |
| AST (U/L) | 88.00 ± 4.39 | 225.4 ± 10.57b | 176.3 ± 15.3d |
| FBG (mmol/L) | 10.45 ± 0.66 | 13.33 ± 0.40b | 7.83 ± 0.30d |
| TG (mmol/L) | 0.57 ± 0.04 | 0.76 ± 0.04b | 0.44 ± 0.02d |
| TC (mmol/L) | 1.13 ± 0.04 | 2.26 ± 0.12b | 1.48 ± 0.04d |
| HDL (mmol/L) | 0.94 ± 0.03 | 0.75 ± 0.05b | 0.74 ± 0.04 |
| LDL (mmol/L) | 0.24 ± 0.02 | 1.37 ± 0.10b | 0.89 ± 0.03d |
| TBA (µmol/L) | 37.5 ± 5.57 | 68 ± 7.49b | 44.17 ± 3.92c |
| Hcy (µmol/L) | 7.35 ± 0.29 | 13.05 ± 0.52b | 11.17 ± 0.42d |
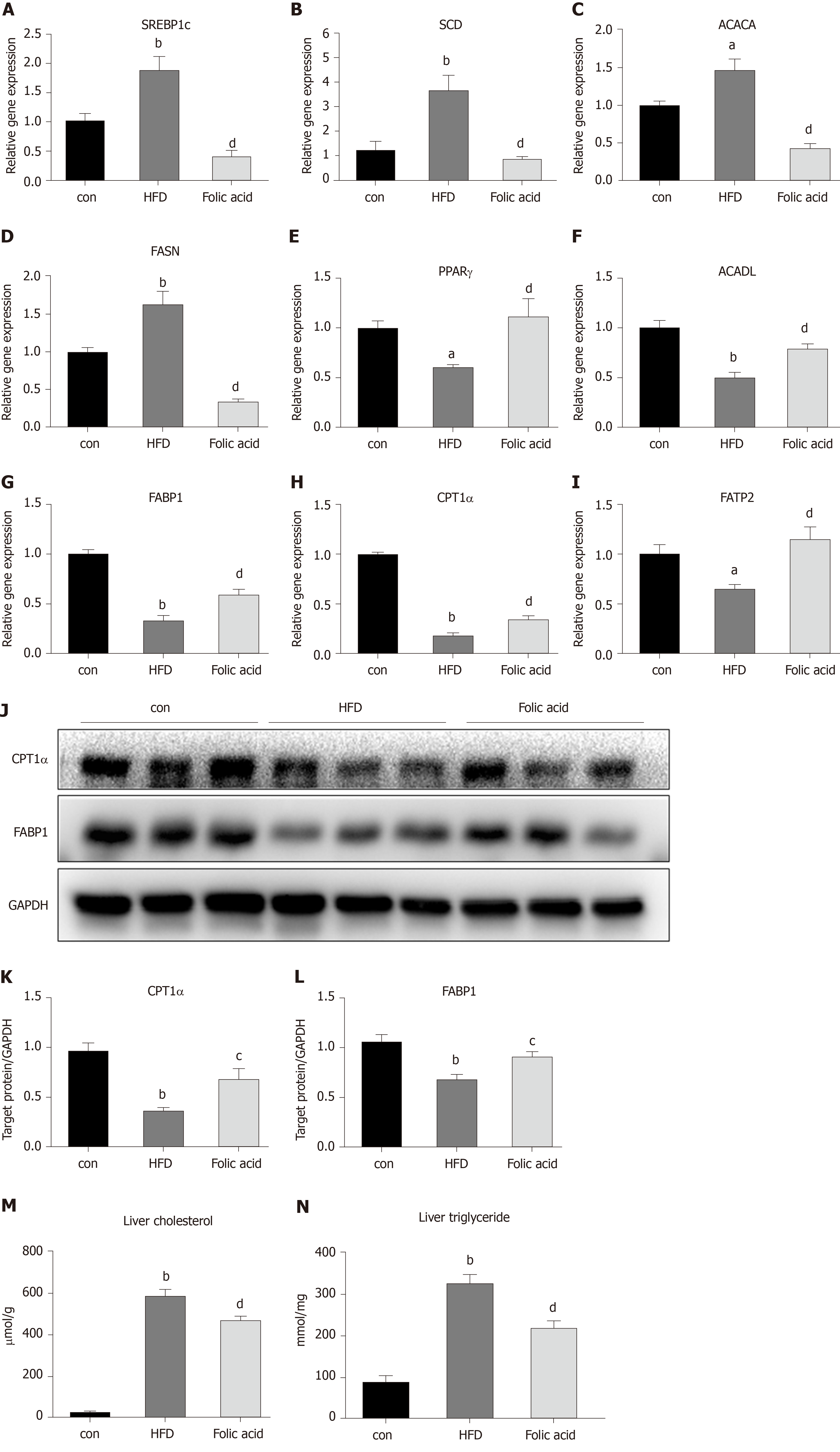
Abnormal hepatic lipid uptake, de novo lipogenesis (DNL), and β-oxidation contribute to the progression of NAFLD[15]. To further characterize the effects of folic acid on hepatic lipid metabolism in HFD-induced NASH rats, we analyzed the expression levels of genes related to DNL, β-oxidation, and lipid uptake. As shown in Figure 3A-D, folic acid significantly downregulated the expression levels of sterol regulatory element binding transcription protein 1c, stearoyl-CoA desaturase, acetyl-CoA carboxylase, and fatty acid synthase. Moreover, genes related to hepatic lipid β-oxidation and lipid uptake such as PPARγ (Figure 3E), long-chain specific acyl-CoA dehydrogenase (Figure 3F), FABP1 (Figure 3G), CPT1α (Figure 3H), and fatty acid transport protein 2 (Figure 3I) were elevated after folic acid administration. To further confirm the ameliorative effect of folic acid on hepatic lipid β-oxidation. We also detected the expression levels of related genes at the protein level. As shown in Figure 3J-L, CPT1α, and FABP1 levels were strikingly reduced by HFD and significantly restored by folic acid intervention. Furthermore, liver cholesterol (Figure 3M) and triglyceride (Figure 3N) levels were reduced in the folic acid group compared with the HFD group. This part of results suggested that folic acid improves abnormal hepatic lipid metabolism and then reduces hepatic lipid accumulation.
