Published online Sep 14, 2019. doi: 10.3748/wjg.v25.i34.5026
Peer-review started: June 25, 2019
First decision: July 21, 2019
Revised: July 26, 2019
Accepted: August 7, 2019
Article in press: August 7, 2019
Published online: September 14, 2019
Processing time: 80 Days and 0.9 Hours
Long non-coding RNAs (lncRNAs) are members of the non-protein coding RNA family longer than 200 nucleotides. They participate in the regulation of gene and protein expression influencing apoptosis, cell proliferation and immune responses, thereby playing a critical role in the development and progression of various cancers, including colorectal cancer (CRC). As CRC is one of the most frequently diagnosed malignancies worldwide with high mortality, its screening and early detection are crucial, so the identification of disease-specific biomarkers is necessary. LncRNAs are promising candidates as they are involved in carcinogenesis, and certain lncRNAs (e.g., CCAT1, CRNDE, CRCAL1-4) show altered expression in adenomas, making them potential early diagnostic markers. In addition to being useful as tissue-specific markers, analysis of circulating lncRNAs (e.g., CCAT1, CCAT2, BLACAT1, CRNDE, NEAT1, UCA1) in peripheral blood offers the possibility to establish minimally invasive, liquid biopsy-based diagnostic tests. This review article aims to describe the origin, structure, and functions of lncRNAs and to discuss their contribution to CRC development. Moreover, our purpose is to summarise lncRNAs showing altered expression levels during tumor formation in both colon tissue and plasma/serum samples and to demonstrate their clinical implications as diagnostic or prognostic biomarkers for CRC.
Core tip: The present review aims to shed light on the complex world of long non-coding RNAs (lncRNAs) by discussing their origin, localization, and functions. By summarizing the constantly growing body of knowledge about lncRNA expression in colorectal tissue and by focusing on potential circulating lncRNA markers, we aim to enhance the understanding of the comprehensive picture of their diagnostic and prognostic potential in precancerous colorectal adenomas and cancer.
- Citation: Galamb O, Barták BK, Kalmár A, Nagy ZB, Szigeti KA, Tulassay Z, Igaz P, Molnár B. Diagnostic and prognostic potential of tissue and circulating long non-coding RNAs in colorectal tumors. World J Gastroenterol 2019; 25(34): 5026-5048
- URL: https://www.wjgnet.com/1007-9327/full/v25/i34/5026.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i34.5026
Colorectal cancer (CRC) is one of the most frequent malignant diseases worldwide with a remarkably high mortality rate[1]. The number of CRC-related deaths can be reduced only by diagnosis at the earliest stage when the disease is more likely to be cured.
Long non-coding RNAs (lncRNAs), a novel family of non-protein coding RNAs (200 nt-10 kb) are of outstanding interest as their expression is often altered in various disease types including malignancies[2]. They are known to have a crucial role in the regulation of gene expression, alternative splicing mechanisms, protein localization and activity, formation of cellular substructures and protein complexes through their diverse interactions with DNA, RNA and proteins[3,4].
In cancers, lncRNAs are involved in every stage of carcinogenesis and tumor progression including tumor initiation, proliferation, apoptosis and migration of cancer cells, angiogenesis, tumor invasion and metastasis formation[5,6]. Their altered expression can influence several oncogenic signaling cascades including the WNT/β-catenin, PI3K/Akt, EGFR, NOTCH, mTOR and TP53 signaling pathways[4,7-20]. Besides local expression changes in cancerous tissue and tumor-related stroma, lncRNAs also remain stable in body fluids due to their resistance to RNases[2,21].
Several lncRNAs showing altered expression in colorectal tumors including precancerous adenomas have potential as early diagnostic markers[22-24]. In this review, we summarize the colorectal tumor-related tissue and circulating lncRNAs, altered lncRNA expression patterns, and technical aspects of their isolation and detection. Our aim is to show their potential as diagnostic and prognostic biomarkers based on recently published data.
Regulatory non-coding RNAs (ncRNAs) were first reported in eukaryotes in the 1980s, of which H19[25] and Xist[26] were the first members of the family[27]. When the Human Genome Project was completed, it became clear that only a minor part of our genome codes proteins and the rest was considered as “junk” DNA[28]. Since then, our knowledge about the non-coding genome was expanded, and the still unexplored regulatory role of the ncRNA world is the focus of several studies and holds a significant clinical potential[29]. Over the past decades, along with the development of explorative molecular biology methods, the importance and function of the complex eukaryotic transcriptome have been recognized, a large proportion of which comprises the actively transcribed lncRNAs[30]. After the discovery and the intensive analysis of the class of small ncRNAs called miRNAs since 1993[31], it became evident, that other ncRNAs also play fundamental role in gene expression regulation, and that their alterations can be responsible for the disrupted molecular pathways in multiple cancers[32].
The major class of ncRNAs are lncRNAs, which are derived from highly diverse genomic context and are classified on the basis of the genomic region of origin[28]. According to the genomic database [Ensembl Release 96 (April 2019)], human lncRNAs are categorized into 3prime overlapping ncRNA, antisense, lincRNA (long interspersed ncRNA), retained intron, sense intronic, sense overlapping and macro lncRNAs. The lncRNAs that are not overlapping with protein-coding genes are called stand-alone lncRNAs including the large intergenic (or intervening) lncRNA (lincRNA) group[28,33] (e.g., XIST, H19, MALAT1, and HOTAIR). Antisense lncRNAs are transcripts overlapping the genomic strand of a protein-coding locus in an antisense direction[34], while sense lncRNAs are overlapped with the sense strand of protein coding genes containing exons[35]. Antisense transcription is widespread in the mammalian genome[36]; the estimated ratio of the genes with antisense transcripts varies from less than 2 to more than 70% of the total genes [37]. XIST/TSIX is a well-known example of the sense-antisense transcript pairs[38]. Pseudogenes are defined as nonfunctional sequences of genomic DNA originally derived from functional genes[39]. Long intronic ncRNAs are transcribed from the intronic sequence of a coding gene. On the basis of their association with functional DNA elements, enhancer- and promoter-associated lncRNAs can be distinguished[40]. The lncRNAs localize in the cytoplasm, nucleus, nucleolus, and also in other subcellular compartments and vesicles (such as nuclear bodies, exosomes) and the localization is related to their molecular functions[41]. Certain sequence motifs in their primary sequence are associated with the subcellular localization[42].
As the largest class of non-coding transcripts, lncRNAs have a wide variety of functions. They can act as RNAs (e.g., ribozymes, riboswitches)[43] and widely as ribonucleoprotein particles (RNP)[44]. They can exert their positive or negative regulating functions either in cis or in trans[45]. One of their functions is the regulation of nuclear organization; lncRNAs can modulate the chromatin architecture (e.g., Xist) and they can also regulate inter- and intrachromosomal interactions (e.g., colorectal cancer associated transcript 1. long isoform (CCAT1-L) modulating interchromatin loops between enhancers and promoters[46]). LncRNAs can regulate other non-coding RNAs (e.g., as miRNA sponges leading to reduced miRNA inhibitory effect on target molecules[47]), and also can be processed into single- or double-stranded siRNAs[3]. Several gene transcription processes can be activated or blocked by lncRNAs by recruiting or inhibiting transcription factors of the target gene promoters[3,44]. Certain lncRNAs are linked to the process of alternative splicing (e.g., LINC001133)[48]. Furthermore, protein activity is regulated by lncRNAs and trafficking between the subcellular compartments can also be influenced by lncRNAs[49].
Nuclear lncRNAs also contribute to chromatin remodeling as they can promote or prevent the recruitment of chromatin modifiers[46]. They are also part of nuclear bodies[50] with scaffold function, so-called architectural lncRNAs[51] [such as nuclear enriched abundant transcript 1 (NEAT1), as a well-characterized lncRNA as a crucial component of paraspeckles[52]] and also as non-architectural lncRNAs [e.g., metastasis associated lung carcinoma transcript 1 (MALAT1) as one of the most abundant lncRNA in nuclear speckles[46]].
Epigenetic mechanisms, such as histone modifications are also influenced by lncRNAs. For instance, lncRNA HOTAIR (homeobox transcript antisense intergenic RNA) interacts with both LSD1/CoREST/REST complex and PRC2 as a modular scaffold that leads to coupled histone H3 lysine 27 methylation and lysine 4 demethylation[53].
By the modulation of all three major mammalian DNA methyltransferases (DNMT1, DNMT3a, DNMT3b), lncRNAs influence DNA methylation levels resulting in altered expression of the target genes[44]. DNMT1-associated Colon Cancer Repressed lncRNA 1 (DACOR1) interacts with both chromatin and DNMT1 and targets DNMT1 protein complex to certain genomic loci, also affecting cellular SAM levels[54,55]. Altogether, the expression alterations of lncRNAs influence many biological functions that contribute to the disturbance of the complex fine-tuning machinery of non-coding RNA regulatory network during cancer formation.
Our knowledge about the posttranscriptional regulation of lncRNAs is limited, however, the stability of transcripts can be an important aspect in gene expression regulation[56,57] as the half-life of ncRNAs correlates with their functional cha-racteristics[58]. Each lncRNA has a unique structure, and these transcripts are characterized by complex secondary and tertiary structures which is crucial to exert their functions[59]. Although the stability of these non-coding transcripts was generally considered to be lower compared to mRNAs[60] on the basis of a genome-wide lncRNA analysis by Clark et al[56], a wide variety in their stability can be observed which is consistent with their functional diversity. LncRNA stability is correlated with genomic location, subcellular localization, splicing, and GC percentage, while in contrast, expression levels are not correlated with stability[56]. The half-life of lncRNAs ranges from < 30 min to > 48 h with median value at 3.5 h, and they can be classified as unstable and to highly stable lncRNAs - the latter represented at a lower per-centage[56]. According to Clark et al., nuclear-enriched lncRNAs displayed significantly lower stability compared to those detected both in nucleus and cytoplasm[56]. It is important to note that lncRNAs with even lower stability have been shown to have fundamental role (e.g., NEAT1 as scaffold lncRNA of paraspeckles, as dynamic nuclear subdomains[61]), furthermore, the existence of highly stable lncRNAs illustrate the biomarker potential of this subclass of non-coding transcripts.
Analysis of lncRNAs is technically challenging due to their relatively low expression level and their tissue-specific expression[62], therefore, the following methods are optimized for studying lncRNAs with high sensitivity and resolution.
High-throughput sequencing serial analysis of gene expression (SAGE) is based on short cDNA sequences containing recognition sites for restriction enzymes at the transcripts’ 3’ end, and it was one of the first transcriptome analysis methods to study lncRNA expression[63,64].
Among whole genome analyses, microarrays are widely used to analyse the RNA expression in a high-throughput manner from the 2000s, however, these systems are limited to studying the known RNAs. Furthermore, cross-hybridization and limited detection range due to background and saturation signals make these analyses more challenging[65]. In parallel, the rapid development of next generation sequencing (NGS) systems revolutionized the experimental field, as RNA-Seq provides a cost-effective and rapid solution for whole transcriptome profiling with the potential to discover novel transcripts[65]. The higher resolution and reproducibility of RNA-Seq compared to microarrays[65] resulted in broad use of this approach. RNA-Seq supports the annotation of novel lncRNAs, RNA editing sites, and alternative splicing sites, as well[62]. Cap analysis of gene expression (CAGE) is an NGS-based approach to map and quantify the expression of 5’ capped RNAs[66] and also to identify tran-scriptionally active promoter regions and Pol II-driven TSSs[64].
The lncRNAs regulate and mediate interactions on different molecular levels and complex networks of these non-coding RNAs remain to be explored. RNA-binding protein immunoprecipitation (RIP) is used to study RNA-protein interactions, where the RNA of interest can be complexed with its interacting proteins, and this fraction can be selectively pulled down[67]. The downstream analysis can be performed by combining with the previously discussed methods, including RIP-Chip and RIP-Seq[68]. Native RIP is suitable for the exploration of strong and direct RNA-protein interactions, whereas the crosslinked immunoprecipitation method (CLIP) is used to study weak or indirect binding[62]. Crosslinking is achieved by ultraviolet light (UV) followed by RNase treatment and stringent washes which increases the specificity of the interaction detection[69]. In order to minimize the disadvantages of CLIP, modified methods, such as individual nucleotide resolution CLIP (iCLIP)[70], and photoactivable ribonucleoside-enhanced CLIP (PAR-CLIP) are also available for the identification of the exact crosslinking sites with single nucleotide resolution[62,69].
Other RNA pull-down methods, such as chromatin isolation by RNA purification (ChIRP)[71], capture hybridization analysis of RNA targets (CHART)[72] and RNA antisense purification (RAP)[73] can be applied to study RNA-DNA interactions to shed light on lncRNAs’ functions and identify trans-genomic interacting sites[62]. During ChIRP experiments, a biotin-labeled antisense probe designed to the selected lncRNA is employed to explore its interacting chromosomal fragments[71]. Different probe design criteria are applied in the case of CHART, as in contrast with ChIRP probes spanning the whole interesting lncRNA, the CHART method uses capture oligos specific for the accessible regions of the lncRNA candidate[72]. The co-purified RNA, DNA or proteins potentially interacting with the selected lncRNA can be analysed with NGS, PCR or Western blotting[62]. RAP can be performed with different crosslinking methods (e.g., psoralens) along with the longer biotinylated probes (> 60 bp) to enhance the RNA-DNA hybrid stability[73] and to reduce the signal-to-noise ratio[64].
LncRNAs are known to exert their function also by binding directly or indirectly to other RNAs[64]. These interactions can be studied by RAP-RNA (applying different chemical cross-linking), as 4’aminomethyltrioxalen: RAP-RNA[AMT], formaldehyde: RAP-RNA[FA], FA and disuccinimidyl glutarate: RAP-RNA[FA-DSG][74] or UV-crosslinked CLASH (cross-linking, ligation and sequencing of hybrids)[75] methods.
It is known that lncRNAs fold into secondary and tertiary structures that are crucial to exert their regulatory effects[59], but the structural domains of the RNA interactome still need to be explored. Structural relationships can be studied by dimethyl sulfate sequencing (DMS-Seq), selective 2’-hydroxyl acylation analysed by primer extension sequencing (SHAPE-Seq), genome-wide fragmentation sequencing (FRAG-Seq), and parallel analysis of RNA structure (PARS) techniques[76]. By the intensive development of subcellular visualization approaches, lncRNAs can be localized within the cell with high sensitivity using special fluorescent in situ hybridization (FISH) applications (single molecule FISH - smFISH, sequential FISH - seqFISH, and multiplexed error-robust FISH - MerFISH)[77-79]. High resolution microscopes, as structured illumination microscopy (SIM[80]) or stochastic optical reconstruction microscopy (STORM[81]) enable the precise detection of certain lncRNAs and investigation of their colocalization partners[46]. The functional investigations of lncRNAs can be performed with antisense oligonucleotides (ASO) and also by siRNAs and shRNAs via binding and affecting the target lncRNA’s functionality[82]. The CRISPR-Cas9 genome editing technique[83] has revolutionized functional studies in the lncRNA world, which can be employed to silence (CRISPRi[84]) and also to overexpress (CRISPRa[85]) the lncRNA of interest[86].
