Published online Apr 21, 2019. doi: 10.3748/wjg.v25.i15.1840
Peer-review started: January 24, 2019
First decision: February 26, 2019
Revised: March 20, 2019
Accepted: March 24, 2019
Article in press: March 25 2019
Published online: April 21, 2019
Processing time: 84 Days and 16.7 Hours
Colorectal cancer (CRC) is one of the main causes of cancer-related deaths in China and around the world. Advanced CRC (ACRC) patients suffer from a low cure rate though treated with targeted therapies. The response rate is about 50% to chemotherapy and cetuximab, a monoclonal antibody targeting epidermal growth factor receptor (EGFR) and used for ACRC with wild-type KRAS. It is important to identify more predictors of cetuximab efficacy to further improve precise treatment. Autophagy, showing a key role in the cancer progression, is influenced by the EGFR pathway. Whether autophagy can predict cetuximab efficacy in ACRC is an interesting topic.
To investigate the effect of autophagy on the efficacy of cetuximab in colon cancer cells and ACRC patients with wild-type KRAS.
ACRC patients treated with cetuximab plus chemotherapy, with detailed data and tumor tissue, at Sun Yat-sen University Cancer Center from January 1, 2005, to October 1, 2015, were studied. Expression of autophagy-related proteins [Beclin1, microtubule-associated protein 1A/B-light chain 3 (LC3), and 4E-binding protein 1 (4E-BP1)] was examined by Western blot in CRC cells and by immunohistochemistry in cancerous and normal tissues. The effect of autophagy on cetuximab-treated cancer cells was confirmed by MTT assay. The associations between Beclin1, LC3, and 4E-BP1 expression in tumor tissue and the efficacy of cetuximab-based therapy were analyzed.
In CACO-2 cells exposed to cetuximab, LC3 and 4E-BP1 were upregulated, and P62 was downregulated. Autophagosome formation was observed, and autophagy increased the efficacy of cetuximab. In 68 ACRC patients, immunohistochemistry showed that Beclin1 levels were significantly correlated with those of LC3 (0.657, P < 0.001) and 4E-BP1 (0.211, P = 0.042) in ACRC tissues. LC3 was significantly overexpressed in tumor tissues compared to normal tissues (P < 0.001). In 45 patients with wild-type KRAS, the expression levels of these three proteins were not related to progression-free survival; however, the expression levels of Beclin1 (P = 0.010) and 4E-BP1 (P = 0.005), pathological grade (P = 0.002), and T stage (P = 0.004) were independent prognostic factors for overall survival (OS).
The effect of cetuximab on colon cancer cells might be improved by autophagy. LC3 is overexpressed in tumor tissues, and Beclin1 and 4E-BP1 could be significant predictors of OS in ACRC patients treated with cetuximab.
Core tip: Considering the response rate to cetuximab in advanced colorectal cancer patients with wild-type KRAS, autophagy might be a novel predictor to exclude patients who will not respond to cetuximab. Accordingly, Beclin1, microtubule-associated protein 1A/B-light chain 3 (LC3), and 4E-binding protein 1 (4E-BP1) were studied in this research. We found that LC3 was overexpressed in tumor tissues, and the expression levels of Beclin1 and 4E-BP1 were related to overall survival.
- Citation: Guo GF, Wang YX, Zhang YJ, Chen XX, Lu JB, Wang HH, Jiang C, Qiu HQ, Xia LP. Predictive and prognostic implications of 4E-BP1, Beclin-1, and LC3 for cetuximab treatment combined with chemotherapy in advanced colorectal cancer with wild-type KRAS: Analysis from real-world data. World J Gastroenterol 2019; 25(15): 1840-1853
- URL: https://www.wjgnet.com/1007-9327/full/v25/i15/1840.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i15.1840
Colorectal cancer (CRC) is one of the main causes of cancer-related deaths both in China and worldwide. In past years, the median overall survival of advanced CRC (ACRC) patients was 20 mo due to the development of new combinations of chemotherapy drugs, including oxaliplatin-based[1] and irinotecan-based regimens[2]. However, drug resistance and/or off-target toxicity limit the efficiency of current chemotherapy drugs. The introduction of new targeted therapies, such as monoclonal antibodies targeting epidermal growth factor receptor (EGFR) (cetuximab and panitumumab) or against vascular endothelial growth factor (bevacizumab, ziv-aflibercept, and ramucirumab), has significantly increased the overall survival of ACRC patients to 30 mo[3]. Currently, the only antibody targeting EGFR that has been approved for use in CRC by the Chinese Food and Drug Administration is cetuximab. Studies have demonstrated that only ACRC patients with wild-type KRAS benefit from cetuximab[4] and thus have better overall survival; this beneficial effect is observed especially in patients with left-sided ACRC[5]. However, the response rate (RR) is only 40%-60% for patients with wild-type KRAS[6]. It is important to identify novel predictors to exclude patients with wild-type KRAS who will not respond to cetuximab, which will further improve precise treatment.