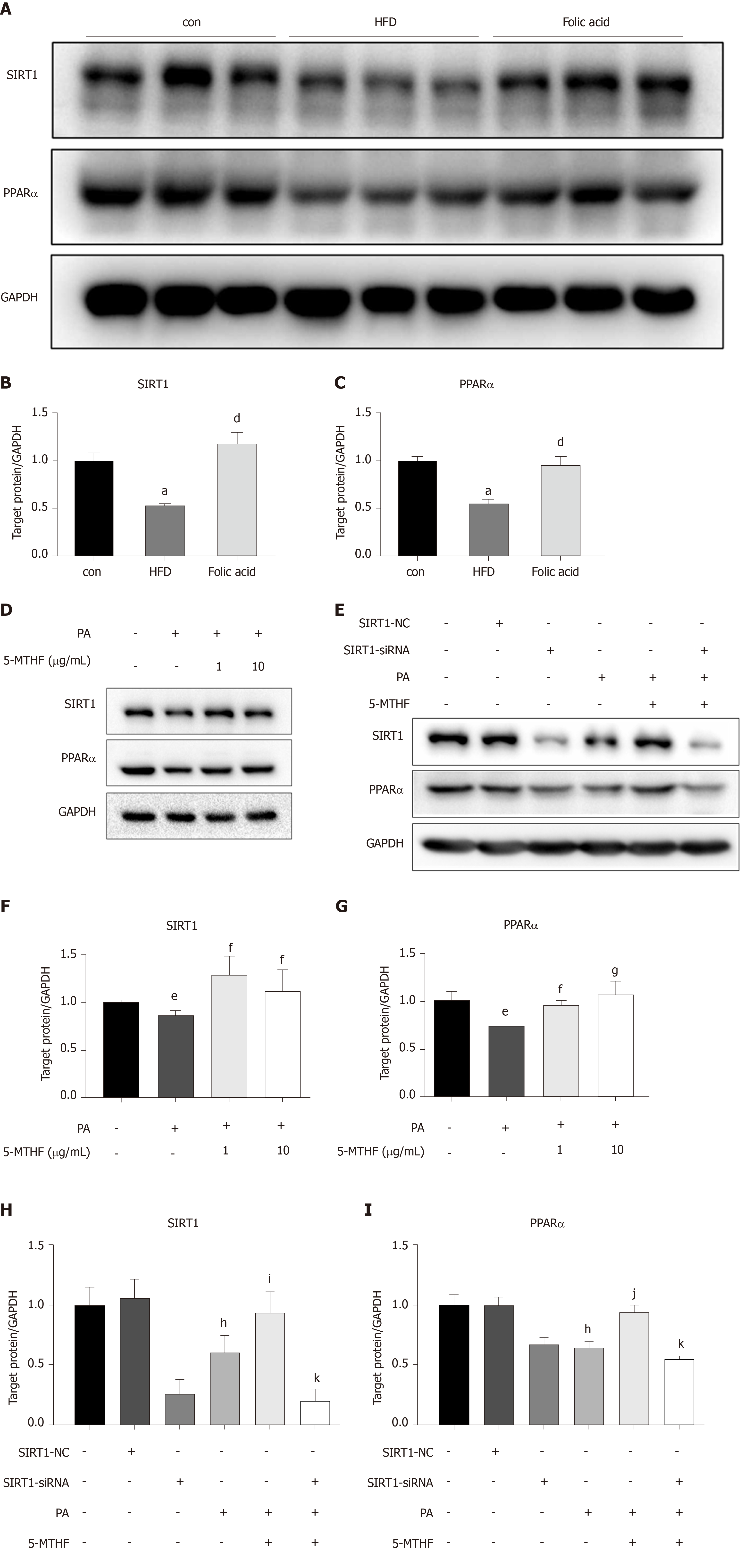
Both PPARs and SIRT1 are key regulators in hepatic lipid β-oxidation. To further determine the effect of folic acid on the remission of hepatic β-oxidation in rats with HFD-induced NASH, we first evaluated the expression levels of SIRT1 and PPARα in animal models. As shown in Figure 4A-C, rats in the HFD group displayed lower levels of SIRT1 and PPARα than controls. Folic acid could strongly restore the expression levels of SIRT1 and increase the expression of PPARα to a certain extent.