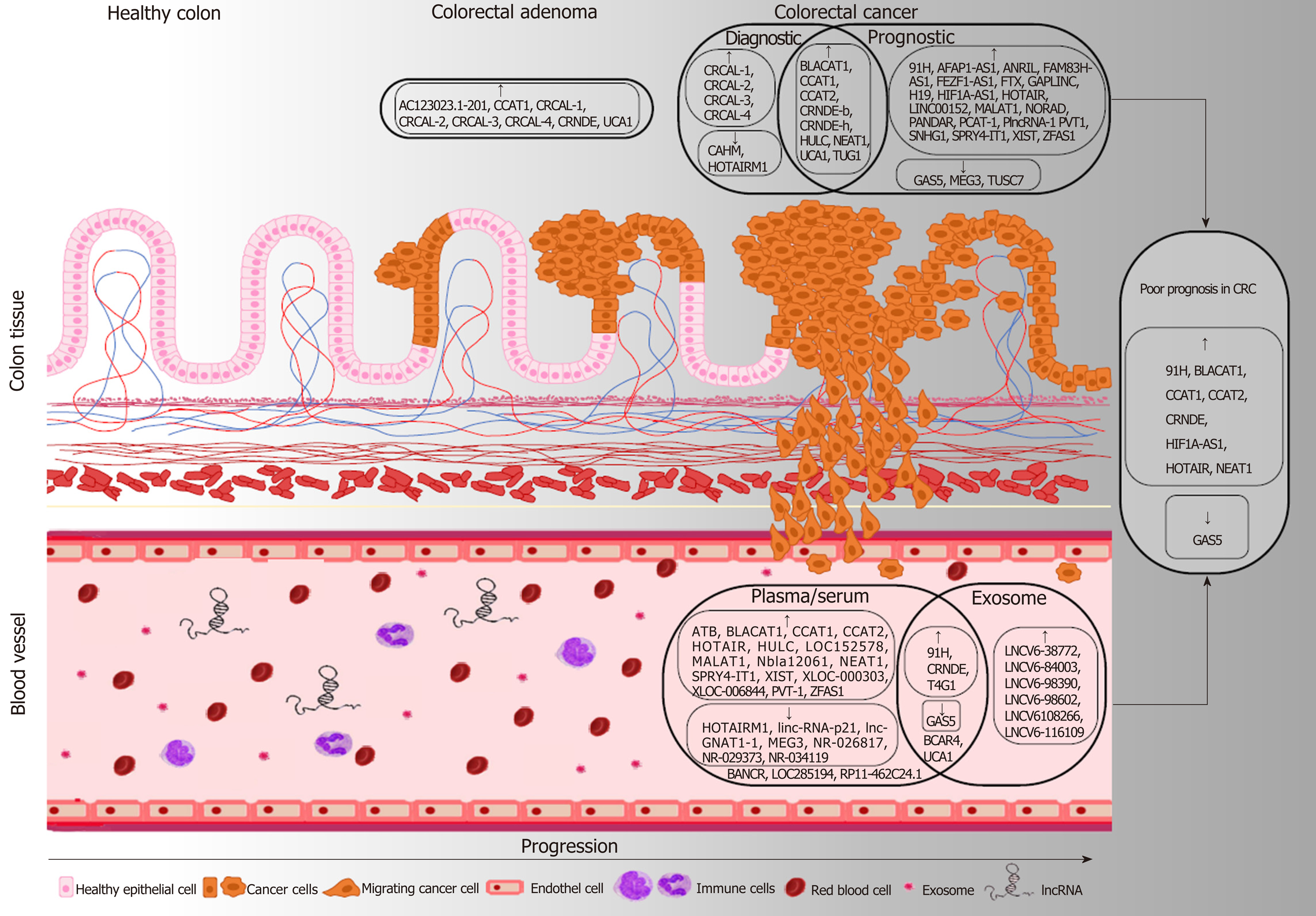
Increasing evidence suggest that lncRNAs are involved in the whole process of CRC development, progression and metastasis formation - similarly to their diverse regulatory role in other types of malignancies - affecting the essential signaling pathways including WNT, TP53, PI3K/Akt, mTOR, EGFR and NOTCH1 in CRC[4-20]. Abnormal expression of numerous lncRNAs including the well-known HOTAIR[87-90], MALAT1[91-93] and H19[94,95] has been described in CRC compared to normal colonic tissue samples (Table 1). From a clinical point of view, lncRNAs - with altered expression in different stages of colorectal carcinogenesis, and disease progression - have a particularly great potential to become early diagnostic and/or prognostic biomarkers.
| lncRNA | Tissue | Plasma/serum | Exosome | Expression in CRC | Ref. | Potentially diagnostic marker | Prognostic role |
| 91H | X | X | X | Up | [130,138,152] | X (comb 1) | Up – poor prognosis |
| ADAMTS9-AS2 | X | Down | [145] | Down – poor prognosis | |||
| AFAP1-AS1 | X | Up | [145,153,154] | Up - poor prognosis | |||
| AK027294 | X | Up | [155] | ||||
| AK123657/BX64820 | X | Down | [156] | Down – poor prognosis | |||
| AK307796 | X | Up | [157] | ||||
| ANRIL | X | Up | [158,159] | Up – poor prognosis | |||
| ATB | X | X | Up | [122,160,161] | Up - poor prognosis | ||
| BA318C17.1 | X | Down | [162] | ||||
| BANCR | X | X | Down/up | [125,163,164] | X (comb 2) | Prognostic | |
| BCAR4/HOXA-AS2 | X | X | X | Up/down | [140,145] | X | Up – poor prognosis |
| BLACAT1 | X | X | Up | [129,151,165] | Up - poor prognosis | ||
| CAHM | X | Down | [131] | X | |||
| CASC11 | X | Up | [166] | ||||
| CASC2 | X | Down | [167] | Down - poor prognosis | |||
| CCAL | X | Up | [4] | Up - poor prognosis | |||
| CCAT1 | X | X | Up | [23,96-98,116,168-170] | X | Up – poor prognosis | |
| CCAT1-L | X | Up | [171] | ||||
| CCAT2 | X | X | Up | [134,172,173] | X | Prognostic | |
| CLMAT3 | X | Up | [174,175] | Up – poor prognosis | |||
| CRNDE | X | X | X | Up | [24,99,103] | X | Up – poor prognosis |
| CRCAL-1/AC021218.2 | X | Up | [100] | X | |||
| CRCAL-2/LINC00858 | X | Up | [100] | X | |||
| CRCAL-3/RP11-138J23.1 | X | Up | [100] | X | |||
| CRCAL-4/RP11-453O5.2 | X | Up | [100] | X | |||
| CTD903 | X | Down | [176] | Down - poor prognosis | |||
| CTNNAP1 | X | Down | [177] | X | |||
| DACOR1 | X | Up | [54] | ||||
| DANCR | X | Up | [178] | Up – poor prognosis | |||
| DQ786243 | X | Up | [179] | Prognostic | |||
| E2F4 antisense | X | Up | [180] | ||||
| ENST00000430471 | X | Up | [181] | ||||
| ENST00000455974/AC123023.1 | X | Up | [111] | X | Up - poor prognosis | ||
| ENST00000465846 | X | Down | [157] | ||||
| FER1L4 | X | X | Down | [123] | Down - poor prognosis | ||
| FEZF1-AS1 | X | Up | [115,182] | Up - poor prognosis | |||
| FTX | X | Up | [183] | Up – poor prognosis | |||
| GAPLINC | X | Up | [184] | Prognostic | |||
| GAS5 | X | X | X | Down | [120,185-188] | Down - poor prognosis | |
| GHET1 | X | Up | [189] | ||||
| lnc-GNAT1-1 | X | [119] | |||||
| H19 | X | Up | [94,143-145,190,191] | Up - poor prognosis | |||
| HIF1-AS1 | X | X | [132] | Up - poor prognosis | |||
| HIF2PUT | X | Up | [192] | ||||
| HOTAIR | X | X | Up | [87-90,116,193,194] | Up - poor prognosis | ||
| HOTAIRM1 | X | X | Down | [117] | Down – poor prognosis | ||
| HOTTIP | X | Up | [195,196] | Up – poor prognosis | |||
| HULC | X | X | Up | [134,197] | Up - poor prognosis | ||
| KCNQ1OT1 | X | Up | [198,199] | ||||
| LINC00152 (CYTOR) | X | Up | [8,11,13] | Up – poor prognosis | |||
| LINC01133 | X | Down | [200] | Down - poor prognosis | |||
| LINC01296 | X | Down | [201] | Down – poor prognosis | |||
| lincRNA-p21 | X | X | Down | [116,202-204] | Down – poor prognosis | ||
| Lnc34a | X | Up | [205] | ||||
| lncRNA-LET/NPNT-IT1 | X | Down | [206] | ||||
| LNCV6_116109 | X | Up | [139] | X | |||
| LNCV6_98390 | X | Up | [139] | X | |||
| LNCV6_38772 | X | Up | [139] | X | |||
| LNCV6_108226 | X | Up | [139] | X | |||
| LNCV6_84003 | X | Up | [139] | X | |||
| LNCV6_98602 | X | Up | [139] | X | |||
| LOC152578 | X | Up | [126] | X (comb 3) | |||
| LOC100287225 | X | Down | [207,208] | ||||
| LOC285194/TUSC7 | X | X | Down/Up | [124,209,210] | X (comb 4) | Down - poor prognosis | |
| Loc554202 | X | Down | [211,212] | Down - poor prognosis | |||
| MALAT1 | X | X | Up | [7,91-93,145,149,213,214] | Up - poor prognosis | ||
| MEG3 | X | X | Down | [100,130,215-218] | X (comb 1) | Down - poor prognosis | |
| Nbla12061 | X | Up | [124] | X (comb 4) | |||
| ncNRFR | X | Up | [219] | ||||
| ncRAN | X | Down | [220,221] | Down – poor prognosis | |||
| ncRuPAR | X | Down | [222] | Down – poor prognosis | |||
| NEAT1 | X | X | Up | [128,223] | Up - poor prognosis | ||
| NORAD | X | [133] | |||||
| NR_026817 | X | Down | [125] | X (comb 2) | Prognostic | ||
| NR_029373 | X | X | Down | [125] | X (comb 2) | Down – poor prognosis | |
| NR_034119 | X | Down | [125] | X (comb 2) | |||
| PANDAR | X | Up | [224] | Up - poor prognosis | |||
| PCAT-1 | X | Up | [225] | Up – poor prognosis | |||
| PRNCR1 | X | Up | [226,227] | ||||
| PVT-1 | X | X | Up | [130,145,228] | X (comb 1) | Up - poor prognosis | |
| RP1-13P20.6 | X | Down | [229] | Down - poor prognosis | |||
| RP11-462C24.1 | X | X | Down/Up | [124,230,231] | X (comb 4) | Down - poor prognosis | |
| SLC25A25-AS1 | X | X | Down | [118] | Down - poor prognosis | ||
| SnaR | X | [232] | |||||
| SNHG20 | X | Up | [233] | Prognostic | |||
| SNHGI2 | X | Up | [234] | Up - poor prognosis | |||
| Sox2ot | X | Up | [235] | ||||
| SPRY4-IT1 | X | X | Up | [121,236] | Up - poor prognosis | ||
| TINCR | X | Down | [100,237] | ||||
| TUG1 | X | X | X | Up | [101,238-240] | Up - poor prognosis | |
| uc.388 | X | Down/Up | [241,242] | X | Prognostic | ||
| uc.73a | X | Down/Up | [241,242] | X | |||
| uc002kmd.1 | X | Up | [243] | ||||
| UCA1 | X | X | X | Up | [100,101,110,244,245] | Up - poor prognosis | |
| UPAT | X | Up | [246] | ||||
| XIST | X | X | Up | [247,248] | Prognostic | ||
| XLOC_000303 | X | Up | [136] | X (comb 3) | |||
| XLOC_006844 | X | X | Up | [136] | X (comb 3) | ||
| ZFAS1 | X | X | Up | [19,249] | Up - poor prognosis | ||
| ZNF582-AS1 | X | Down | [250] | X |
Several studies reported the altered expression of certain lncRNAs including colon cancer associated transcript-1 (CCAT1), colorectal neoplasia differentially expressed (CRNDE-L), colorectal cancer associated lncRNA (CRCAL) 1, -2, -3 and -4 and urothelial carcinoma-associated 1 (UCA1) already in precancerous ade-nomas[23,24,37,96-102].
Nissan et al[96] in their comprehensive RT-qPCR study were the first to demonstrate the massive (often more than 100-fold) upregulation of CCAT1 in CRC and premalignant adenoma tissue samples compared to normal colonic mucosa. Furthermore, elevated CCAT1 levels could be detected in lymph node and distant liver metastases, as well as in peripheral blood mononuclear cells (PBMCs) of CRC patients[96]. Alaiyan et al[23] have confirmed the overexpression of CCAT1 in precancerous conditions and through all CRC stages using RT-qPCR and in situ hybridization (ISH). These data suggest its essential role in both early carcinogenesis and metastatic processes, moreover, in vitro studies revealed that the c-Myc oncogene could facilitate the transcription of CCAT1 by binding to its promoter[97]. CRNDE also becomes activated already in the initial steps of tumor development as its elevated expression was observed in > 90% of neoplastic colon tissue including adenoma and adenocarcinoma samples using both microarray and RT-PCR technology[24]. Liu et al[103] found significant upregulation of CRNDE-h splice variant both in adenoma and CRC tissues compared to control groups containing normal adjacent, inflammatory bowel disease and hyperplastic polyp samples. Moreover, within the CRC group, increased expression of CRNDE-h showed significant correlation with tumor size, lymph node, and distant metastasis. It was observed in in vitro studies, that lncRNA CRNDE can promote CRC development and progression through epigenetic silencing of dual-specificity phosphatase 5 (DUSP5) and cyclin-dependent kinase inhibitor 1A (CDKN1A)[104] or via activating Ras/MAPK[105] and WNT/β-catenin[106,107] signaling pathways. Furthermore, it can contribute to chemoresistance by sponging microRNAs (miR-136[108], miR-181a-5p[107]) in CRC.