Autophagy, an important intracellular homeostatic pathway for the degradation of dysfunctional organelles and proteins, can provide energy for survival under conditions of diverse cellular stresses[7]. Despite accumulating evidence that autophagy plays a critical role in cancer metastasis, autophagy can also suppress pathological processes, including cancer metastasis[8]. Multiple autophagy-related genes and proteins are involved in metastatic CRC[9]. Mammalian target of rapamycin (mTOR), a member of the phosphatidylinositol-3-kinase family, plays a major role in inhibiting the autophagic process and is associated with the proliferation, survival, invasion, and metastasis of CRC cells[10]. Several mTOR signaling components, including mTOR, p70-S6 Kinase 1 (S6K), and eukaryotic initiation factor 4E-binding protein 1 (4E-BP1), are highly expressed and activated in glandular elements of CRC[11]. The current paradigm suggests that mTOR phosphorylates 4E-BP1 at multiple sites, which promotes the dissociation of eIF4E from 4E-BP1, reducing the inhibitory effect of 4E-BP1 on eIF4E-dependent translation initiation and decreasing the translational activity[12]. At the molecular level, the complex autophagy machinery is orchestrated in several stages. One well-characterized regulatory event is the interaction between Beclin1 and Beclin2, which disrupts the interaction of Beclin1 with the class III phosphoinositide 3-kinase (PI3K), a core regulator of autophagy[13]. Autophagy-related genes (Atgs) control autophagosome formation through Atg12-Atg5 and microtubule-associated protein 1A/B-light chain 3 (LC3) complexes, and the modified form of LC3 is attached to the autophagosome membrane[14].
It has been reported that autophagy was induced by EGFR siRNA in cancer cells[15]. Few reports describe autophagy and the anti-cancer role of cetuximab, antibody targeting EGFR, and most of these studies have been performed on cancer cells. In this study, we examined the critical autophagic proteins, Beclin-1, 4E-BP1, and LC3, in CRC cells and investigated whether autophagy determines cell sensitivity to cetuximab in vitro. We also investigated the relationships between clinicopathological characteristics and the expression of Beclin-1, LC3, and 4E-BP1 in ACRC tissue specimens, and evaluated the influence of these autophagy-related proteins on ACRC prognosis. We previously studied the association between Beclin-1 and LC3 expression levels and the effect of cetuximab on ACRC[16], but cetuximab was not given as a first-line therapy in all patients, and the KRAS status of those patients was unknown because KRAS was not used as a predictor of the efficacy of cetuximab at that time. The present study focused on patients with ACRC with wild-type KRAS who were administered with cetuximab plus chemotherapy as the first-line therapy to explore the predictive value of the expression levels of Beclin-1, LC3, and 4E-BP1. The differences in their expression levels between cancerous and normal tissues as well as the correlation of their expression with clinical and pathological grade, progression-free survival (PFS), and overall survival (OS) were also examined in a group of patients wild-type KRAS who were initially or later treated with cetuximab and chemotherapy.
The CRC cell line CACO-2, which has the wild-type KRAS gene, was obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cells were maintained at 37 °C in an atmosphere containing 5% CO2.