Next, we constructed a PA-induced steatosis cell model using the Huh7 cell line. 5-MTHF, a predominant form of folic acid, was used as an intervention drug. After 12 h of treatment with PA solution, the expression levels of SIRT1 (P < 0.05) and PPARα (P < 0.05) were significantly downregulated. 5-MTHF strongly elevated the expression levels of SIRT1 (1.45-fold in the 1 μg/mL and 1.26-fold in 10 μg/mL 5-MTHF group compared with the levels in the PA treatment group, Figure 4D and F) and PPARα (1.29-fold in the 1 μg/mL and 1.44-fold in 10 μg/mL 5-MTHF group compared with the levels in the PA treatment group, Figure 4D and G). The upregulating effect of PPARα by 5-MTHF was dramatically blocked after knockdown of SIRT1 with a siRNA (Figure 4E, H, and I). Overall, folic acid restores hepatic PPARα levels via a SIRT1-dependent mechanism and then improves hepatic lipid metabolism under HFD-feeding conditions.
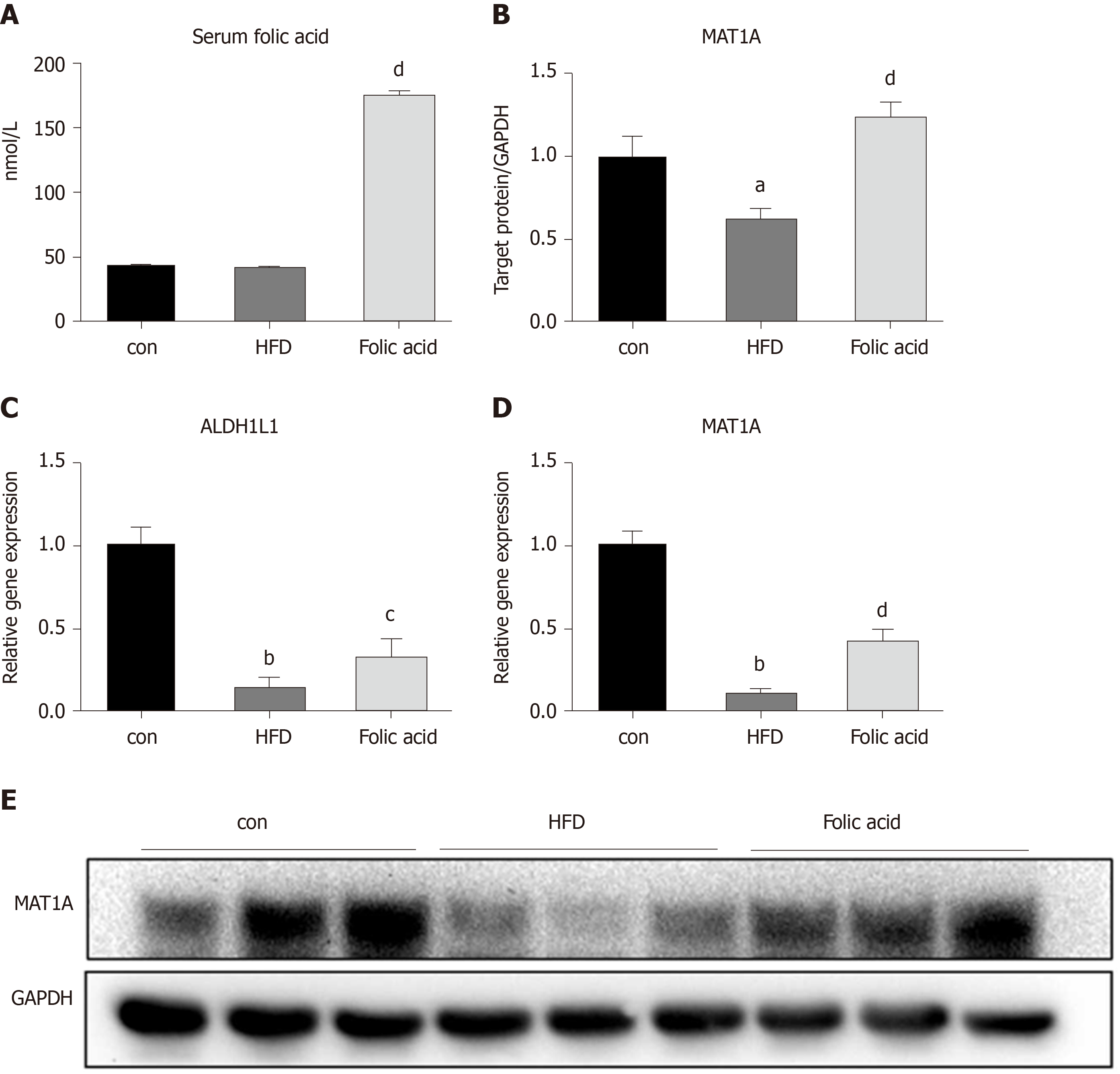
We measured the serum folic acid level in each group to further characterize the effect of the folic acid intervention. As shown in Figure 5A, folic acid intragastric administration significantly increased serum folic acid levels, although there was no difference in serum folic acid levels between the control and HFD groups. To further evaluate the effect of folic acid on one-carbon metabolism under HFD conditions, we detected the expression levels of key enzymes involved in one-carbon metabolism. qRT-PCR showed decreased ALDH1L1 (0.15-fold, P < 0.01, Figure 4C) and MAT1A (0.10-fold, P < 0.01, Figure 4D) levels in the HFD group than controls. Western blot confirmed the lower MAT1A level as well (Figure 4B and 4B). Folic acid supplementation could increase the expression of those genes at the transcription (Figure 4C and D) or translation levels (Figure 4B and E). The results above implied that folic acid could partially restore depleted one-carbon metabolism in HFD rats and suggested that folic acid has a direct effect on the liver.