Some colorectal cancer associated lncRNAs [CRCALs: CRCAL-1 (AC021218.2), CRCAL-2 (LINC00858), CRCAL-3 (RP11-138J23.1) and CRCAL-4 (RP11-435O5.2)] were identified as overexpressed and novel CRC biomarkers using RNA-sequencing techniques[100]. These lncRNAs may “be involved in the very early steps of the neoplastic process” as the expression levels of all four CRCALs were found to be elevated in colorectal adenoma samples, as well. RNA interference-mediated knockdown experiments and gene ontology analysis of The Cancer Genome Atlas (TCGA) dataset suggest the involvement of CRCAL-3 and CRCAL-4 in cell cycle regulation[100].
Several studies have also indicated the tumor-promoting role of UCA1 lncRNA in CRC[101,102,109]. Intensive UCA1 expression was found to be correlated with larger tumor size, depth of invasion, and a less differentiated histology[110]. Moreover, elevated UCA1 levels could be detected in precancerous adenomas which increase in CRC[102].
In a recent publication, Lao et al. have described the gradual elevation of expression of a novel lncRNA, AC123023.1-201 (ENST0000455974) along the colonic normal-adenoma-dysplasia-carcinoma-metastasis sequence[111]. High levels of this lncRNA were found to be significantly associated with poor survival of DNA mismatch repair proficient (pMMR) CRC patients. In vitro studies suggest that AC123023.1-201 might exert an oncogenic role in the pathomechanism of pMMR CRC via promoting JAG2-mediated Notch signaling[111].
LncRNA molecules can cross the cell membrane, and hence can be found in different body fluids, such as blood, plasma/serum or urine[112]. They can be derived from apoptotic and necrotic cells, or from living cells by an active manner. These molecules occur in association with RNA-binding proteins or lipoprotein complexes, however, extracellular vesicles are reported to be the primary source of plasma lncRNAs[113]. These forms contribute to the relative resistance to degradation by RNase enzymes that make circulating lncRNAs promising markers for the prognosis, diagnosis, or screening of various diseases, including CRC[114]. The altered expression levels of several lncRNAs were reported in tumor tissues of CRC patients, and recently, additional articles have been published describing their presence in plasma or serum samples[115]. CCAT1 and HOTAIR are among the first markers reported to have significantly elevated expression in the plasma of CRC patients compared to healthy controls[116]. It was also observed that after surgical treatment of CRC patients, the serum levels of these lncRNAs decreased in comparison with pre-operative samples. HOTAIR expression was also reported in peripheral blood mononuclear cells (PBMC) of CRC blood donors as compared with controls; of note, patients with right-sided CRC had lower levels of HOTAIR lncRNA than those with left-sided cancers[90]. In contrast, HOX antisense intergenic RNA myeloid 1 (HOTAIRM1) showed reduced expression in tumor tissue, and low levels were reported in plasma of CRC patients compared to healthy controls using nested TaqMan RT‐PCR method[117]. It has been assumed that this lncRNA can inhibit intense cell division and therefore, it may function as a tumor suppressor. The expression of lncRNA SLC25A25-AS1 was also significantly decreased in both tumor tissue and serum samples, and based on in vitro measurements, it was observed that downregulation of SLC25A25-AS1 has an impact on chemoresistance and induces the epithelial-mesenchymal transition (EMT) process[118]. Low levels of lnc-GNAT1-1 were detected in the plasma of CRC patients, and with advanced TNM stages, the level of this lncRNA decreased in the peripheral blood[119]. LncRNA growth arrest specific transcript 5 (GAS5) had diminished expression in serum samples of 109 CRC patients compared with 99 healthy controls[120]. Further experiments highlighted that low level of GAS5 was correlated with advanced TNM stages and larger tumor size. LncRNAs that can enhance cell proliferation were also described in some reports. For instance, lncRNA SPRY4-IT1 was found to be significantly upregulated in CRC tissue and serum samples, and its increased expression was associated with late TNM stages. It influences proliferation, migration, and invasion of CRC cells, and has an effect on the expression of EMT-related genes[121]. Long non-coding RNA-activated by TGF-β (lncRNA-ATB) has been analysed in 50 preoperative and postoperative plasma samples of cancer patients and in 50 healthy volunteers, and its overexpression was reported in 70% (35/50) of CRC cases one month after surgery[122]. Moreover, lncRNA-ATB levels were found to be significantly higher in postoperative plasma in comparison with preoperative samples, suggesting that lncRNA might be released by other mechanisms than by the primary tumor. This research group described another lncRNA, fer-1-like protein 4 (FER1L4) that showed decreased expression level in postoperative blood samples compared with the matched preoperative ones in contrast to the above-mentioned lncRNA-ATB[123]. Wang et al. compiled a panel of lncRNA containing 3 RNAs (LOC285194, RP11-462C24.1, and Nbla12061) that were upregulated in 61 CRC serum samples compared to healthy controls (n = 60)[124]. Another study selected four lncRNAs (BANCR, NR_026817, NR_029373, and NR_034119) for further experiments after high-throughput microarray analysis, and concluded that this panel was dysregulated in tissue and serum samples of colon carcinoma patients[125]. Shi et al[126] also performed microarray analysis on the circulating plasma lncRNA fraction using Human LncRNA Array v3.0, and 8 transcripts were further examined with RT-qPCR technique. From these candidates, expression of three (XLOC_006844, LOC152578, and XLOC_000303) lncRNAs were found to be significantly higher in CRC plasma samples (n = 220) compared to cancer-free controls (n = 180). Another lncRNA, nuclear-enriched abundant transcript 1 (NEAT1) was identified based on microarray results as the most significantly upregulated gene in whole blood samples of CRC patients[127]. Two variants of this lncRNA, NEAT1_v1 and NEAT1_v2 were studied separately, and high levels of both two transcripts were observed[128]. Moreover, Wu et al[129] showed that knockdown of NEAT1_v1 caused inhibition of cell invasion and proliferation in vitro, while in case of NEAT1_v2, the knockdown of the transcript could induce cell growth. Similarly to the previous studies, lncRNA bladder cancer associated transcript 1 (BLACAT1) was also found to be overexpressed using microarray analysis and the increased expression was confirmed using RT-PCR in CRC serum samples. Liu et al. selected 3 lncRNAs, H19 antisense (91H), plas-mocytoma variant translocation 1 (PVT-1) and maternally expressed gene 3 (MEG3) and reported increased levels in plasma of CRC patients compared to non-cancerous controls[130]. Our knowledge on the regulation of lncRNA gene expression is incomplete; however, a study by Pedersen et al[131] demonstrated reduced level of lncRNA CAHM in CRC patients coupled with elevated methylation of CAHM gene, which was detectable also in plasma samples.
Additional circulating lncRNAs have been described as potential biomarkers for CRC detection (e.g., HIF1A-AS1, NORAD, CCAT2 or HULC), and more are expected to be identified in the near future [132-134]. The most promising lncRNAs to date are summarised in Table 1.
Exosomes are a subgroup of extracellular vesicles (EVs) that can be found in different body fluids, including blood, serum/plasma, urine or saliva. The particles range from 30 to 100 nm in diameter, and around 2 × 1015 exosomes have been identified in the blood of healthy people; however, in case of cancer, the exosome numbers can increase, and reaching 4 × 1015[135,136]. Recent studies highlighted that exosomes secreted by tumor cells contain DNAs, proteins, lipids, different small molecules and RNAs including lncRNAs, and these molecules may also be taken by target cells. Therefore, the contents of exosomes can influence the biological functions of the recipient cells and play an important part in long distance cell-cell communication[137].
Several differentially expressed lncRNAs in exosomes were reported in plasma/serum samples of CRC patients. According to Liu et al[103], colorectal neoplasia differentially expressed-h (CRNDE-h) showed elevated expression in isolated exosomes of 148 CRC patients compared to benign colorectal disease patients and healthy controls. Moreover, it was observed that a high exosomal level of this lncRNA correlated with both lymph node and distant metastasis and was related to low overall survival rates. Expression of exosomal lncRNA 91H also increased in CRC serum samples, which occurs at a higher level in the vesicles, than in exosome-free sera[138]. It has been also reported that the elevated expression was decreased after surgery. Based on real-time PCR results, Barbagallo et al[101] demonstrated that UCA1 in serum exosomes of cancerous patients was downregulated, while taurine up-regulated 1 (TUG1) was overexpressed. Another study constructed a six-member (LNCV6_116109, LNCV6_98390, LNCV6_38772, LNCV_108266, LNCV6_84003 and LNCV6_98602) panel of plasma exosomal lncRNAs based on microarray analysis that indicated overexpression in CRC patients compared to healthy individuals[139]. The increased level was already observed in the early stages of CRC suggesting that these lncRNAs are potential markers for early detection of cancer. Dong et al[140] showed that two mRNAs (KRTAP5-4 and MAGEA3) and one lncRNA (BCAR4) extracted from sera exosomes are present at a lower level in colorectal adenoma and carcinoma patients compared to healthy individuals, and the combination of these RNAs could be used as CRC biomarkers. Interestingly, according to Li et al[120] lncRNA GAS5 was found to be downregulated in CRC sera samples and acts as a tumor suppressor in cancer development, however, another study revealed that this lncRNA was upregulated in tissues, plasma and exosomes of CRC patients and its expression was related to TNM stage, Dukes stage, lymph node metastasis, local recurrence rate and distant metastasis rate[141].
Analysis of lncRNAs in exosomes is ongoing, and because altered levels of lncRNAs can serve as a potential markers for CRC detection, clarification of their function in cancer development is also a crucial step. The exosomal lncRNAs with altered expression in CRC are listed in Table 1.
Biomarkers - as objectively measurable molecules suitable for monitoring phy-siological and pathological processes and the effect of treatments - have a crucial role in the clinical workup of tumors, enhancing the early diagnosis, classification of tumors, monitoring therapy response, and supporting the evolvement of personalized therapies, as well[21]. LncRNAs can serve as diagnostic, prognostic and predictive biomarkers in malignant diseases including CRC[22]. Principally, lncRNAs with altered levels in different stages of tumorigenesis and progression have a great potential to become early diagnostic and/or prognostic biomarkers. Besides the remarkable expression difference associated with disease stages, the important aspect of their presence and stability in the circulatory system are opening a new path for noninvasive diagnostic applications[21,142]. CCAT1 can serve as a promising marker for early CRC recognition due to its high expression in malignant and benign colorectal tumors compared to normal controls[23,96], and its detection both in PBMC and plasma samples, as well[96,116]. Increased plasma CCAT1 could predict the presence of CRC with 75.7% sensitivity and 85.3% specificity[116]. Almost all splice variants of CRNDE lncRNA, (except for CRNDE-d), and particularly CRDNME-b and CRNDE-h, were found to be intensively (approximately 5- to 100-fold) upregulated in both benign and malignant neoplastic colorectal tissue[24]. On the basis of CRNDE-h expression levels, CRC and normal tissue samples could be discriminated with 85% sensitivity and 96% specificity, which was also proven to be a highly sensitive and specific marker in adenoma vs normal tissue comparison (sensitivity: 95%, specificity: 96%)[24]. Based on CRNDE-h levels in tissue, CRC could be differentiated from adenoma and healthy tissues with 70.4% sensitivity and 70.8% specificity[99]. Its strong diagnostic potential was also supported by the circulating CRNDE-h RT-qPCR results at a reported 87% sensitivity and 93% specificity between CRC vs healthy controls[24]. Moreover, the analysis of exosomal CRC-related CRNDE-h of serum also allowed separation of CRC samples from benign and healthy controls (AUC = 0.892, sensitivity: 70.3%, specificity: 94.4%)[103]. The newly identified upregulated CRCAL1-4 lncRNAs might be suitable for early recognition of colorectal neoplasias, however, only marginal significance could be observed between adenoma and CRC[100]. Potential utilization in CRC screening and diagnostics of several other differentially expressed lncRNAs including BLACAT1[129], CCAT2[134], HULC[134], NEAT1[128], UCA1[101,109] and HOTAIRM1[117] has also emerged in RT-qPCR studies analyzing circulating lncRNAs resulting in various specificity (43%-96%) and sensitivity (55%-100%) values. In addition to the altered expression levels, the DNA methylation changes of lncRNAs can hold a discriminative ability, as the amount of methylated CAHM DNA molecules in the circulatory system depends on the CRC stages; hence it can serve as a promising marker for CRC screening[131].
In addition to single lncRNA marker candidates, lncRNA marker combinations and multi-marker lncRNA panels have also been identified as a potential diagnostic approach. By testing the CRC diagnostic efficacy of circulating HOTAIR and CCAT1, the combined measurement of their plasma/serum levels resulted in higher sensitivity and specificity values (84.3% and 80.2%, respectively) than the above-mentioned markers alone[116]. This marker combination could provide an effective CRC diagnosis performance, moreover, it could detect CRC efficiently already at an early stage (85%). Analysis of Barbagallo et al[101] revealed that diagnostic accuracy of serum exosome UCA1 levels for CRC (sensitivity: 100%, specificity: 43%) could be enhanced by applying it in combination with TUG1 lncRNA (sensitivity: 93%, specificity: 64%) or with circHIPK3 circular non-coding RNA (sensitivity: 100%, specificity: 70%). A promising lncRNA panel containing three lncRNAs (LOC152578, XLOC_000303, and XLOC_0006844) upregulated in CRC was identified and validated on a large independent plasma sample cohort (220 CRCs, 180 controls) (positive predictive value: 0.80, negative predictive value: 0.84, AUC = 0.975)[126]. The double-blind test on another 100 plasma samples (50 CRC, 50 cancer-free controls) also confirmed that the above-mentioned biomarker set is suitable for indicating the occurrence of CRC with 85% accuracy[126]. CRC and healthy normal cases could be distinguished based on the increased serum levels of LOC285194, RP11-46C24.1, and Nbla12061 lncRNAs (AUC = 0.793, sensitivity: 68.33%, specificity: 86.89%)[124]. The predictive value of this lncRNA signature was significantly higher than of the conventional clinical serum protein markers (CEA, CA199, CA125, and CA724) (AUC values were 0.633, 0.567, 0.517 and 0.592, respectively)[124]. Microarray analysis of CRC-NAT tissue sample pairs revealed a four-lncRNA panel (upregulated BANCR and downregulated NR_026817, NR_029373, NR_034119) which had consistently altered pattern both in CRC tissue and serum samples compared to normal controls[125]. The high AUC, specificity and sensitivity values for both the training and validation sample sets support the reliable diagnostic ability of this biomarker set (AUC: 0.891 and 0.881; specificity: 80% and 75.83%; sensitivity: 81.67% and 89.17%) which even exceeded the diagnostic power of CEA[125]. A pilot study of Liu et al. revealed a new promising diagnostic plasma ncRNA biomarker set (H91, PVT-1, MEG3) for early-stage CRCs as the panel could differentiate CRC samples from controls with 82.76% sensitivity and 78.57% specificity[130].