MTT assays were performed to determine the anti-proliferative effect of cetuximab on CACO-2 cells. After treatment with cetuximab for 24, 48, and 72 h, 20 μg (5 mg/mL) of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (Sigma, United States) was added to 96-well plates and incubated at 37 °C for 5 h; then, 150 μL of dimethyl sulfoxide (DMSO) was added to each well and incubated for 10 min at room temperature to dissolve the formazan crystals. The absorbance of each well was measured with an ELISA reader (BIO-TEK, United States) at a wavelength of 562 nm. The experiment was performed in triplicate, and the data were analyzed by comparison to DMSO-treated control cells.
The tissue samples were homogenized in sodium dodecyl sulphate (SDS) buffer containing the protease inhibitor PMSF. The homogenates were incubated on ice for 20 min and then centrifuged at 12000 rpm for 30 min at 4 °C. The supernatant was collected and equal volume of 2 × SDS buffer was added. The mixture was boiled for 10 min and preserved at -20 °C. The protein extracts (50 μg) were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto polyvinylidene difluoride membranes (Millipore, United States). The membranes were blocked with 5% nonfat milk in Tris-buffered saline containing 0.1% Tween-20 at room temperature for 90 min and then incubated with primary antibodies against Beclin-1 (3 μg/mL; ab55878, Abcam, Cambridge, United Kingdom), LC3 (2 μg/mL; ab48394, Abcam), 4E-BP1 (2 μg/mL; ab2606, Abcam), and actin (0.5 μg/mL, ab3280, Abcam). The protein bands were detected with secondary antibodies conjugated to horseradish peroxidase (1:5000, Abcam, United Kingdom) and visualized with enhanced chemiluminescence reagents. Each band was quantified through densitometry, and the results are presented as the relative expression of each protein from different samples.
For fluorescence microscopy, 500 μL of cultured cells were removed from the flask at the desired time points, centrifuged for 3 min at 5000 × g, and resuspended in an appropriate volume of water. A total of 4 μL of each sample was spotted on a microscope slide and viewed using an Olympus 1 × 51 inverted fluorescence microscope.
In this retrospective study, ACRC patients with definitive pathological diagnoses, paraffin-embedded pathological specimens, and complete clinicopathologic information who received cetuximab combined with first-line or later chemotherapy at our institution (Sun Yat-sen University Cancer Center) between January 1, 2005 and October 1, 2015 were enrolled.
Our primary study endpoints were OS, which was defined as the time from the date of the first cycle of front-line therapy to the date of death from any cause, and first-line PFS, which was defined as the time from the initial therapy to tumor progression, death from any cause, or the last follow-up before the initiation of second-line therapy. In addition, the objective RR (ORR) and disease RR (DCR) to cetuximab combined with chemotherapy as a first-line treatment were also studied. The study was approved by the ethics committee of our institution.
CRC tissues were surgical or biopsy specimens taken from primary colorectal tumors, and the negative margins of the surgical specimens were considered as normal tissues. All tissue samples were acquired from the sample bank of the pathology department at our cancer center, and all formalin-fixed paraffin-embedded samples in this study were preserved intact. All pathological diagnoses of tissues were adenocarcinoma.
The expression of Beclin-1, LC3, and 4E-BP1 in primary tumors was detected through immunohistochemical staining (IHC) and then compared with the expression in the normal tissue samples. The formalin-fixed paraffin-embedded tissue was cut into 4-μm thick sections, dewaxed, rehydrated, and blocked with hydrogen peroxide. After incubation with primary antibodies against 4E-BP1 (1:200), Beclin-1 (1:100), and LC3 (1:400) (all from Cell Signaling Technology, United States) diluted in 1% PBS, the sections were incubated with an anti-rabbit secondary antibody (Invitrogen, United States) for 30 min. After an additional two washes in PBS, the sections were incubated with diaminobenzidine (DAB, Invitrogen, United States) for 10 min to detect signals. Finally, the sections were counterstained with hematoxylin (Invitrogen, United States). Negative controls were obtained by substituting nonimmune rabbit serum for the primary antibodies.