Fecal samples were collected and subjected to 16S rRNA sequencing to detect the effect of folic acid on the gut microbiota. As shown in Figure 6A, folic acid restored the alpha diversity based on PD_whole_tree measurement, which demonstrated that folic acid could restore the HFD-induced depletion of the gut microbiota abundance. Principal coordinates analysis showed that folic acid could alter the composition of the gut microbiota in HFD-fed rats (Figure 6B). The unweighted pair-group method with arithmetic mean analysis showed that folic acid partially restored the alteration in the overall structure of the gut microbiota induced by the HFD (Figure 6C and D). Besides, compared to the control group, lower abundance of Bacteroidetes was detected in the HFD group, and folic acid administration could partially increase levels of Bacteroidetes. Compared with the HFD group, an increase in several genera such as Pseudomonadaceae and Leptotrichiaceae was observed (data not shown). Moreover, HE and Occludin immunohistochemical staining of the ileum showed that folic acid could restore the villus structure and the abundance of the expression of tight junctions (Figure 6E). Moreover, serum endotoxin levels were significantly reduced in the folic acid group (Figure 6F). Then, the expression levels of proinflammatory factors such as tumor necrosis factor alpha (Figure 6G), interleukin-6 (Figure 6H), and interleukin-1 beta (Figure 6I); chemokine receptor C-C chemokine receptor type 2 (Figure 6J); and oxidative stress-related factors such as neutrophil cytosol factor 1 (Figure 6K), neutrophil cytosolic factor 2 (Figure 6L), cytochrome b-245 alpha chain (Figure 6M), and cytochrome b-245 beta chain (Figure 6N) were greatly decreased by folic acid treatment. Overall, folic acid could restore the depleted diversity and the intestinal barrier, ameliorate endotoxemia, and decrease hepatic inflammatory reactions under HFD conditions.
We demonstrated that folic acid attenuated hepatic lipid metabolism in rats with HFD-induced steatohepatitis, increased PPARα levels through a SIRT1-dependent mechanism in vivo and vitro, ameliorated HFD-induced depleted hepatic one-carbon metabolism, and restored the diversity of the gut microbiota, thus contributing to the improvements of HFD-induced NASH in rats.
One of important findings in our present study is that folic acid plays an important role in regulating hepatic lipid metabolism in the HFD-induced NASH model. Lipid metabolism disorder is one of the most important pathophysiological changes in individuals with NAFLD. Either the “two-hit” or “multiple parallel hits” hypothesis confirms that abnormal lipid metabolism is one of the core causes of steatosis[16,17]. Both increased DNL[18] and impaired fatty acid oxidation[19] contribute to the pathogenesis of NAFLD. Previous studies confirmed that folic acid could reduce lipid accumulation in primary chicken hepatocytes[12] and alter lipid metabolism genes in male rat offspring[20]. Studies also indicated that folic acid may alleviate abnormal lipid metabolism and cholesterol deposition in the liver through the LKB1-AMPK pathway[11]. However, the further mechanism for the effect of folic acid in regulating hepatic fatty acid oxidation is still rarely known. PPARs belong to the nuclear hormone receptor superfamily, and of the PPARs, PPARα regulates hepatic lipid metabolism, glucose metabolism, and liver inflammation[21,22]. Numerous rate-limited enzymes associated with fatty acid uptake[23] and mitochondrial β-oxidation[24,25] are regulated by PPARα. Hepatocyte-specific PPARα deletion impaired fatty acid homeostasis and promoted the progression of NAFLD[26]. SIRT1 is an NAD+-dependent deacetylase in mammalian cells that plays a key role in metabolic diseases[27] and regulates the transcription network in free fatty acid oxidation[28]. Microarray analysis confirmed that SIRT1, PPARα, and peroxisome proliferator-activated receptorγcoactivator-1 (PGC1α) played a core role in the regulation of genes responsible for β-oxidation[29]. Hepatic deletion of SIRT1 could impair PPARα signaling, and overexpression of SIRT1 could restore the expression levels of PPARα and its target genes[30]. We confirmed that folic acid restores hepatic PPARα via a SIRT1 dependent pathway, which further reveals the effect of folic acid on hepatic lipid metabolism.
Significantly lower FBG levels in the folic acid group indicated that folic acid may play a role in glucose metabolism in metabolic diseases including NAFLD. Studies showed that chronic folic acid deficiency induced glucose metabolism disorder[31]. Folic acid treatment decreased serum glucose levels in a diabetic rat model[32]. Administration of folic acid improved insulin resistance by altering the DNA methylation profile in HFD-fed mice[33]. This indicated that folic acid could improve glucose metabolism in NASH conditions, but specific mechanisms need further research.