According to the Lnc2Cancer 2.0 database (http://www.bio-bigdata.com/lnc2cancer), the most frequently described lncRNAs with prognostic value in CRC are H19[95,143-145], CRNDE[99,103,105,107,146], HOTAIR[89,90,147,148] and MALAT1[92,145,149] (Supplemental Table 1). In silico lncRNA expression analysis of CRC data from The Cancer Genome Atlas (TCGA) database (n = 534) showed that H19 was the lncRNA mostly associated with the overall survival (OS) of CRC patients (P = 0.0005), independently from tumor stages[143]. Elevated H19 levels were found to be correlated with tumor differentiation and advanced TNM stage[144], and its expression could be considered as an independent predictor for OS and disease-free survival (DFS). Other studies also confirmed that overexpression of H19 lncRNA could predict the unfavorable prognosis in CRC[145]. CRNDE-h can serve as a promising early diagnostic biomarker for CRC, and it also has a prognostic capability due to its high tissue and serum exosome levels significantly correlated with tumor size, lymph node, and distant metastasis[99,103]. In addition, increased exosomal CRNDE-h levels were proven to be a negative predictor of OS of CRC patients [34.6% (high CRNDE-h) vs 68.2% (low CRNDE-h), P < 0.001)][103]. Similar associations with CRC stages were reported for CRNDE-p, another overexpressed transcript variant of CRNDE[146]. HOTAIR lncRNA was also observed to be a negative prognostic factor in CRC, as its upregulated expression in primary tumor tissue, even more in blood of CRC patients were found to be associated with higher mortality [Cox's proportional hazard, hazard ratio (HR) (tissue) = 4.4, HR (blood) = 5.9][90]. Significant differences in clinicopathological parameters such as less differentiated histology, greater tumor depth, and liver metastasis were observed in CRC cases with high HOTAIR expression (n = 20) compared CRCs with low HOTAIR levels (n = 80) (P < 0.05)[89]. Results of several other studies verified the correlation of higher HOTAIR levels with poorer OS[89,148]. With the RT-qPCR analysis of tissue samples from 146 stage II/III CRC patients, it was observed that patients with more intense MALAT1 lncRNA expression had a significantly worse prognosis with a HR of 2.863 for DFS and 3.968 for OS[92]. Moreover, high MALAT1 levels were found to be associated with decreased patient survival and poor response to oxaliplatin-based chemotherapy in advanced CRC patients suggesting its utility as a prognostic marker and therapeutic target in CRC[149].
In addition to CRNDE[103,146] and HOTAIR[90,116], among the 31 potentially prognostic lncRNAs published in at least two independent studies, CCAT2[150], GAS5[141], BLACAT1[129], CCAT1[96,116], NEAT1[128], 91H[138] and BANCR[125] lncRNAs were also detectable in the circulation suggesting their application as minimally invasive markers for CRC prognosis (Table 1 and Supplemental Table 1). As reported by Ozawa et al[150] in a study involving two independent cohorts, the evaluation of CCAT2 expression in combination with CCAT1 may be a powerful tool for predicting tumor recurrence and prognosis in CRC patients. According to the expression analysis in tissue, plasma and exosome samples, GAS5 had a prognostic value in CRC based on its expression that was negatively correlated with TNM status, Dukes stage, and lymph node metastasis (LNM), local recurrence and distant metastasis rate, while its level was in positive relation with differentiation degree and the 3-year OS rate[141]. On the other hand, elevated BLACAT1 expression could be considered as an independent unfavorable prognostic indicator for CRC, as it was observed to be associated with advanced CRC stages and shorter OS[151]. The predictive potential of lncRNA transcript variants can differ, as the OS of CRC patients with intensive NEAT1_v1 expression was worse, while high levels of the other isoform, NEAT1_v2 was correlated with better OS[128]. Determination of clinical significance of elevated exosomal H91 lncRNA expression suggested that it might be an early minimal invasive biomarker for CRC recurrence or metastasis[138]. Gong et al[132] evaluated the diagnostic and prognostic value of increased serum HIF1A-AS1 levels in 151 CRC and 160 healthy control samples by RT-PCR, and reported a high diagnostic efficacy (86.8% sensitivity and 92.5% specificity); moreover, it was described as a predictor for worse prognosis in CRC.
In addition to the diagnostic and prognostic utility of lncRNAs with altered expression, ongoing research focused on the role of lncRNAs in chemoresistance and therapy response prediction are revealing several lncRNAs which could be promising therapeutic targets in CRC. Similarly to the above-mentioned MALAT1 whose increased levels were found to be associated with poor response to oxaliplatin (OXA)-based chemotherapy[149], CRNDE can also contribute to oxaliplatin resistance in CRC[107,108]. According to a recent in vitro study, CRNDE facilitates the resistance against OXA or 5-fluorouracil (5FU) treatment via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling[107]. Association between high HOTAIR expression and poor response to 5FU treatment was assessed[147]. HOTAIR can contribute to 5FU resistance through suppressing miR-218 and activating NF-κB signaling in CRC[147]. HOTAIR was observed to be upregulated in drug-resistant cisplatin- or paclitaxel-treated SW620 and Colo205 CRC cells, as well[148] and could affect the chemoresistance of CRC via miR-203a-3p-mediated modulation of Wnt/β-Catenin pathway[148]. The most important tissue and circulating lncRNAs with diagnostic and prognostic potential in colorectal tumors are represented in Figure 1.
The increasing number of genome-wide expression analysis studies have led to the identification of a number of long non-coding RNAs with altered expression patterns in cancers including CRC. LncRNAs are proven to contribute to each step of the colorectal carcinogenesis and tumor progression by influencing the key cancer-related signal transduction pathways such as WNT/β-catenin, PI3K/Akt, EGFR, NOTCH, mTOR and TP53 signaling. Dysregulated lncRNAs can appear in the pre-malignant adenoma stage of CRC and the expression alterations of a relatively large number of lncRNAs were found to be associated with clinicopathological parameters indicating CRC progression. Furthermore, lncRNAs are stable and detectable in body fluids facilitating their utilization as early detection and prognostic biomarkers. In order to open the door for implementation of minimally invasive lncRNA-based tests in the clinical practice, certain relevant technical aspects should be considered: (1) Standardization of the pre-processing and sample preparation procedure including the applied blood collection tubes, sample storage conditions and time, optimized lncRNA isolation protocols from liquid biopsy samples; (2) Selection of appropriate quantification, quality checking and sensitive techniques allowing the precise detection of cancer-related alterations; and (3) Application of proper universal endogenous controls for increasing the reliability and the accuracy of RT-qPCR measurements. For the development of adequately sensitive and CRC-specific, clinically applicable diagnostic and prognostic tests based on lncRNA markers/marker panels, validation studies with large sample cohorts are essential. On the other hand, as recent studies shed light on the potential role of lncRNAs as novel therapeutic targets, the specific lncRNA expression alterations in liquid biopsy samples may contribute to the improved early recognition, prognosis prediction and therapy monitoring in CRC. Moreover, lncRNAs as druggable targets might represent the basis of novel therapeutic methods in the fight against cancer.
We thank Ramani Gopal PhD and Theo deVos PhD for their careful language assistance.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Hungary
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Fiori E, Gkekas I, Zouiten-Mekki L S-Editor: Ma RY L-Editor: A E-Editor: Zhang YL
| 1. | Ferlay J, Colombet M, Soerjomataram I, Dyba T, Randi G, Bettio M, Gavin A, Visser O, Bray F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer. 2018;103:356-387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1625] [Cited by in RCA: 1660] [Article Influence: 237.1] [Reference Citation Analysis (0)] |
| 2. | Shi T, Gao G, Cao Y. Long Noncoding RNAs as Novel Biomarkers Have a Promising Future in Cancer Diagnostics. Dis Markers. 2016;2016:9085195. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 199] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 3. | Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703-719. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1254] [Cited by in RCA: 1504] [Article Influence: 115.7] [Reference Citation Analysis (0)] |
| 4. | Ma Y, Yang Y, Wang F, Moyer MP, Wei Q, Zhang P, Yang Z, Liu W, Zhang H, Chen N, Wang H, Wang H, Qin H. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/β-catenin signalling pathway via suppression of activator protein 2α. Gut. 2016;65:1494-1504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 268] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 5. | Zhang H, Chen Z, Wang X, Huang Z, He Z, Chen Y. Long non-coding RNA: a new player in cancer. J Hematol Oncol. 2013;6:37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 276] [Cited by in RCA: 360] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 6. | Qiu MT, Hu JW, Yin R, Xu L. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol. 2013;34:613-620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 320] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 7. | Ji Q, Liu X, Fu X, Zhang L, Sui H, Zhou L, Sun J, Cai J, Qin J, Ren J, Li Q. Resveratrol inhibits invasion and metastasis of colorectal cancer cells via MALAT1 mediated Wnt/β-catenin signal pathway. PLoS One. 2013;8:e78700. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 305] [Cited by in RCA: 303] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 8. | Yue B, Liu C, Sun H, Liu M, Song C, Cui R, Qiu S, Zhong M. A Positive Feed-Forward Loop between LncRNA-CYTOR and Wnt/β-Catenin Signaling Promotes Metastasis of Colon Cancer. Mol Ther. 2018;26:1287-1298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 159] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 9. | Xu S, Sui S, Zhang J, Bai N, Shi Q, Zhang G, Gao S, You Z, Zhan C, Liu F, Pang D. Downregulation of long noncoding RNA MALAT1 induces epithelial-to-mesenchymal transition via the PI3K-AKT pathway in breast cancer. Int J Clin Exp Pathol. 2015;8:4881-4891. [PubMed] |
| 10. | Zhou J, Zhi X, Wang L, Wang W, Li Z, Tang J, Wang J, Zhang Q, Xu Z. Linc00152 promotes proliferation in gastric cancer through the EGFR-dependent pathway. J Exp Clin Cancer Res. 2015;34:135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 105] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 11. | Yue B, Cai D, Liu C, Fang C, Yan D. Linc00152 Functions as a Competing Endogenous RNA to Confer Oxaliplatin Resistance and Holds Prognostic Values in Colon Cancer. Mol Ther. 2016;24:2064-2077. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 155] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 12. | Cai Q, Wang ZQ, Wang SH, Li C, Zhu ZG, Quan ZW, Zhang WJ. Upregulation of long non-coding RNA LINC00152 by SP1 contributes to gallbladder cancer cell growth and tumor metastasis via PI3K/AKT pathway. Am J Transl Res. 2016;8:4068-4081. [PubMed] |
| 13. | Bian Z, Zhang J, Li M, Feng Y, Yao S, Song M, Qi X, Fei B, Yin Y, Hua D, Huang Z. Correction: Long non-coding RNA LINC00152 promotes cell proliferation, metastasis, and confers 5-FU resistance in colorectal cancer by inhibiting miR-139-5p. Oncogenesis. 2018;7:63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 14. | Yang X, Duan B, Zhou X. Long non-coding RNA FOXD2-AS1 functions as a tumor promoter in colorectal cancer by regulating EMT and Notch signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21:3586-3591. [PubMed] |
| 15. | Lu S, Dong W, Zhao P, Liu Z. lncRNA FAM83H-AS1 is associated with the prognosis of colorectal carcinoma and promotes cell proliferation by targeting the Notch signaling pathway. Oncol Lett. 2018;15:1861-1868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 16. | Ji J, Tang J, Deng L, Xie Y, Jiang R, Li G, Sun B. LINC00152 promotes proliferation in hepatocellular carcinoma by targeting EpCAM via the mTOR signaling pathway. Oncotarget. 2015;6:42813-42824. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 122] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 17. | Jahangiri B, Khalaj-Kondori M, Asadollahi E, Sadeghizadeh M. Cancer-associated fibroblasts enhance cell proliferation and metastasis of colorectal cancer SW480 cells by provoking long noncoding RNA UCA1. J Cell Commun Signal. 2019;13:53-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 61] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 18. | Sun L, Jiang C, Xu C, Xue H, Zhou H, Gu L, Liu Y, Xu Q. Down-regulation of long non-coding RNA RP11-708H21.4 is associated with poor prognosis for colorectal cancer and promotes tumorigenesis through regulating AKT/mTOR pathway. Oncotarget. 2017;8:27929-27942. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 19. | Thorenoor N, Faltejskova-Vychytilova P, Hombach S, Mlcochova J, Kretz M, Svoboda M, Slaby O. Long non-coding RNA ZFAS1 interacts with CDK1 and is involved in p53-dependent cell cycle control and apoptosis in colorectal cancer. Oncotarget. 2016;7:622-637. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 155] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 20. | Yang P, Yang Y, An W, Xu J, Zhang G, Jie J, Zhang Q. The long noncoding RNA-ROR promotes the resistance of radiotherapy for human colorectal cancer cells by targeting the p53/miR-145 pathway. J Gastroenterol Hepatol. 2017;32:837-845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 81] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 21. | Qi P, Zhou XY, Du X. Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol Cancer. 2016;15:39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 191] [Cited by in RCA: 245] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 22. | Yang Y, Junjie P, Sanjun C, Ma Y. Long non-coding RNAs in Colorectal Cancer: Progression and Future Directions. J Cancer. 2017;8:3212-3225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 59] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 23. | Alaiyan B, Ilyayev N, Stojadinovic A, Izadjoo M, Roistacher M, Pavlov V, Tzivin V, Halle D, Pan H, Trink B, Gure AO, Nissan A. Differential expression of colon cancer associated transcript1 (CCAT1) along the colonic adenoma-carcinoma sequence. BMC Cancer. 2013;13:196. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 127] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 24. | Graham LD, Pedersen SK, Brown GS, Ho T, Kassir Z, Moynihan AT, Vizgoft EK, Dunne R, Pimlott L, Young GP, Lapointe LC, Molloy PL. Colorectal Neoplasia Differentially Expressed (CRNDE), a Novel Gene with Elevated Expression in Colorectal Adenomas and Adenocarcinomas. Genes Cancer. 2011;2:829-840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 221] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 25. | Pachnis V, Belayew A, Tilghman SM. Locus unlinked to alpha-fetoprotein under the control of the murine raf and Rif genes. Proc Natl Acad Sci USA. 1984;81:5523-5527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 244] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 26. | Borsani G, Tonlorenzi R, Simmler MC, Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzny D, Lawrence C, Willard HF, Avner P, Ballabio A. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351:325-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 409] [Cited by in RCA: 426] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 27. | Jarroux J, Morillon A, Pinskaya M. History, Discovery, and Classification of lncRNAs. Adv Exp Med Biol. 2017;1008:1-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 461] [Cited by in RCA: 613] [Article Influence: 76.6] [Reference Citation Analysis (0)] |