Protein expression was evaluated via an Olympus CX31 microscope (Olympus, Center Valley, PA, United States) by two individuals who were blinded to the patient outcomes and other clinical findings. The immunoreactivity was evaluated according to the intensity of staining and the percentage of stained cells. Immunostaining intensity was rated as follows: 0, pale yellow or no staining; 1, yellow; 2, deep yellow; and 3, brown. The percentage of stained cells positive for Beclin-1 and LC3 was graded as follows: grade 0 (0%-10%); 1 (10%-25%); 2 (25%-50%); and 3 (50%-100%). The percentage of stained cells positive for 4E-BP1 was graded as grade 0, 0%; 1 (1%-20%); 2 (20%-40%); 3 (40%-60%); 4 (60%-80%); and 5 (80%-100%). The staining intensity and percentage of stained tumor cells were scored for at least 10 high-power fields (400×). The mean score was calculated as [(intensityreader 1 × percentagereader 1) + (intensityreader 2 × percentagereader 2)] / 2. Then, the total scores for Beclin1, LC3, and 4E-BP1 were categorized as low expression (=0) or high expression (>0).
All statistical analyses were performed with SPSS for Windows, version 24.0. The associations between Beclin1, LC3, and 4E-BP1 expression and clinicopathological characteristics were assessed using the chi-square test or Fisher’s exact test, as appropriate. The chi-square test with continuity correction was used to analyze data derived from small cohorts. The relationships among the expression of Beclin1, LC3, and 4E-BP1 were assessed by Spearman’s correction analysis. The Kaplan-Meier method was used to estimate tumor progression in mCRC, and the log-rank test was used to determine the statistical significance. The groups were compared with respect to survival using Cox regression analysis. Hazard ratios were determined using univariate and multivariate Cox regression. P < 0.05 indicated a significant difference.
Autophagy was induced by cetuximab treatment in CACO-2 cells with wild-type KRAS, which were sensitive to cetuximab (Figure 1A). Abundant characteristic autophagosomes in CACO-2 cells were present 72 h after cetuximab treatment; in contrast, autophagosomes were scarce in untreated cells (Figure 1B). We also observed the conversion of LC3/Atg8 from the cytoplasmic form, LC3-1 (18 kDa), to the autophagosomic form, LC3-2 (16 kDa), in the cells exposed to cetuximab (Figure 1C) and decreased expression of P62, which indicated autophagosome formation. Moreover, we observed increased expression of 4E-BP1, indicating that the mTOR pathway was inactive (Figure 1C). MTT assay showed that autophagy increased the inhibitory effect of cetuximab on cell proliferation (Figure 1D).
Initially, 235 ACRC patients treated with cetuximab plus chemotherapy were found in the clinical database of our center, but only 68 patients with detailed data and well-preserved tumor specimens were finally enrolled in this study. The age of the patients ranged from 23 to 77 years, with a median age of 54.5 years. The Beclin1, LC3, and 4E-BP1 expression levels were evaluated in all patients treated with cetuximab combined with chemotherapy; the basic characteristics of those patients and the correlations of those characteristics with the expression of Beclin1, LC3, and 4E-BP1 for all patients are reported in Table 1. Forty-five patients were confirmed to have the wild-type KRAS gene, 25 were given cetuximab as first-line therapy, and 20 were given cetuximab after having already received a different first-line therapy. The number of patients with pathological grades 3, 2, and 1 was 18, 41, and 3, respectively.