Another finding in our present study is that folic acid could restore one-carbon metabolism in rats with HFD-induced NASH. Several studies have reported that folic acid and other methyl donors have an alleviating effect on chronic liver diseases, such as liver fibrosis[34], cholestasis[34], drug-induced liver injury[6,35], alcoholic liver disease[13], obesity[36], and NAFLD[37,38]. However, serum folic acid level in NAFLD patients is still controversial in recent studies. Some researchers[39] believed that there was no significant difference in serum folic acid and vitamin B12 levels between NAFLD and healthy control groups and that neither folic acid nor vitamin B12 levels were associated with pathological severity. Other studies have found varying degrees of positive results. Hirsch et al[40] found a lower serum folic acid concentration in female obese patients with NAFLD than in healthy controls; Mahamid et al[41] posited that lower folate or vitamin B12 levels were associated with the histological severity of NASH. An association between serum folic acid levels and the severity of liver steatosis was also found by research on Chinese adult NAFLD patients[42]. The serum folic acid levels in the NAFLD patients from the abovementioned literature varied from 9.3 to 12.6 ng/mL on average, all of which were normal levels. Therefore, we believed that HFD had little effect on folate absorption or serum folate levels. This result was consistent with the lack of a significant difference in serum folic acid levels between the control and HFD groups in our study. However, as a co-enzymatic substrate, folic acid serves a core role in on-carbon transfer reactions. Folic acid-dependent one-carbon metabolism is important for methylation reactions in mammal cells[43]. It has been well demonstrated that differential DNA methylation occurs in individuals with NAFLD[44-46]. Genes involved in one-carbon metabolism showed abnormal DNA methylation, and of these genes, MAT1A and ALDH1L1 showed hypermethylated levels and downregulation[46]. MAT1A participates in the synthesis of S-adenosylmethionine[47] and ALDH1L1 is involved in metabolism in the carbon pathway[48]. Both of them are required for lipid homeostasis. We found strong downregulation of MAT1A and ALDH1L1 in HFD-fed rats in our present study, and additional folic acid supplementation was effective in restoring their expression levels. These findings indicated that folic acid supplementation is required for NASH individuals to improve hepatic lipid metabolism through restoring one-carbon metabolism.
An increasing number of studies have demonstrated that NAFLD has a disease-specific gut microbiome signature[49], of which the depleted diversity of the microbiota and microbial gene richness and differential bacterial clusters were most commonly reported[50,51]. Additionally, HFD consumption disturbed gut permeability by reducing tight-junction proteins such as Occludin and ZO-1, which leads to endotoxemia and chronic systemic inflammation[52] and promotes the progression of NASH. We notably found that folic acid could stabilize the intestinal barrier and the diversity of the gut microbiota, which partially explained the calming effect on the whole body and hepatic inflammation.
There are still some limitations that deserve further study. First, we have not demonstrated the effect of folic acid on the SIRT1-PPARα pathway in vivo. SIRT1 conditional knock-out mice should be used in future study to further evaluate the molecular mechanism of folic acid in the improvement of NAFLD. Second, studies have confirmed that several genes related to lipid metabolism, such as PGC1α[53], ZNF274, and SREBP2[44], had enriched DNA methylation in individuals with NAFLD. Therefore, whether folic acid could influence the balance of acetylation and methylation in genes related to free fatty acid oxidation, especially PPARα and PGC1α, remains an interesting question. Finally, a drug-dose gradient in vivo could be considered to evaluate the optimal intervention dose for clinical guidance.
In conclusion, we have confirmed the improvement effect of folic acid on HFD-induced NASH in rats. We demonstrated that folic acid improves hepatic lipid metabolism by upregulating PPARα via a SIRT1-dependent mechanism. Meanwhile, folic acid administration restores depleted hepatic one-carbon metabolism and the diversity of gut microbiota in HFD-fed rats. These results further clarify the therapeutic role of folic acid in NAFLD and its possible mechanism, suggesting that folic acid may become a therapeutic drug to treat NAFLD in the future.
Non-alcoholic fatty liver disease has become a global burden, but there is still a lack of convinced drug therapy strategies for non-alcoholic steatohepatitis (NASH). As one of essential water-soluble vitamins for the human body, folic acid may become one of the drug targets for treatment of NASH, but the specific mechanism is not fully understood.
As one of essential vitamins absorbed by the intestine mainly, food-sourced folic acid improved high-fat diet (HFD)-induced steatohepatitis in previous studies, but further mechanism of folic acid on host hepatic lipid metabolism and the effect of folic acid on lipid one-carbon metabolism and gut microbiota remains rarely understood.
We aimed to evaluated the effect of folic acid on HFD-fed rat models and further clarify the mechanism of folic acid on hepatic lipid metabolism and gut microbiota.
An HFD-induced rat model of NASH was used in the present study. Treatment of folic acid by oral administration lasted for 8 wk. Hepatic lipid metabolism was evaluated by real-time quantitative polymerase chain reaction (qRT-PCR). Expression levels of silence information regulation factor 1 (SIRT1) and peroxisome proliferator-activated receptor alpha (PPARα) were measured by Western blot analysis in HFD-induced rat models and palmitic acid-induced Huh7 cells. SIRT1 siRNA was transfected in Huh7 cells to examine whether folic acid restored PPARα levels through a SIRT1-dependent mechanism. Genes and proteins related to hepatic one-carbon metabolism were detected by qRT-PCR and Western blot. 16S rDNA sequencing was used to evaluate the effect of folic acid on gut microbiota profile.
Administration of folic acid ameliorated HFD-induced steatohepatitis. Folic acid repaired impaired hepatic lipid β-oxidation and hepatic one-carbon metabolism. SIRT1 and PPARα levels were restored by folic acid treatment. The restoration effect of PPARα by folic acid was blocked after SIRT1 knockdown in vitro. Furthermore, folic acid restored the diversity and altered the overall structure of gut microbiota profile.