| 28. | Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651-669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1443] [Cited by in RCA: 1470] [Article Influence: 122.5] [Reference Citation Analysis (0)] |
| 29. | Gloss BS, Dinger ME. Realizing the significance of noncoding functionality in clinical genomics. Exp Mol Med. 2018;50:97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 67] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 30. | Cheng L, Wang P, Tian R, Wang S, Guo Q, Luo M, Zhou W, Liu G, Jiang H, Jiang Q. LncRNA2Target v2.0: a comprehensive database for target genes of lncRNAs in human and mouse. Nucleic Acids Res. 2019;47:D140-D144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 210] [Cited by in RCA: 250] [Article Influence: 50.0] [Reference Citation Analysis (0)] |
| 31. | Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-854. [PubMed] |
| 32. | Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3924] [Cited by in RCA: 4442] [Article Influence: 277.6] [Reference Citation Analysis (0)] |
| 33. | Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1840] [Cited by in RCA: 2121] [Article Influence: 176.8] [Reference Citation Analysis (0)] |
| 34. | Pelechano V, Steinmetz LM. Gene regulation by antisense transcription. Nat Rev Genet. 2013;14:880-893. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 454] [Cited by in RCA: 496] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 35. | Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:925-933. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 755] [Cited by in RCA: 980] [Article Influence: 81.7] [Reference Citation Analysis (0)] |
| 36. | Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engström PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C; RIKEN Genome Exploration Research Group; Genome Science Group (Genome Network Project Core Group); FANTOM Consortium. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564-1566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1240] [Cited by in RCA: 1286] [Article Influence: 64.3] [Reference Citation Analysis (0)] |
| 37. | He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855-1857. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 423] [Cited by in RCA: 420] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 38. | Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21:400-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 584] [Cited by in RCA: 593] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 39. | Balakirev ES, Ayala FJ. Pseudogenes: are they "junk" or functional DNA? Annu Rev Genet. 2003;37:123-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 343] [Cited by in RCA: 349] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 40. | St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet. 2015;31:239-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 688] [Cited by in RCA: 880] [Article Influence: 88.0] [Reference Citation Analysis (0)] |
| 41. | Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699-712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 1155] [Article Influence: 96.3] [Reference Citation Analysis (0)] |
| 42. | Gudenas BL, Wang L. Prediction of LncRNA Subcellular Localization with Deep Learning from Sequence Features. Sci Rep. 2018;8:16385. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 105] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 43. | Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1528] [Cited by in RCA: 1762] [Article Influence: 160.2] [Reference Citation Analysis (0)] |
| 44. | Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs regulate transcription? Sci Adv. 2017;3:eaao2110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 405] [Cited by in RCA: 525] [Article Influence: 65.6] [Reference Citation Analysis (0)] |
| 45. | Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11:59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 488] [Cited by in RCA: 563] [Article Influence: 46.9] [Reference Citation Analysis (0)] |
| 46. | Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol. 2019;21:542-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 693] [Cited by in RCA: 1041] [Article Influence: 173.5] [Reference Citation Analysis (0)] |
| 47. | Yang S, Sun Z, Zhou Q, Wang W, Wang G, Song J, Li Z, Zhang Z, Chang Y, Xia K, Liu J, Yuan W. MicroRNAs, long noncoding RNAs, and circular RNAs: potential tumor biomarkers and targets for colorectal cancer. Cancer Manag Res. 2018;10:2249-2257. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 72] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 48. | Romero-Barrios N, Legascue MF, Benhamed M, Ariel F, Crespi M. Splicing regulation by long noncoding RNAs. Nucleic Acids Res. 2018;46:2169-2184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 216] [Cited by in RCA: 218] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 49. | Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570-1573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 579] [Cited by in RCA: 602] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 50. | Staněk D, Fox AH. Nuclear bodies: news insights into structure and function. Curr Opin Cell Biol. 2017;46:94-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 100] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 51. | Chujo T, Yamazaki T, Hirose T. Architectural RNAs (arcRNAs): A class of long noncoding RNAs that function as the scaffold of nuclear bodies. Biochim Biophys Acta. 2016;1859:139-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 124] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 52. | Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717-726. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1198] [Cited by in RCA: 1142] [Article Influence: 71.4] [Reference Citation Analysis (0)] |
| 53. | Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689-693. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2754] [Cited by in RCA: 2649] [Article Influence: 176.6] [Reference Citation Analysis (0)] |
| 54. | Merry CR, Forrest ME, Sabers JN, Beard L, Gao XH, Hatzoglou M, Jackson MW, Wang Z, Markowitz SD, Khalil AM. DNMT1-associated long non-coding RNAs regulate global gene expression and DNA methylation in colon cancer. Hum Mol Genet. 2015;24:6240-6253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 134] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 55. | Zhao Y, Sun H, Wang H. Long noncoding RNAs in DNA methylation: new players stepping into the old game. Cell Biosci. 2016;6:45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 85] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 56. | Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, Dinger ME, Mattick JS. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885-898. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 380] [Cited by in RCA: 436] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 57. | Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik N, Ko MS. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 2009;16:45-58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 489] [Cited by in RCA: 444] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 58. | Tani H, Mizutani R, Salam KA, Tano K, Ijiri K, Wakamatsu A, Isogai T, Suzuki Y, Akimitsu N. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 2012;22:947-956. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 303] [Cited by in RCA: 318] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 59. | Zampetaki A, Albrecht A, Steinhofel K. Long Non-coding RNA Structure and Function: Is There a Link? Front Physiol. 2018;9:1201. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 166] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 60. | Dinger ME, Amaral PP, Mercer TR, Mattick JS. Pervasive transcription of the eukaryotic genome: functional indices and conceptual implications. Brief Funct Genomic Proteomic. 2009;8:407-423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 124] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 61. | Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13:95-101. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 407] [Cited by in RCA: 375] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 62. | Charles Richard JL, Eichhorn PJA. Platforms for Investigating LncRNA Functions. SLAS Technol. 2018;23:493-506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 133] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 63. | Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484-487. [PubMed] |
| 64. | Kashi K, Henderson L, Bonetti A, Carninci P. Discovery and functional analysis of lncRNAs: Methodologies to investigate an uncharacterized transcriptome. Biochim Biophys Acta. 2016;1859:3-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 141] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 65. | Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57-63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10137] [Cited by in RCA: 8286] [Article Influence: 517.9] [Reference Citation Analysis (0)] |
| 66. | Shiraki T, Kondo S, Katayama S, Waki K, Kasukawa T, Kawaji H, Kodzius R, Watahiki A, Nakamura M, Arakawa T, Fukuda S, Sasaki D, Podhajska A, Harbers M, Kawai J, Carninci P, Hayashizaki Y. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci USA. 2003;100:15776-15781. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 574] [Cited by in RCA: 522] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 67. | Ma J, Chen Q, Ma D. Biological Functions and Research Methods of Long Noncoding RNAs. Shengzhi Yu Biyun. 2017;1:23-29. |
| 68. | Ferrè F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform. 2016;17:106-116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 323] [Cited by in RCA: 512] [Article Influence: 51.2] [Reference Citation Analysis (0)] |
| 69. | Uren PJ, Bahrami-Samani E, Burns SC, Qiao M, Karginov FV, Hodges E, Hannon GJ, Sanford JR, Penalva LO, Smith AD. Site identification in high-throughput RNA-protein interaction data. Bioinformatics. 2012;28:3013-3020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 243] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 70. | Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP--transcriptome-wide mapping of protein-RNA interactions with individual nucleotide resolution. J Vis Exp. 2011;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 135] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 71. | Chu C, Quinn J, Chang HY. Chromatin isolation by RNA purification (ChIRP). J Vis Exp. 2012;pii:3912. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 215] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 72. | Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko AA, Borowsky ML, Kuroda MI, Kingston RE. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci USA. 2011;108:20497-20502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 320] [Cited by in RCA: 343] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 73. | Engreitz J, Lander ES, Guttman M. RNA antisense purification (RAP) for mapping RNA interactions with chromatin. Methods Mol Biol. 2015;1262:183-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 68] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 74. | Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M, Lander ES. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 2014;159:188-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 340] [Cited by in RCA: 393] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 75. | Kudla G, Granneman S, Hahn D, Beggs JD, Tollervey D. Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc Natl Acad Sci USA. 2011;108:10010-10015. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 245] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 76. | Wan Y, Kertesz M, Spitale RC, Segal E, Chang HY. Understanding the transcriptome through RNA structure. Nat Rev Genet. 2011;12:641-655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 351] [Cited by in RCA: 353] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 77. | Shah S, Takei Y, Zhou W, Lubeck E, Yun J, Eng CL, Koulena N, Cronin C, Karp C, Liaw EJ, Amin M, Cai L. Dynamics and Spatial Genomics of the Nascent Transcriptome by Intron seqFISH. Cell. 2018;174:363-376.e16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 218] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 78. | Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1151] [Cited by in RCA: 1738] [Article Influence: 173.8] [Reference Citation Analysis (0)] |
| 79. | Cabili MN, Dunagin MC, McClanahan PD, Biaesch A, Padovan-Merhar O, Regev A, Rinn JL, Raj A. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015;16:20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 457] [Cited by in RCA: 512] [Article Influence: 51.2] [Reference Citation Analysis (0)] |
| 80. | Gustafsson MG. Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. Proc Natl Acad Sci U S A. 2005;102:13081-13086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1727] [Cited by in RCA: 1175] [Article Influence: 58.8] [Reference Citation Analysis (0)] |
| 81. | Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Methods. 2006;3:793-795. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6999] [Cited by in RCA: 4863] [Article Influence: 255.9] [Reference Citation Analysis (0)] |
| 82. | Dias N, Stein CA. Potential roles of antisense oligonucleotides in cancer therapy. The example of Bcl-2 antisense oligonucleotides. Eur J Pharm Biopharm. 2002;54:263-269. [PubMed] |
| 83. | Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816-821. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11979] [Cited by in RCA: 11018] [Article Influence: 847.5] [Reference Citation Analysis (0)] |
| 84. | Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen Y, Mandegar MA, Olvera MP, Gilbert LA, Conklin BR, Chang HY, Weissman JS, Lim DA. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2017;355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 435] [Cited by in RCA: 541] [Article Influence: 60.1] [Reference Citation Analysis (0)] |
| 85. | Bester AC, Lee JD, Chavez A, Lee YR, Nachmani D, Vora S, Victor J, Sauvageau M, Monteleone E, Rinn JL, Provero P, Church GM, Clohessy JG, Pandolfi PP. An Integrated Genome-wide CRISPRa Approach to Functionalize lncRNAs in Drug Resistance. Cell. 2018;173:649-664.e20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 239] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 86. | Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, Qi LS, Kampmann M, Weissman JS. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647-661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1983] [Cited by in RCA: 1999] [Article Influence: 181.7] [Reference Citation Analysis (0)] |
| 87. | Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071-1076. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4202] [Cited by in RCA: 4231] [Article Influence: 282.1] [Reference Citation Analysis (0)] |
| 88. | Wu ZH, Wang XL, Tang HM, Jiang T, Chen J, Lu S, Qiu GQ, Peng ZH, Yan DW. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol Rep. 2014;32:395-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 173] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 89. | Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, Miyano S, Mori M. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320-6326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 922] [Cited by in RCA: 1039] [Article Influence: 74.2] [Reference Citation Analysis (0)] |
| 90. | Svoboda M, Slyskova J, Schneiderova M, Makovicky P, Bielik L, Levy M, Lipska L, Hemmelova B, Kala Z, Protivankova M, Vycital O, Liska V, Schwarzova L, Vodickova L, Vodicka P. HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis. 2014;35:1510-1515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 205] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 91. | Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: a long non-coding RNA and its important 3' end functional motif in colorectal cancer metastasis. Int J Oncol. 2011;39:169-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 183] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 92. | Zheng HT, Shi DB, Wang YW, Li XX, Xu Y, Tripathi P, Gu WL, Cai GX, Cai SJ. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7:3174-3181. [PubMed] |