| Variable | n = 68 | Beclin1 expression | LC3 expression | 4E-BP1 expression | ||||||
| Low | High | P-value | Low | High | P-value | Low | High | P-value | ||
| Sex | 0.061 | 0.372 | 0.571 | |||||||
| Male | 44 (64.7) | 13 | 24 | 12 | 25 | 8 | 28 | |||
| Female | 24 (35.3) | 2 | 17 | 4 | 15 | 3 | 16 | |||
| Age (yr) | 0.119 | 0.167 | 0.865 | |||||||
| < 65 | 55 (80.9) | 10 | 35 | 11 | 34 | 9 | 35 | |||
| ≥ 65 | 13 (19.1) | 5 | 6 | 5 | 6 | 2 | 9 | |||
| Family history of cancer | 0.593 | 0.151 | 0.382 | |||||||
| Yes | 13 (19.1) | 2 | 8 | 1 | 9 | 1 | 9 | |||
| No | 55 (80.9) | 13 | 33 | 15 | 31 | 10 | 35 | |||
| Location1 | 0.862 | 1.000 | 0.634 | |||||||
| Left | 50 (73.5) | 11 | 31 | 12 | 30 | 2 | 33 | |||
| Right | 18 (26.5) | 4 | 10 | 4 | 10 | 9 | 11 | |||
| Pathological grade | 0.349 | 0.349 | 8 | 0.897 | ||||||
| 1 + 2 | 44 (64.7) | 8 | 30 | 8 | 30 | 8 | 29 | |||
| 3 | 18 (26.4) | 5 | 10 | 5 | 10 | 3 | 12 | |||
| NA | 6 (8.9) | |||||||||
| T stage | 0.618 | 0.618 | 0.911 | |||||||
| T1 + T2 + T3 | 41 (60.3) | 9 | 24 | 9 | 24 | 6 | 26 | |||
| T4 | 22 (32.4) | 4 | 15 | 4 | 15 | 4 | 16 | |||
| NA | 5 (7.3) | |||||||||
| N stage | 0.058 | 0.058 | 0.654 | |||||||
| No | 22 (32.4) | 6 | 9 | 6 | 9 | 4 | 12 | |||
| N1 + N2 | 36 (52.9) | 5 | 28 | 5 | 28 | 6 | 25 | |||
| NA | 10 (14.7) | |||||||||
| Synchronous/metachronous metastasis | 0.020 | 0.058 | 0.162 | |||||||
| Synchronous metastasis | 24 (35.3) | 1 | 18 | 2 | 17 | 5 | 12 | |||
| Metachronous metastasis | 43 (63.2) | 13 | 23 | 13 | 23 | 5 | 32 | |||
| NA | 1 (1.5) | |||||||||
| KRAS | 0.044 | 0.044 | 0.651 | |||||||
| Wild type | 45 (66.2) | 13 | 24 | 13 | 24 | 7 | 30 | |||
| Mutant type | 10 (14.7) | 0 | 9 | 0 | 9 | 2 | 6 | |||
| NA | 13 (19.1) | |||||||||
For the 45 patients with wild-type KRAS, the ORR and DCR to first-line treatment with cetuximab combined with chemotherapy were 45.5% and 72.7%, respectively, and the median OS was 31 mo (2.2-137).
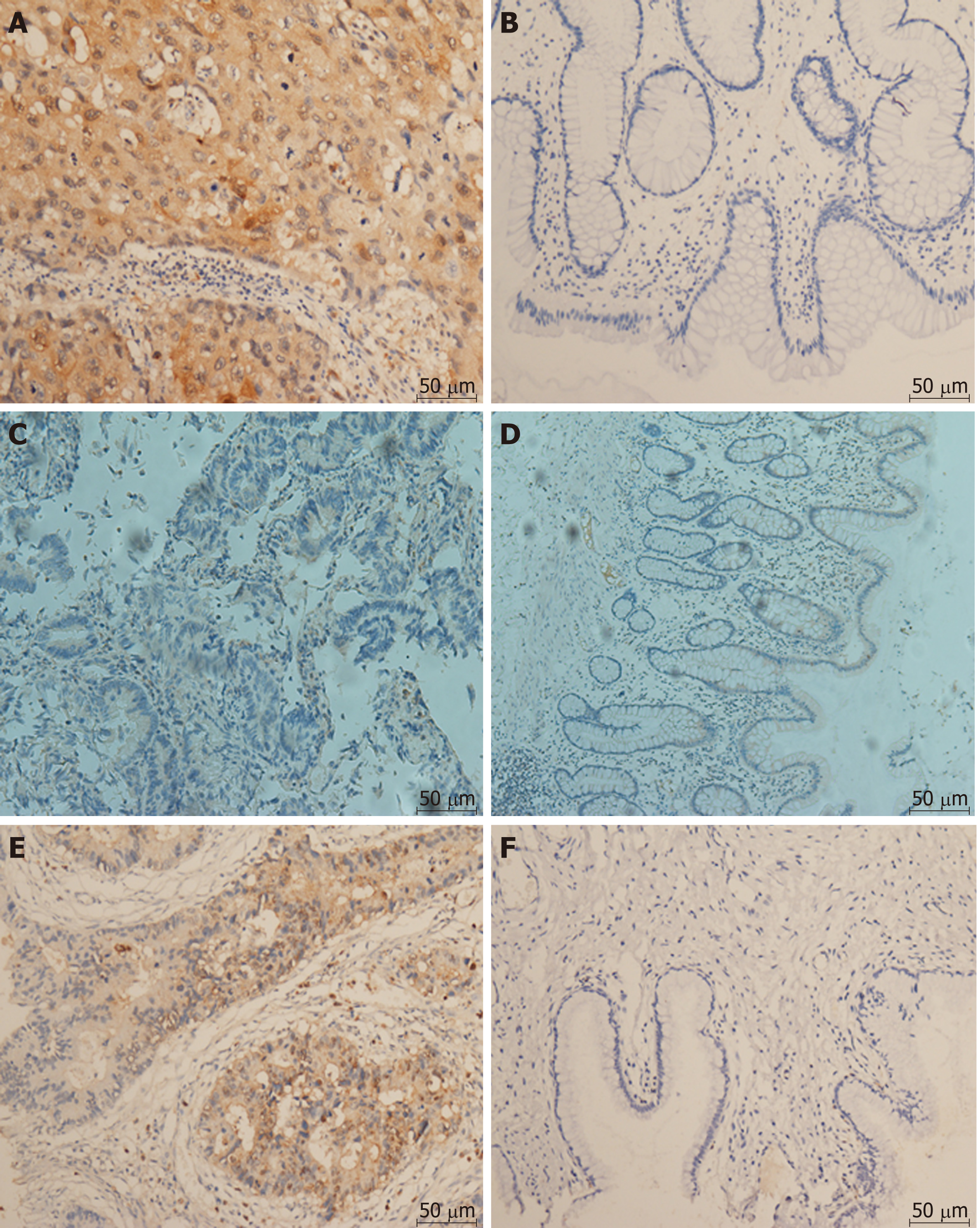