Folic acid restores PPARα levels via a SIRT1-dependent mechanism, ameliorates HFD-induced impaired hepatic lipid metabolism and hepatic one-carbon metabolism, and improves the diversity of gut microbiota, thus acting a protective role in HFD-induced NASH in rats.
Folic acid may become one of drug targets for treatment of NASH. Research about folic acid in epigenetic regulation may further clarify the mechanism of folic acid on NASH.
Manuscript source: Unsolicited Manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Shimizu Y S-Editor: Wang YQ L-Editor: Wang TQ E-Editor: Ma YJ
| 1. | Fan JG, Kim SU, Wong VW. New trends on obesity and NAFLD in Asia. J Hepatol. 2017;67:862-873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 848] [Cited by in RCA: 801] [Article Influence: 100.1] [Reference Citation Analysis (2)] |
| 2. | Lee HW, Wong VW. Changing NAFLD Epidemiology in China. Hepatology. 2019;70:1095-1098. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 3. | Zhou F, Zhou J, Wang W, Zhang XJ, Ji YX, Zhang P, She ZG, Zhu L, Cai J, Li H. Unexpected Rapid Increase in the Burden of NAFLD in China From 2008 to 2018: A Systematic Review and Meta-Analysis. Hepatology. 2019;70:1119-1133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 431] [Cited by in RCA: 420] [Article Influence: 70.0] [Reference Citation Analysis (0)] |
| 4. | Younossi ZM. Non-alcoholic fatty liver disease - A global public health perspective. J Hepatol. 2019;70:531-544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 943] [Cited by in RCA: 1456] [Article Influence: 242.7] [Reference Citation Analysis (1)] |
| 5. | Leoni S, Tovoli F, Napoli L, Serio I, Ferri S, Bolondi L. Current guidelines for the management of non-alcoholic fatty liver disease: A systematic review with comparative analysis. World J Gastroenterol. 2018;24:3361-3373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 412] [Cited by in RCA: 393] [Article Influence: 56.1] [Reference Citation Analysis (7)] |
| 6. | Ebaid H, Bashandy SA, Alhazza IM, Rady A, El-Shehry S. Folic acid and melatonin ameliorate carbon tetrachloride-induced hepatic injury, oxidative stress and inflammation in rats. Nutr Metab (Lond). 2013;10:20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 7. | Ebara S. Nutritional role of folate. Congenit Anom (Kyoto). 2017;57:138-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 88] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 8. | Visentin M, Diop-Bove N, Zhao R, Goldman ID. The intestinal absorption of folates. Annu Rev Physiol. 2014;76:251-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 139] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 9. | Ojeda ML, Rua RM, Nogales F, Díaz-Castro J, Murillo ML, Carreras O. The Benefits of Administering Folic Acid in Order to Combat the Oxidative Damage Caused by Binge Drinking in Adolescent Rats. Alcohol Alcohol. 2016;51:235-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 10. | Sid V, Shang Y, Siow YL, Hewage SM, House JD, O K. Folic Acid Supplementation Attenuates Chronic Hepatic Inflammation in High-Fat Diet Fed Mice. Lipids. 2018;53:709-716. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 11. | Sid V, Wu N, Sarna LK, Siow YL, House JD, O K. Folic acid supplementation during high-fat diet feeding restores AMPK activation via an AMP-LKB1-dependent mechanism. Am J Physiol Regul Integr Comp Physiol. 2015;309:R1215-R1225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 12. | Liu Y, Shen J, Yang X, Sun Q, Yang X. Folic Acid Reduced Triglycerides Deposition in Primary Chicken Hepatocytes. J Agric Food Chem. 2018;66:13162-13172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 13. | Lee SJ, Kang MH, Min H. Folic acid supplementation reduces oxidative stress and hepatic toxicity in rats treated chronically with ethanol. Nutr Res Pract. 2011;5:520-526. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 14. | Bedossa P, Poitou C, Veyrie N, Bouillot JL, Basdevant A, Paradis V, Tordjman J, Clement K. Histopathological algorithm and scoring system for evaluation of liver lesions in morbidly obese patients. Hepatology. 2012;56:1751-1759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 526] [Cited by in RCA: 648] [Article Influence: 49.8] [Reference Citation Analysis (0)] |
| 15. | Geisler CE, Renquist BJ. Hepatic lipid accumulation: cause and consequence of dysregulated glucoregulatory hormones. J Endocrinol. 2017;234:R1-R21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 133] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 16. | Musso G, Cassader M, Gambino R. Non-alcoholic steatohepatitis: emerging molecular targets and therapeutic strategies. Nat Rev Drug Discov. 2016;15:249-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 342] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 17. | Fang YL, Chen H, Wang CL, Liang L. Pathogenesis of non-alcoholic fatty liver disease in children and adolescence: From "two hit theory" to "multiple hit model". World J Gastroenterol. 2018;24:2974-2983. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 191] [Cited by in RCA: 267] [Article Influence: 38.1] [Reference Citation Analysis (5)] |
| 18. | Sanders FW, Griffin JL. De novo lipogenesis in the liver in health and disease: more than just a shunting yard for glucose. Biol Rev Camb Philos Soc. 2016;91:452-468. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 222] [Cited by in RCA: 367] [Article Influence: 36.7] [Reference Citation Analysis (0)] |