| 93. | Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z, Sui H, Tang Y, Wang Y, Liu N, Ren J, Hou F, Li Q. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. 2014;111:736-748. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 246] [Cited by in RCA: 323] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 94. | Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ, Kwok TT. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31:350-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 340] [Cited by in RCA: 374] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 95. | Yang W, Ning N, Jin X. The lncRNA H19 Promotes Cell Proliferation by Competitively Binding to miR-200a and Derepressing β-Catenin Expression in Colorectal Cancer. Biomed Res Int. 2017;2017:2767484. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 96. | Nissan A, Stojadinovic A, Mitrani-Rosenbaum S, Halle D, Grinbaum R, Roistacher M, Bochem A, Dayanc BE, Ritter G, Gomceli I, Bostanci EB, Akoglu M, Chen YT, Old LJ, Gure AO. Colon cancer associated transcript-1: a novel RNA expressed in malignant and pre-malignant human tissues. Int J Cancer. 2012;130:1598-1606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 240] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 97. | He X, Tan X, Wang X, Jin H, Liu L, Ma L, Yu H, Fan Z. C-Myc-activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumour Biol. 2014;35:12181-12188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 191] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 98. | Xie X, Tang B, Xiao YF, Xie R, Li BS, Dong H, Zhou JY, Yang SM. Long non-coding RNAs in colorectal cancer. Oncotarget. 2016;7:5226-5239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 121] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 99. | Liu T, Zhang X, Yang YM, Du LT, Wang CX. Increased expression of the long noncoding RNA CRNDE-h indicates a poor prognosis in colorectal cancer, and is positively correlated with IRX5 mRNA expression. Onco Targets Ther. 2016;9:1437-1448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 100. | Yamada A, Yu P, Lin W, Okugawa Y, Boland CR, Goel A. A RNA-Sequencing approach for the identification of novel long non-coding RNA biomarkers in colorectal cancer. Sci Rep. 2018;8:575. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 80] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 101. | Barbagallo C, Brex D, Caponnetto A, Cirnigliaro M, Scalia M, Magnano A, Caltabiano R, Barbagallo D, Biondi A, Cappellani A, Basile F, Di Pietro C, Purrello M, Ragusa M. LncRNA UCA1, Upregulated in CRC Biopsies and Downregulated in Serum Exosomes, Controls mRNA Expression by RNA-RNA Interactions. Mol Ther Nucleic Acids. 2018;12:229-241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 167] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 102. | Kalmar A, Nagy ZB, Galamb O, Csabai I, Bodor A, Wichmann B, Valcz G, Bartak BK, Tulassay Z, Igaz P, Molnar B. Genome-wide expression profiling in colorectal cancer focusing on lncRNAs in the adenoma-carcinoma transition. BMC Cancer. 2019;In press. |
| 103. | Liu T, Zhang X, Gao S, Jing F, Yang Y, Du L, Zheng G, Li P, Li C, Wang C. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget. 2016;7:85551-85563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 196] [Cited by in RCA: 259] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 104. | Ding J, Li J, Wang H, Tian Y, Xie M, He X, Ji H, Ma Z, Hui B, Wang K, Ji G. Long noncoding RNA CRNDE promotes colorectal cancer cell proliferation via epigenetically silencing DUSP5/CDKN1A expression. Cell Death Dis. 2017;8:e2997. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 132] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 105. | Jiang H, Wang Y, Ai M, Wang H, Duan Z, Wang H, Zhao L, Yu J, Ding Y, Wang S. Long noncoding RNA CRNDE stabilized by hnRNPUL2 accelerates cell proliferation and migration in colorectal carcinoma via activating Ras/MAPK signaling pathways. Cell Death Dis. 2017;8:e2862. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 80] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 106. | Yu B, Ye X, Du Q, Zhu B, Zhai Q, Li XX. The Long Non-Coding RNA CRNDE Promotes Colorectal Carcinoma Progression by Competitively Binding miR-217 with TCF7L2 and Enhancing the Wnt/β-Catenin Signaling Pathway. Cell Physiol Biochem. 2017;41:2489-2502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 80] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 107. | Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu XY, Yu ZW, Jia YH, Bai XF, Li L, Liu YL, Cui BB. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol Cancer. 2017;16:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 286] [Cited by in RCA: 406] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 108. | Gao H, Song X, Kang T, Yan B, Feng L, Gao L, Ai L, Liu X, Yu J, Li H. Long noncoding RNA CRNDE functions as a competing endogenous RNA to promote metastasis and oxaliplatin resistance by sponging miR-136 in colorectal cancer. Onco Targets Ther. 2017;10:205-216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 77] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 109. | Tao K, Yang J, Hu Y, Sun Y, Tan Z, Duan J, Zhang F, Yan H, Deng A. Clinical significance of urothelial carcinoma associated 1 in colon cancer. Int J Clin Exp Med. 2015;8:21854-21860. [PubMed] |
| 110. | Han Y, Yang YN, Yuan HH, Zhang TT, Sui H, Wei XL, Liu L, Huang P, Zhang WJ, Bai YX. UCA1, a long non-coding RNA up-regulated in colorectal cancer influences cell proliferation, apoptosis and cell cycle distribution. Pathology. 2014;46:396-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 195] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 111. | Lao Y, Li Q, Li N, Liu H, Liu K, Jiang G, Wei N, Wang C, Wang Y, Wu J. Long noncoding RNA ENST00000455974 plays an oncogenic role through up-regulating JAG2 in human DNA mismatch repair-proficient colon cancer. Biochem Biophys Res Commun. 2019;508:339-347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 112. | Sole C, Arnaiz E, Manterola L, Otaegui D, Lawrie CH. The circulating transcriptome as a source of cancer liquid biopsy biomarkers. Semin Cancer Biol. 2019;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 87] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 113. | Li Q, Shao Y, Zhang X, Zheng T, Miao M, Qin L, Wang B, Ye G, Xiao B, Guo J. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol. 2015;36:2007-2012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 320] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 114. | Viereck J, Thum T. Circulating Noncoding RNAs as Biomarkers of Cardiovascular Disease and Injury. Circ Res. 2017;120:381-399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 302] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 115. | Chen D, Sun Q, Cheng X, Zhang L, Song W, Zhou D, Lin J, Wang W. Genome-wide analysis of long noncoding RNA (lncRNA) expression in colorectal cancer tissues from patients with liver metastasis. Cancer Med. 2016;5:1629-1639. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 61] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 116. | Zhao W, Song M, Zhang J, Kuerban M, Wang H. Combined identification of long non-coding RNA CCAT1 and HOTAIR in serum as an effective screening for colorectal carcinoma. Int J Clin Exp Pathol. 2015;8:14131-14140. [PubMed] |
| 117. | Wan L, Kong J, Tang J, Wu Y, Xu E, Lai M, Zhang H. HOTAIRM1 as a potential biomarker for diagnosis of colorectal cancer functions the role in the tumour suppressor. J Cell Mol Med. 2016;20:2036-2044. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 71] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 118. | Li Y, Huang S, Li Y, Zhang W, He K, Zhao M, Lin H, Li D, Zhang H, Zheng Z, Huang C. Decreased expression of LncRNA SLC25A25-AS1 promotes proliferation, chemoresistance, and EMT in colorectal cancer cells. Tumour Biol. 2016;37:14205-14215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 119. | Ye C, Shen Z, Wang B, Li Y, Li T, Yang Y, Jiang K, Ye Y, Wang S. A novel long non-coding RNA lnc-GNAT1-1 is low expressed in colorectal cancer and acts as a tumor suppressor through regulating RKIP-NF-κB-Snail circuit. J Exp Clin Cancer Res. 2016;35:187. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 120. | Li Y, Li Y, Huang S, He K, Zhao M, Lin H, Li D, Qian J, Zhou C, Chen Y, Huang C. Long non-coding RNA growth arrest specific transcript 5 acts as a tumour suppressor in colorectal cancer by inhibiting interleukin-10 and vascular endothelial growth factor expression. Oncotarget. 2017;8:13690-13702. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 52] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 121. | Cao D, Ding Q, Yu W, Gao M, Wang Y. Long noncoding RNA SPRY4-IT1 promotes malignant development of colorectal cancer by targeting epithelial-mesenchymal transition. Onco Targets Ther. 2016;9:5417-5425. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 42] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 122. | Yue B, Qiu S, Zhao S, Liu C, Zhang D, Yu F, Peng Z, Yan D. LncRNA-ATB mediated E-cadherin repression promotes the progression of colon cancer and predicts poor prognosis. J Gastroenterol Hepatol. 2016;31:595-603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 142] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 123. | Yue B, Sun B, Liu C, Zhao S, Zhang D, Yu F, Yan D. Long non-coding RNA Fer-1-like protein 4 suppresses oncogenesis and exhibits prognostic value by associating with miR-106a-5p in colon cancer. Cancer Sci. 2015;106:1323-1332. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 101] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 124. | Wang C, Yu J, Han Y, Li L, Li J, Li T, Qi P. Long non-coding RNAs LOC285194, RP11-462C24.1 and Nbla12061 in serum provide a new approach for distinguishing patients with colorectal cancer from healthy controls. Oncotarget. 2016;7:70769-70778. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 125. | Wang R, Du L, Yang X, Jiang X, Duan W, Yan S, Xie Y, Zhu Y, Wang Q, Wang L, Yang Y, Wang C. Identification of long noncoding RNAs as potential novel diagnosis and prognosis biomarkers in colorectal cancer. J Cancer Res Clin Oncol. 2016;142:2291-2301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 126. | Shi J, Li X, Zhang F, Zhang C, Guan Q, Cao X, Zhu W, Zhang X, Cheng Y, Ou K, Chen Q, Hu S. Circulating lncRNAs associated with occurrence of colorectal cancer progression. Am J Cancer Res. 2015;5:2258-2265. [PubMed] |
| 127. | Xu Y, Xu Q, Yang L, Ye X, Liu F, Wu F, Ni S, Tan C, Cai G, Meng X, Cai S, Du X. Identification and validation of a blood-based 18-gene expression signature in colorectal cancer. Clin Cancer Res. 2013;19:3039-3049. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 128. | Wu Y, Yang L, Zhao J, Li C, Nie J, Liu F, Zhuo C, Zheng Y, Li B, Wang Z, Xu Y. Nuclear-enriched abundant transcript 1 as a diagnostic and prognostic biomarker in colorectal cancer. Mol Cancer. 2015;14:191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 123] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 129. | Dai M, Chen X, Mo S, Li J, Huang Z, Huang S, Xu J, He B, Zou Y, Chen J, Dai S. Meta-signature LncRNAs serve as novel biomarkers for colorectal cancer: integrated bioinformatics analysis, experimental validation and diagnostic evaluation. Sci Rep. 2017;7:46572. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 55] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 130. | Liu H, Ye D, Chen A, Tan D, Zhang W, Jiang W, Wang M, Zhang X. A pilot study of new promising non-coding RNA diagnostic biomarkers for early-stage colorectal cancers. Clin Chem Lab Med. 2019;57:1073-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 131. | Pedersen SK, Mitchell SM, Graham LD, McEvoy A, Thomas ML, Baker RT, Ross JP, Xu ZZ, Ho T, LaPointe LC, Young GP, Molloy PL. CAHM, a long non-coding RNA gene hypermethylated in colorectal neoplasia. Epigenetics. 2014;9:1071-1082. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 132. | Gong W, Tian M, Qiu H, Yang Z. Elevated serum level of lncRNA-HIF1A-AS1 as a novel diagnostic predictor for worse prognosis in colorectal carcinoma. Cancer Biomark. 2017;20:417-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 133. | Wang L, Du L, Duan W, Yan S, Xie Y, Wang C. Overexpression of long noncoding RNA NORAD in colorectal cancer associates with tumor progression. Onco Targets Ther. 2018;11:6757-6766. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 134. | Shaker OG, Senousy MA, Elbaz EM. Association of rs6983267 at 8q24, HULC rs7763881 polymorphisms and serum lncRNAs CCAT2 and HULC with colorectal cancer in Egyptian patients. Sci Rep. 2017;7:16246. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 73] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 135. | Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208-1215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 944] [Cited by in RCA: 1418] [Article Influence: 157.6] [Reference Citation Analysis (0)] |
| 136. | Shao Y, Shen Y, Chen T, Xu F, Chen X, Zheng S. The functions and clinical applications of tumor-derived exosomes. Oncotarget. 2016;7:60736-60751. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 137. | Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 2013;32:623-642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 922] [Cited by in RCA: 900] [Article Influence: 75.0] [Reference Citation Analysis (0)] |
| 138. | Gao T, Liu X, He B, Nie Z, Zhu C, Zhang P, Wang S. Exosomal lncRNA 91H is associated with poor development in colorectal cancer by modifying HNRNPK expression. Cancer Cell Int. 2018;18:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 91] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 139. | Hu D, Zhan Y, Zhu K, Bai M, Han J, Si Y, Zhang H, Kong D. Plasma Exosomal Long Non-Coding RNAs Serve as Biomarkers for Early Detection of Colorectal Cancer. Cell Physiol Biochem. 2018;51:2704-2715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 73] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 140. | Dong L, Lin W, Qi P, Xu MD, Wu X, Ni S, Huang D, Weng WW, Tan C, Sheng W, Zhou X, Du X. Circulating Long RNAs in Serum Extracellular Vesicles: Their Characterization and Potential Application as Biomarkers for Diagnosis of Colorectal Cancer. Cancer Epidemiol Biomarkers Prev. 2016;25:1158-1166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 178] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 141. | Liu L, Meng T, Yang XH, Sayim P, Lei C, Jin B, Ge L, Wang HJ. Prognostic and predictive value of long non-coding RNA GAS5 and mircoRNA-221 in colorectal cancer and their effects on colorectal cancer cell proliferation, migration and invasion. Cancer Biomark. 2018;22:283-299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 95] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 142. | Molnár B, Galamb O, Kalmár A, Barták BK, Nagy ZB, Tóth K, Tulassay Z, Igaz P, Dank M. Circulating cell-free nucleic acids as biomarkers in colorectal cancer screening and diagnosis - an update. Expert Rev Mol Diagn. 2019;19:477-498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 143. | Ohtsuka M, Ling H, Ivan C, Pichler M, Matsushita D, Goblirsch M, Stiegelbauer V, Shigeyasu K, Zhang X, Chen M, Vidhu F, Bartholomeusz GA, Toiyama Y, Kusunoki M, Doki Y, Mori M, Song S, Gunther JR, Krishnan S, Slaby O, Goel A, Ajani JA, Radovich M, Calin GA. H19 Noncoding RNA, an Independent Prognostic Factor, Regulates Essential Rb-E2F and CDK8-β-Catenin Signaling in Colorectal Cancer. EBioMedicine. 2016;13:113-124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 98] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 144. | Han D, Gao X, Wang M, Qiao Y, Xu Y, Yang J, Dong N, He J, Sun Q, Lv G, Xu C, Tao J, Ma N. Long noncoding RNA H19 indicates a poor prognosis of colorectal cancer and promotes tumor growth by recruiting and binding to eIF4A3. Oncotarget. 2016;7:22159-22173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 167] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 145. | Li Q, Dai Y, Wang F, Hou S. Differentially expressed long non-coding RNAs and the prognostic potential in colorectal cancer. Neoplasma. 2016;63:977-983. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 82] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 146. | Yu B, Du Q, Li H, Liu HY, Ye X, Zhu B, Zhai Q, Li XX. Diagnostic potential of serum exosomal colorectal neoplasia differentially expressed long non-coding RNA (CRNDE-p) and microRNA-217 expression in colorectal carcinoma. Oncotarget. 2017;8:83745-83753. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 52] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 147. | Li P, Zhang X, Wang L, Du L, Yang Y, Liu T, Li C, Wang C. lncRNA HOTAIR Contributes to 5FU Resistance through Suppressing miR-218 and Activating NF-κB/TS Signaling in Colorectal Cancer. Mol Ther Nucleic Acids. 2017;8:356-369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 108] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 148. | Xiao Z, Qu Z, Chen Z, Fang Z, Zhou K, Huang Z, Guo X, Zhang Y. LncRNA HOTAIR is a Prognostic Biomarker for the Proliferation and Chemoresistance of Colorectal Cancer via MiR-203a-3p-Mediated Wnt/ß-Catenin Signaling Pathway. Cell Physiol Biochem. 2018;46:1275-1285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 123] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 149. | Li P, Zhang X, Wang H, Wang L, Liu T, Du L, Yang Y, Wang C. MALAT1 Is Associated with Poor Response to Oxaliplatin-Based Chemotherapy in Colorectal Cancer Patients and Promotes Chemoresistance through EZH2. Mol Cancer Ther. 2017;16:739-751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 211] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 150. | Ozawa T, Matsuyama T, Toiyama Y, Takahashi N, Ishikawa T, Uetake H, Yamada Y, Kusunoki M, Calin G, Goel A. CCAT1 and CCAT2 long noncoding RNAs, located within the 8q.24.21 'gene desert', serve as important prognostic biomarkers in colorectal cancer. Ann Oncol. 2017;28:1882-1888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 144] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 151. | Su J, Zhang E, Han L, Yin D, Liu Z, He X, Zhang Y, Lin F, Lin Q, Mao P, Mao W, Shen D. Long noncoding RNA BLACAT1 indicates a poor prognosis of colorectal cancer and affects cell proliferation by epigenetically silencing of p15. Cell Death Dis. 2017;8:e2665. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 87] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 152. | Deng Q, He B, Gao T, Pan Y, Sun H, Xu Y, Li R, Ying H, Wang F, Liu X, Chen J, Wang S. Up-regulation of 91H promotes tumor metastasis and predicts poor prognosis for patients with colorectal cancer. PLoS One. 2014;9:e103022. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 71] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 153. | Han X, Wang L, Ning Y, Li S, Wang Z. Long non-coding RNA AFAP1-AS1 facilitates tumor growth and promotes metastasis in colorectal cancer. Biol Res. 2016;49:36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 154. | Wang F, Ni H, Sun F, Li M, Chen L. Overexpression of lncRNA AFAP1-AS1 correlates with poor prognosis and promotes tumorigenesis in colorectal cancer. Biomed Pharmacother. 2016;81:152-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 78] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 155. | Niu H, Hu Z, Liu H, Hu G, Yang B, Wu S, Li F. Long non-coding RNA AK027294 involves in the process of proliferation, migration, and apoptosis of colorectal cancer cells. Tumour Biol. 2016;37:10097-10105. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 156. | Hu Y, Chen HY, Yu CY, Xu J, Wang JL, Qian J, Zhang X, Fang JY. A long non-coding RNA signature to improve prognosis prediction of colorectal cancer. Oncotarget. 2014;5:2230-2242. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 135] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 157. | Han J, Rong LF, Shi CB, Dong XG, Wang J, Wang BL, Wen H, He ZY. Screening of lymph nodes metastasis associated lncRNAs in colorectal cancer patients. World J Gastroenterol. 2014;20:8139-8150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 23] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 158. | Sun Y, Zheng ZP, Li H, Zhang HQ, Ma FQ. ANRIL is associated with the survival rate of patients with colorectal cancer, and affects cell migration and invasion in vitro. Mol Med Rep. 2016;14:1714-1720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 58] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 159. | Naemura M, Tsunoda T, Inoue Y, Okamoto H, Shirasawa S, Kotake Y. ANRIL regulates the proliferation of human colorectal cancer cells in both two- and three-dimensional culture. Mol Cell Biochem. 2016;412:141-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 160. | Iguchi T, Uchi R, Nambara S, Saito T, Komatsu H, Hirata H, Ueda M, Sakimura S, Takano Y, Kurashige J, Shinden Y, Eguchi H, Sugimachi K, Maehara Y, Mimori K. A long noncoding RNA, lncRNA-ATB, is involved in the progression and prognosis of colorectal cancer. Anticancer Res. 2015;35:1385-1388. [PubMed] |
| 161. | Li W, Kang Y. A new Lnc in metastasis: long noncoding RNA mediates the prometastatic functions of TGF-β. Cancer Cell. 2014;25:557-559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 74] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 162. | Davison EJ, Tarpey PS, Fiegler H, Tomlinson IP, Carter NP. Deletion at chromosome band 20p12.1 in colorectal cancer revealed by high resolution array comparative genomic hybridization. Genes Chromosomes Cancer. 2005;44:384-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 163. | Shi Y, Liu Y, Wang J, Jie D, Yun T, Li W, Yan L, Wang K, Feng J. Downregulated Long Noncoding RNA BANCR Promotes the Proliferation of Colorectal Cancer Cells via Downregualtion of p21 Expression. PLoS One. 2015;10:e0122679. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 97] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 164. | Guo Q, Zhao Y, Chen J, Hu J, Wang S, Zhang D, Sun Y. BRAF-activated long non-coding RNA contributes to colorectal cancer migration by inducing epithelial-mesenchymal transition. Oncol Lett. 2014;8:869-875. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 97] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 165. | Gao X, Wen J, Gao P, Zhang G, Zhang G. Overexpression of the long non-coding RNA, linc-UBC1, is associated with poor prognosis and facilitates cell proliferation, migration, and invasion in colorectal cancer. Onco Targets Ther. 2017;10:1017-1026. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 166. | Zhang Z, Zhou C, Chang Y, Zhang Z, Hu Y, Zhang F, Lu Y, Zheng L, Zhang W, Li X, Li X. Long non-coding RNA CASC11 interacts with hnRNP-K and activates the WNT/β-catenin pathway to promote growth and metastasis in colorectal cancer. Cancer Lett. 2016;376:62-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 193] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 167. | Huang G, Wu X, Li S, Xu X, Zhu H, Chen X. The long noncoding RNA CASC2 functions as a competing endogenous RNA by sponging miR-18a in colorectal cancer. Sci Rep. 2016;6:26524. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 128] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 168. | Xin Y, Li Z, Shen J, Chan MT, Wu WK. CCAT1: a pivotal oncogenic long non-coding RNA in human cancers. Cell Prolif. 2016;49:255-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 93] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 169. | McCleland ML, Mesh K, Lorenzana E, Chopra VS, Segal E, Watanabe C, Haley B, Mayba O, Yaylaoglu M, Gnad F, Firestein R. CCAT1 is an enhancer-templated RNA that predicts BET sensitivity in colorectal cancer. J Clin Invest. 2016;126:639-652. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 184] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 170. | Ye Z, Zhou M, Tian B, Wu B, Li J. Expression of lncRNA-CCAT1, E-cadherin and N-cadherin in colorectal cancer and its clinical significance. Int J Clin Exp Med. 2015;8:3707-3715. [PubMed] |
| 171. | Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z, Zhang S, Wang HB, Ge J, Lu X, Yang L, Chen LL. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24:513-531. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 457] [Cited by in RCA: 579] [Article Influence: 52.6] [Reference Citation Analysis (0)] |
| 172. | Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, Nishida N, Gafà R, Song J, Guo Z, Ivan C, Barbarotto E, De Vries I, Zhang X, Ferracin M, Churchman M, van Galen JF, Beverloo BH, Shariati M, Haderk F, Estecio MR, Garcia-Manero G, Patijn GA, Gotley DC, Bhardwaj V, Shureiqi I, Sen S, Multani AS, Welsh J, Yamamoto K, Taniguchi I, Song MA, Gallinger S, Casey G, Thibodeau SN, Le Marchand L, Tiirikainen M, Mani SA, Zhang W, Davuluri RV, Mimori K, Mori M, Sieuwerts AM, Martens JW, Tomlinson I, Negrini M, Berindan-Neagoe I, Foekens JA, Hamilton SR, Lanza G, Kopetz S, Fodde R, Calin GA. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23:1446-1461. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 456] [Cited by in RCA: 513] [Article Influence: 42.8] [Reference Citation Analysis (0)] |
| 173. | Kasagi Y, Oki E, Ando K, Ito S, Iguchi T, Sugiyama M, Nakashima Y, Ohgaki K, Saeki H, Mimori K, Maehara Y. The Expression of CCAT2, a Novel Long Noncoding RNA Transcript, and rs6983267 Single-Nucleotide Polymorphism Genotypes in Colorectal Cancers. Oncology. 2017;92:48-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 174. | Ye LC, Ren L, Qiu JJ, Zhu DX, Chen T, Chang WJ, Lv SX, Xu J. Aberrant expression of long noncoding RNAs in colorectal cancer with liver metastasis. Tumour Biol. 2015;36:8747-8754. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 175. | Ye LC, Chen T, Zhu DX, Lv SX, Qiu JJ, Xu J, Yuan FL, Wei Y. Downregulated long non-coding RNA CLMAT3 promotes the proliferation of colorectal cancer cells by targeting regulators of the cell cycle pathway. Oncotarget. 2016;7:58931-58938. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 176. | Yuan Z, Yu X, Ni B, Chen D, Yang Z, Huang J, Wang J, Chen D, Wang L. Overexpression of long non-coding RNA-CTD903 inhibits colorectal cancer invasion and migration by repressing Wnt/β-catenin signaling and predicts favorable prognosis. Int J Oncol. 2016;48:2675-2685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 61] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 177. | Chen X, Zhu H, Wu X, Xie X, Huang G, Xu X, Li S, Xing C. Downregulated pseudogene CTNNAP1 promote tumor growth in human cancer by downregulating its cognate gene CTNNA1 expression. Oncotarget. 2016;7:55518-55528. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 178. | Liu Y, Zhang M, Liang L, Li J, Chen YX. Over-expression of lncRNA DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2015;8:11480-11484. [PubMed] |
| 179. | Sun L, Xue H, Jiang C, Zhou H, Gu L, Liu Y, Xu C, Xu Q. LncRNA DQ786243 contributes to proliferation and metastasis of colorectal cancer both in vitro and in vivo. Biosci Rep. 2016;36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 180. | Yochum GS, Cleland R, McWeeney S, Goodman RH. An antisense transcript induced by Wnt/beta-catenin signaling decreases E2F4. J Biol Chem. 2007;282:871-878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 48] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 181. | Yang P, Xu ZP, Chen T, He ZY. Long noncoding RNA expression profile analysis of colorectal cancer and metastatic lymph node based on microarray data. Onco Targets Ther. 2016;9:2465-2478. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 13] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 182. | Chen N, Guo D, Xu Q, Yang M, Wang D, Peng M, Ding Y, Wang S, Zhou J. Long non-coding RNA FEZF1-AS1 facilitates cell proliferation and migration in colorectal carcinoma. Oncotarget. 2016;7:11271-11283. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 90] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 183. | Guo XB, Hua Z, Li C, Peng LP, Wang JS, Wang B, Zhi QM. Biological significance of long non-coding RNA FTX expression in human colorectal cancer. Int J Clin Exp Med. 2015;8:15591-15600. [PubMed] |
| 184. | Yang P, Chen T, Xu Z, Zhu H, Wang J, He Z. Long noncoding RNA GAPLINC promotes invasion in colorectal cancer by targeting SNAI2 through binding with PSF and NONO. Oncotarget. 2016;7:42183-42194. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 61] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 185. | Yin D, He X, Zhang E, Kong R, De W, Zhang Z. Long noncoding RNA GAS5 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Med Oncol. 2014;31:253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 153] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 186. | Ma C, Shi X, Zhu Q, Li Q, Liu Y, Yao Y, Song Y. The growth arrest-specific transcript 5 (GAS5): a pivotal tumor suppressor long noncoding RNA in human cancers. Tumour Biol. 2016;37:1437-1444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 145] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 187. | Krell J, Frampton AE, Mirnezami R, Harding V, De Giorgio A, Roca Alonso L, Cohen P, Ottaviani S, Colombo T, Jacob J, Pellegrino L, Buchanan G, Stebbing J, Castellano L. Growth arrest-specific transcript 5 associated snoRNA levels are related to p53 expression and DNA damage in colorectal cancer. PLoS One. 2014;9:e98561. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 188. | Zheng Y, Song D, Xiao K, Yang C, Ding Y, Deng W, Tong S. LncRNA GAS5 contributes to lymphatic metastasis in colorectal cancer. Oncotarget. 2016;7:83727-83734. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 66] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 189. | Zhou J, Li X, Wu M, Lin C, Guo Y, Tian B. Knockdown of Long Noncoding RNA GHET1 Inhibits Cell Proliferation and Invasion of Colorectal Cancer. Oncol Res. 2016;23:303-309. [PubMed] |
| 190. | Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF, Waye MM. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513-22525. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 398] [Cited by in RCA: 503] [Article Influence: 55.9] [Reference Citation Analysis (0)] |
| 191. | Wu KF, Liang WC, Feng L, Pang JX, Waye MM, Zhang JF, Fu WM. H19 mediates methotrexate resistance in colorectal cancer through activating Wnt/β-catenin pathway. Exp Cell Res. 2017;350:312-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 91] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 192. | Yao J, Li J, Geng P, Li Y, Chen H, Zhu Y. Knockdown of a HIF-2α promoter upstream long noncoding RNA impairs colorectal cancer stem cell properties in vitro through HIF-2α downregulation. Onco Targets Ther. 2015;8:3467-3474. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 193. | Dou J, Ni Y, He X, Wu D, Li M, Wu S, Zhang R, Guo M, Zhao F. Decreasing lncRNA HOTAIR expression inhibits human colorectal cancer stem cells. Am J Transl Res. 2016;8:98-108. [PubMed] |
| 194. | Luo ZF, Zhao D, Li XQ, Cui YX, Ma N, Lu CX, Liu MY, Zhou Y. Clinical significance of HOTAIR expression in colon cancer. World J Gastroenterol. 2016;22:5254-5259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 52] [Cited by in RCA: 55] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 195. | Ren YK, Xiao Y, Wan XB, Zhao YZ, Li J, Li Y, Han GS, Chen XB, Zou QY, Wang GC, Lu CM, Xu YC, Wang YC. Association of long non-coding RNA HOTTIP with progression and prognosis in colorectal cancer. Int J Clin Exp Pathol. 2015;8:11458-11463. [PubMed] |