| 19. | Khan RS, Bril F, Cusi K, Newsome PN. Modulation of Insulin Resistance in Nonalcoholic Fatty Liver Disease. Hepatology. 2019;70:711-724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 298] [Article Influence: 49.7] [Reference Citation Analysis (0)] |
| 20. | Cuthbert CE, Foster JE, Ramdath DD. A maternal high-fat, high-sucrose diet alters insulin sensitivity and expression of insulin signalling and lipid metabolism genes and proteins in male rat offspring: effect of folic acid supplementation. Br J Nutr. 2017;118:580-588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 21. | Kersten S, Desvergne B, Wahli W. Roles of PPARs in health and disease. Nature. 2000;405:421-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1452] [Cited by in RCA: 1448] [Article Influence: 57.9] [Reference Citation Analysis (0)] |
| 22. | Rakhshandehroo M, Knoch B, Müller M, Kersten S. Peroxisome proliferator-activated receptor alpha target genes. PPAR Res. 2010;2010. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 513] [Cited by in RCA: 593] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 23. | Hashimoto T, Fujita T, Usuda N, Cook W, Qi C, Peters JM, Gonzalez FJ, Yeldandi AV, Rao MS, Reddy JK. Peroxisomal and mitochondrial fatty acid beta-oxidation in mice nullizygous for both peroxisome proliferator-activated receptor alpha and peroxisomal fatty acyl-CoA oxidase. Genotype correlation with fatty liver phenotype. J Biol Chem. 1999;274:19228-19236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 183] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 24. | Leone TC, Weinheimer CJ, Kelly DP. A critical role for the peroxisome proliferator-activated receptor alpha (PPARalpha) in the cellular fasting response: the PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc Natl Acad Sci USA. 1999;96:7473-7478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 762] [Cited by in RCA: 763] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 25. | Yamazaki K, Kuromitsu J, Tanaka I. Microarray analysis of gene expression changes in mouse liver induced by peroxisome proliferator- activated receptor alpha agonists. Biochem Biophys Res Commun. 2002;290:1114-1122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 83] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 26. | Montagner A, Polizzi A, Fouché E, Ducheix S, Lippi Y, Lasserre F, Barquissau V, Régnier M, Lukowicz C, Benhamed F, Iroz A, Bertrand-Michel J, Al Saati T, Cano P, Mselli-Lakhal L, Mithieux G, Rajas F, Lagarrigue S, Pineau T, Loiseau N, Postic C, Langin D, Wahli W, Guillou H. Liver PPARα is crucial for whole-body fatty acid homeostasis and is protective against NAFLD. Gut. 2016;65:1202-1214. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 397] [Cited by in RCA: 562] [Article Influence: 62.4] [Reference Citation Analysis (0)] |
| 27. | Chalasani N, Vuppalanchi R, Rinella M, Middleton MS, Siddiqui MS, Barritt AS 4th, Kolterman O, Flores O, Alonso C, Iruarrizaga-Lejarreta M, Gil-Redondo R, Sirlin CB, Zemel MB. Randomised clinical trial: a leucine-metformin-sildenafil combination (NS-0200) vs placebo in patients with non-alcoholic fatty liver disease. Aliment Pharmacol Ther. 2018;47:1639-1651. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 28. | Guarente L. Sirtuins as potential targets for metabolic syndrome. Nature. 2006;444:868-874. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 354] [Cited by in RCA: 344] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 29. | Kim TH, Yang YM, Han CY, Koo JH, Oh H, Kim SS, You BH, Choi YH, Park TS, Lee CH, Kurose H, Noureddin M, Seki E, Wan YY, Choi CS, Kim SG. Gα12 ablation exacerbates liver steatosis and obesity by suppressing USP22/SIRT1-regulated mitochondrial respiration. J Clin Invest. 2018;128:5587-5602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 30. | Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327-338. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 912] [Cited by in RCA: 879] [Article Influence: 54.9] [Reference Citation Analysis (0)] |
| 31. | Zhao M, Yuan MM, Yuan L, Huang LL, Liao JH, Yu XL, Su C, Chen YH, Yang YY, Yu H, Xu X. Chronic folate deficiency induces glucose and lipid metabolism disorders and subsequent cognitive dysfunction in mice. PLoS One. 2018;13:e0202910. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 32. | Mutavdzin S, Gopcevic K, Stankovic S, Jakovljevic Uzelac J, Labudovic Borovic M, Djuric D. The Effects of Folic Acid Administration on Cardiac Oxidative Stress and Cardiovascular Biomarkers in Diabetic Rats. Oxid Med Cell Longev. 2019;2019:1342549. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 33. | Li W, Tang R, Ma F, Ouyang S, Liu Z, Wu J. Folic acid supplementation alters the DNA methylation profile and improves insulin resistance in high-fat-diet-fed mice. J Nutr Biochem. 2018;59:76-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 48] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 34. | Mohammadian Z, Eidi A, Mortazavi P, Tavangar MM, Asghari A. Effects of folic acid on dyslipidemia and serum homocysteine in a rat model of cholestasis and hepatic fibrosis. Pol J Pathol. 2015;66:49-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 35. | Majumdar S, Maiti A, Karmakar S, Das AS, Mukherjee S, Das D, Mitra C. Antiapoptotic efficacy of folic acid and vitamin B₁₂ against arsenic-induced toxicity. Environ Toxicol. 