| 196. | Lian Y, Ding J, Zhang Z, Shi Y, Zhu Y, Li J, Peng P, Wang J, Fan Y, De W, Wang K. The long noncoding RNA HOXA transcript at the distal tip promotes colorectal cancer growth partially via silencing of p21 expression. Tumour Biol. 2016;37:7431-7440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 197. | Yang XJ, Huang CQ, Peng CW, Hou JX, Liu JY. Long noncoding RNA HULC promotes colorectal carcinoma progression through epigenetically repressing NKD2 expression. Gene. 2016;592:172-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 56] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 198. | Tanaka K, Shiota G, Meguro M, Mitsuya K, Oshimura M, Kawasaki H. Loss of imprinting of long QT intronic transcript 1 in colorectal cancer. Oncology. 2001;60:268-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 31] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 199. | Nakano S, Murakami K, Meguro M, Soejima H, Higashimoto K, Urano T, Kugoh H, Mukai T, Ikeguchi M, Oshimura M. Expression profile of LIT1/KCNQ1OT1 and epigenetic status at the KvDMR1 in colorectal cancers. Cancer Sci. 2006;97:1147-1154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 76] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 200. | Kong J, Sun W, Li C, Wan L, Wang S, Wu Y, Xu E, Zhang H, Lai M. Long non-coding RNA LINC01133 inhibits epithelial-mesenchymal transition and metastasis in colorectal cancer by interacting with SRSF6. Cancer Lett. 2016;380:476-484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 155] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 201. | Qiu JJ, Yan JB. Long non-coding RNA LINC01296 is a potential prognostic biomarker in patients with colorectal cancer. Tumour Biol. 2015;36:7175-7183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 54] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 202. | Zhai H, Fesler A, Schee K, Fodstad O, Flatmark K, Ju J. Clinical significance of long intergenic noncoding RNA-p21 in colorectal cancer. Clin Colorectal Cancer. 2013;12:261-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 90] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 203. | Wang J, Lei ZJ, Guo Y, Wang T, Qin ZY, Xiao HL, Fan LL, Chen DF, Bian XW, Liu J, Wang B. miRNA-regulated delivery of lincRNA-p21 suppresses β-catenin signaling and tumorigenicity of colorectal cancer stem cells. Oncotarget. 2015;6:37852-37870. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 72] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 204. | Wang G, Li Z, Zhao Q, Zhu Y, Zhao C, Li X, Ma Z, Li X, Zhang Y. LincRNA-p21 enhances the sensitivity of radiotherapy for human colorectal cancer by targeting the Wnt/β-catenin signaling pathway. Oncol Rep. 2014;31:1839-1845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 121] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 205. | Wang L, Bu P, Ai Y, Srinivasan T, Chen HJ, Xiang K, Lipkin SM, Shen X. A long non-coding RNA targets microRNA miR-34a to regulate colon cancer stem cell asymmetric division. Elife. 2016;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 88] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 206. | Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083-1096. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 415] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
| 207. | Kazemzadeh M, Safaralizadeh R, Feizi MA, Ravanbakhsh R, Somi MH, Hashemzadeh S. LOC100287225, novel long intergenic non-coding RNA, misregulates in colorectal cancer. Cancer Biomark. 2016;16:499-505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 208. | Kazemzadeh M, Safaralizadeh R, Feizi MA, Somi MH, Shokoohi B. Misregulation of the dependence receptor DCC and its upstream lincRNA, LOC100287225, in colorectal cancer. Tumori. 2017;103:40-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 209. | Qi P, Xu MD, Ni SJ, Huang D, Wei P, Tan C, Zhou XY, Du X. Low expression of LOC285194 is associated with poor prognosis in colorectal cancer. J Transl Med. 2013;11:122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 120] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 210. | Xu J, Zhang R, Zhao J. The Novel Long Noncoding RNA TUSC7 Inhibits Proliferation by Sponging MiR-211 in Colorectal Cancer. Cell Physiol Biochem. 2017;41:635-644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 83] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 211. | Ding J, Lu B, Wang J, Wang J, Shi Y, Lian Y, Zhu Y, Wang J, Fan Y, Wang Z, De W, Wang K. Long non-coding RNA Loc554202 induces apoptosis in colorectal cancer cells via the caspase cleavage cascades. J Exp Clin Cancer Res. 2015;34:100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 61] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 212. | Yang L, Wei H, Xiao HJ. Long non-coding RNA Loc554202 expression as a prognostic factor in patients with colorectal cancer. Eur Rev Med Pharmacol Sci. 2016;20:4243-4247. [PubMed] |
| 213. | Yang MH, Hu ZY, Xu C, Xie LY, Wang XY, Chen SY, Li ZG. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim Biophys Acta. 2015;1852:166-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 165] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 214. | Wang Y, Xue D, Li Y, Pan X, Zhang X, Kuang B, Zhou M, Li X, Xiong W, Li G, Zeng Z, Yang T. The Long Noncoding RNA MALAT-1 is A Novel Biomarker in Various Cancers: A Meta-analysis Based on the GEO Database and Literature. J Cancer. 2016;7:991-1001. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 98] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 215. | Li L, Shang J, Zhang Y, Liu S, Peng Y, Zhou Z, Pan H, Wang X, Chen L, Zhao Q. MEG3 is a prognostic factor for CRC and promotes chemosensitivity by enhancing oxaliplatin-induced cell apoptosis. Oncol Rep. 2017;38:1383-1392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 216. | Yin DD, Liu ZJ, Zhang E, Kong R, Zhang ZH, Guo RH. Decreased expression of long noncoding RNA MEG3 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Tumour Biol. 2015;36:4851-4859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 132] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 217. | Kong H, Wu Y, Zhu M, Zhai C, Qian J, Gao X, Wang S, Hou Y, Lu S, Zhu H. Long non-coding RNAs: novel prognostic biomarkers for liver metastases in patients with early stage colorectal cancer. Oncotarget. 2016;7:50428-50436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 218. | Cao X, Zhuang S, Hu Y, Xi L, Deng L, Sheng H, Shen W. Associations between polymorphisms of long non-coding RNA MEG3 and risk of colorectal cancer in Chinese. Oncotarget. 2016;7:19054-19059. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 58] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 219. | Franklin JL, Rankin CR, Levy S, Snoddy JR, Zhang B, Washington MK, Thomson JM, Whitehead RH, Coffey RJ. Malignant transformation of colonic epithelial cells by a colon-derived long noncoding RNA. Biochem Biophys Res Commun. 2013;440:99-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 220. | Qi P, Xu MD, Ni SJ, Shen XH, Wei P, Huang D, Tan C, Sheng WQ, Zhou XY, Du X. Down-regulation of ncRAN, a long non-coding RNA, contributes to colorectal cancer cell migration and invasion and predicts poor overall survival for colorectal cancer patients. Mol Carcinog. 2015;54:742-750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 221. | Smolle M, Uranitsch S, Gerger A, Pichler M, Haybaeck J. Current status of long non-coding RNAs in human cancer with specific focus on colorectal cancer. Int J Mol Sci. 2014;15:13993-14013. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 49] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 222. | Yan B, Gu W, Yang Z, Gu Z, Yue X, Gu Q, Liu L. Downregulation of a long noncoding RNA-ncRuPAR contributes to tumor inhibition in colorectal cancer. Tumour Biol. 2014;35:11329-11335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 223. | Li Y, Li Y, Chen W, He F, Tan Z, Zheng J, Wang W, Zhao Q, Li J. NEAT expression is associated with tumor recurrence and unfavorable prognosis in colorectal cancer. Oncotarget. 2015;6:27641-27650. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 124] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 224. | Lu M, Liu Z, Li B, Wang G, Li D, Zhu Y. The high expression of long non-coding RNA PANDAR indicates a poor prognosis for colorectal cancer and promotes metastasis by EMT pathway. J Cancer Res Clin Oncol. 2017;143:71-81. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 70] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 225. | Ge X, Chen Y, Liao X, Liu D, Li F, Ruan H, Jia W. Overexpression of long noncoding RNA PCAT-1 is a novel biomarker of poor prognosis in patients with colorectal cancer. Med Oncol. 2013;30:588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 159] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 226. | Yang L, Qiu M, Xu Y, Wang J, Zheng Y, Li M, Xu L, Yin R. Upregulation of long non-coding RNA PRNCR1 in colorectal cancer promotes cell proliferation and cell cycle progression. Oncol Rep. 2016;35:318-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 227. | Li L, Sun R, Liang Y, Pan X, Li Z, Bai P, Zeng X, Zhang D, Zhang L, Gao L. Association between polymorphisms in long non-coding RNA PRNCR1 in 8q24 and risk of colorectal cancer. J Exp Clin Cancer Res. 2013;32:104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 75] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 228. | Takahashi Y, Sawada G, Kurashige J, Uchi R, Matsumura T, Ueo H, Takano Y, Eguchi H, Sudo T, Sugimachi K, Yamamoto H, Doki Y, Mori M, Mimori K. Amplification of PVT-1 is involved in poor prognosis via apoptosis inhibition in colorectal cancers. Br J Cancer. 2014;110:164-171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 219] [Cited by in RCA: 261] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 229. | Liu J, Wang J, Song Y, Yang Y, Hu Y, Gao P, Sun J, Chen X, Xu Y, Wang Z. A potential biomarker for colorectal cancer: long non-coding RNA RP1-13P20.6. Neoplasma. 2016;63:984-990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 230. | Chen H, Xu J, Hong J, Tang R, Zhang X, Fang JY. Long noncoding RNA profiles identify five distinct molecular subtypes of colorectal cancer with clinical relevance. Mol Oncol. 2014;8:1393-1403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 52] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 231. | Shi D, Zheng H, Zhuo C, Peng J, Li D, Xu Y, Li X, Cai G, Cai S. Low expression of novel lncRNA RP11-462C24.1 suggests a biomarker of poor prognosis in colorectal cancer. Med Oncol. 2014;31:31. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 67] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 232. | Lee H, Kim C, Ku JL, Kim W, Yoon SK, Kuh HJ, Lee JH, Nam SW, Lee EK. A long non-coding RNA snaR contributes to 5-fluorouracil resistance in human colon cancer cells. Mol Cells. 2014;37:540-546. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 70] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 233. | Li C, Zhou L, He J, Fang XQ, Zhu SW, Xiong MM. Increased long noncoding RNA SNHG20 predicts poor prognosis in colorectal cancer. BMC Cancer. 2016;16:655. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 69] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 234. | Wang JZ, Xu CL, Wu H, Shen SJ. LncRNA SNHG12 promotes cell growth and inhibits cell apoptosis in colorectal cancer cells. Braz J Med Biol Res. 2017;50:e6079. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 85] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 235. | Liu S, Xu B, Yan D. Enhanced expression of long non-coding RNA Sox2ot promoted cell proliferation and motility in colorectal cancer. Minerva Med. 2016;107:279-286. [PubMed] |
| 236. | Shen F, Cai WS, Feng Z, Chen JW, Feng JH, Liu QC, Fang YP, Li KP, Xiao HQ, Cao J, Xu B. Long non-coding RNA SPRY4-IT1 pormotes colorectal cancer metastasis by regulate epithelial-mesenchymal transition. Oncotarget. 2017;8:14479-14486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 237. | Zhang ZY, Lu YX, Zhang ZY, Chang YY, Zheng L, Yuan L, Zhang F, Hu YH, Zhang WJ, Li XN. Loss of TINCR expression promotes proliferation, metastasis through activating EpCAM cleavage in colorectal cancer. Oncotarget. 2016;7:22639-22649. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 55] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 238. | Sun J, Ding C, Yang Z, Liu T, Zhang X, Zhao C, Wang J. The long non-coding RNA TUG1 indicates a poor prognosis for colorectal cancer and promotes metastasis by affecting epithelial-mesenchymal transition. J Transl Med. 2016;14:42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 130] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 239. | Wang L, Zhao Z, Feng W, Ye Z, Dai W, Zhang C, Peng J, Wu K. Long non-coding RNA TUG1 promotes colorectal cancer metastasis via EMT pathway. Oncotarget. 2016;7:51713-51719. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 88] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 240. | Zhai HY, Sui MH, Yu X, Qu Z, Hu JC, Sun HQ, Zheng HT, Zhou K, Jiang LX. Overexpression of Long Non-Coding RNA TUG1 Promotes Colon Cancer Progression. Med Sci Monit. 2016;22:3281-3287. [PubMed] |
| 241. | Sana J, Hankeova S, Svoboda M, Kiss I, Vyzula R, Slaby O. Expression levels of transcribed ultraconserved regions uc.73 and uc.388 are altered in colorectal cancer. Oncology. 2012;82:114-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 59] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 242. | Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, Shimizu M, Tili E, Rossi S, Taccioli C, Pichiorri F, Liu X, Zupo S, Herlea V, Gramantieri L, Lanza G, Alder H, Rassenti L, Volinia S, Schmittgen TD, Kipps TJ, Negrini M, Croce CM. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 564] [Cited by in RCA: 557] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 243. | Wu X, He X, Li S, Xu X, Chen X, Zhu H. Long Non-Coding RNA ucoo2kmd.1 Regulates CD44-Dependent Cell Growth by Competing for miR-211-3p in Colorectal Cancer. PLoS One. 2016;11:e0151287. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 244. | Bian Z, Jin L, Zhang J, Yin Y, Quan C, Hu Y, Feng Y, Liu H, Fei B, Mao Y, Zhou L, Qi X, Huang S, Hua D, Xing C, Huang Z. LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci Rep. 2016;6:23892. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 290] [Cited by in RCA: 309] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 245. | Ni B, Yu X, Guo X, Fan X, Yang Z, Wu P, Yuan Z, Deng Y, Wang J, Chen D, Wang L. Increased urothelial cancer associated 1 is associated with tumor proliferation and metastasis and predicts poor prognosis in colorectal cancer. Int J Oncol. 2015;47:1329-1338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 69] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 246. | Taniue K, Kurimoto A, Sugimasa H, Nasu E, Takeda Y, Iwasaki K, Nagashima T, Okada-Hatakeyama M, Oyama M, Kozuka-Hata H, Hiyoshi M, Kitayama J, Negishi L, Kawasaki Y, Akiyama T. Long noncoding RNA UPAT promotes colon tumorigenesis by inhibiting degradation of UHRF1. Proc Natl Acad Sci USA. 2016;113:1273-1278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 127] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 247. | Lassmann S, Weis R, Makowiec F, Roth J, Danciu M, Hopt U, Werner M. Array CGH identifies distinct DNA copy number profiles of oncogenes and tumor suppressor genes in chromosomal- and microsatellite-unstable sporadic colorectal carcinomas. J Mol Med (Berl). 2007;85:293-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 116] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 248. | Xiao Y, Yurievich UA, Yosypovych SV. Long noncoding RNA XIST is a prognostic factor in colorectal cancer and inhibits 5-fluorouracil-induced cell cytotoxicity through promoting thymidylate synthase expression. Oncotarget. 2017;8:83171-83182. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 249. | Wang W, Xing C. Upregulation of long noncoding RNA ZFAS1 predicts poor prognosis and prompts invasion and metastasis in colorectal cancer. Pathol Res Pract. 2016;212:690-695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 62] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 250. | Kumegawa K, Maruyama R, Yamamoto E, Ashida M, Kitajima H, Tsuyada A, Niinuma T, Kai M, Yamano HO, Sugai T, Tokino T, Shinomura Y, Imai K, Suzuki H. A genomic screen for long noncoding RNA genes epigenetically silenced by aberrant DNA methylation in colorectal cancer. Sci Rep. 2016;6:26699. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (1)] |