2012;27:351-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 36. | Cordero P, Milagro FI, Campion J, Martinez JA. Supplementation with methyl donors during lactation to high-fat-sucrose-fed dams protects offspring against liver fat accumulation when consuming an obesogenic diet. J Dev Orig Health Dis. 2014;5:385-395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 37. | da Silva RP, Kelly KB, Al Rajabi A, Jacobs RL. Novel insights on interactions between folate and lipid metabolism. Biofactors. 2014;40:277-283. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 132] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 38. | Bakir MB, Salama MA, Refaat R, Ali MA, Khalifa EA, Kamel MA. Evaluating the therapeutic potential of one-carbon donors in nonalcoholic fatty liver disease. Eur J Pharmacol. 2019;847:72-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 39. | Polyzos SA, Kountouras J, Patsiaoura K, Katsiki E, Zafeiriadou E, Zavos C, Deretzi G, Tsiaousi E, Slavakis A. Serum vitamin B12 and folate levels in patients with non-alcoholic fatty liver disease. Int J Food Sci Nutr. 2012;63:659-666. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 40. | Hirsch S, Poniachick J, Avendaño M, Csendes A, Burdiles P, Smok G, Diaz JC, de la Maza MP. Serum folate and homocysteine levels in obese females with non-alcoholic fatty liver. Nutrition. 2005;21:137-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 73] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 41. | Mahamid M, Mahroum N, Bragazzi NL, Shalaata K, Yavne Y, Adawi M, Amital H, Watad A. Folate and B12 Levels Correlate with Histological Severity in NASH Patients. Nutrients. 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 42. | Xia MF, Bian H, Zhu XP, Yan HM, Chang XX, Zhang LS, Lin HD, Hu XQ, Gao X. Serum folic acid levels are associated with the presence and severity of liver steatosis in Chinese adults. Clin Nutr. 2018;37:1752-1758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 43. | Sid V, Siow YL, O K. Role of folate in nonalcoholic fatty liver disease. Can J Physiol Pharmacol. 2017;95:1141-1148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 67] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 44. | Ahrens M, Ammerpohl O, von Schönfels W, Kolarova J, Bens S, Itzel T, Teufel A, Herrmann A, Brosch M, Hinrichsen H, Erhart W, Egberts J, Sipos B, Schreiber S, Häsler R, Stickel F, Becker T, Krawczak M, Röcken C, Siebert R, Schafmayer C, Hampe J. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013;18:296-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 335] [Cited by in RCA: 417] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 45. | Gerhard GS, Malenica I, Llaci L, Chu X, Petrick AT, Still CD, DiStefano JK. Differentially methylated loci in NAFLD cirrhosis are associated with key signaling pathways. Clin Epigenetics. 2018;10:93. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 46. | Murphy SK, Yang H, Moylan CA, Pang H, Dellinger A, Abdelmalek MF, Garrett ME, Ashley-Koch A, Suzuki A, Tillmann HL, Hauser MA, Diehl AM. Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology. 2013;145:1076-1087. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 260] [Cited by in RCA: 312] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 47. | Cano A, Buqué X, Martínez-Uña M, Aurrekoetxea I, Menor A, García-Rodríguez JL, Lu SC, Martínez-Chantar ML, Mato JM, Ochoa B, Aspichueta P. Methionine adenosyltransferase 1A gene deletion disrupts hepatic very low-density lipoprotein assembly in mice. Hepatology. 2011;54:1975-1986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 75] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 48. | Oleinik NV, Krupenko NI, Krupenko SA. Epigenetic Silencing of ALDH1L1, a Metabolic Regulator of Cellular Proliferation, in Cancers. Genes Cancer. 2011;2:130-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 49. | Grabherr F, Grander C, Effenberger M, Adolph TE, Tilg H. Gut Dysfunction and Non-alcoholic Fatty Liver Disease. Front Endocrinol (Lausanne). 2019;10:611. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 68] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 50. | Zhou D, Pan Q, Xin FZ, Zhang RN, He CX, Chen GY, Liu C, Chen YW, Fan JG. Sodium butyrate attenuates high-fat diet-induced steatohepatitis in mice by improving gut microbiota and gastrointestinal barrier. World J Gastroenterol. 2017;23:60-75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 342] [Cited by in RCA: 329] [Article Influence: 41.1] [Reference Citation Analysis (6)] |
| 51. | Zhao ZH, Xin FZ, Xue Y, Hu Z, Han Y, Ma F, Zhou D, Liu XL, Cui A, Liu Z, Liu Y, Gao J, Pan Q, Li Y, Fan JG. Indole-3-propionic acid inhibits gut dysbiosis and endotoxin leakage to attenuate steatohepatitis in rats. Exp Mol Med. 2019;51:1-14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 166] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 52. | Sanduzzi Zamparelli M, Compare D, Coccoli P, Rocco A, Nardone OM, Marrone G, Gasbarrini A, Grieco A, Nardone G, Miele L. The Metabolic Role of Gut Microbiota in the Development of Nonalcoholic Fatty Liver Disease and Cardiovascular Disease. Int J Mol Sci. 2016;17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 45] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 53. | Sookoian S, Rosselli MS, Gemma C, Burgueño AL, Fernández Gianotti T, Castaño GO, Pirola CJ. Epigenetic regulation of insulin resistance in nonalcoholic fatty liver disease: impact of liver methylation of the peroxisome proliferator-activated receptor γ coactivator 1α promoter. Hepatology. 2010;52:1992-2000. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 251] [Article Influence: 16.7] [Reference Citation Analysis (0)] |