There were significant correlations between the levels of Beclin1 and those of LC3 and 4E-BP1 in CRC tissues and that LC3 was significantly overexpressed in tumor tissues compared to normal tissues (Figure 2). According to the quantitative scoring method described above, the expression levels of Beclin1, LC3, and 4E-BP1 were classified as either high or low. The expression levels of Beclin1, LC3, and 4E-BP1 in cancerous and normal tissues are shown in Figure 3. LC3 was significantly upregulated in tumor specimens compared to normal specimens (64.87% vs 2.00%, P < 0.001); the sensitivity was 71.4%, with a specificity of 91.3%. The expression levels of Beclin1 (64.87% vs 81.81%, P = 0.248) and 4E-BP1 (81.09% vs 87.10%, P = 0.793) were not different between CRC and normal tissues.
The expression levels of Beclin1 were correlated with those of LC3 and 4E-BP1, with coefficients of 0.657 (P < 0.001) and 0.211 (P = 0.042), respectively. However, LC3 expression was not correlated with that of 4E-BP1 (P = 0.251). Compared to patients with the wild-type KRAS gene, patients with the mutated KRAS gene expressed higher levels of Beclin1 (100% vs 64.9%, P = 0.044) and LC3 (100% vs 64.9%, P = 0.044). The associations of the expression levels of Beclin1, LC,3 and 4E-BP1 with the clinicopathologic factors in all 68 patients are shown in Table 1. Patients with metachronous metastasis had higher expression levels of Beclin1 than those with synchronous metastasis (94.7% vs 63.9%, P = 0.020).
For the 25 patients with the wild-type KRAS gene who received cetuximab-containing chemotherapy as the first-line treatment, the ORRs and DCRs for patients with low and high Beclin1, LC3, and 4E-BP1 expression levels are shown in Table 2. There were no associations between the expression levels of Beclin1 (P = 1.000, P = 0.325), LC3 (P = 0.362, P = 0.325), or 4E-BP1 (P = 0.303, P = 0.560) and the ORR or DCR. In the whole group, the median PFS was 8.81 mo (0.72-32.43). The expression levels of Beclin1, LC3, and 4E-BP1 did not have any relationship with PFS [8.80 mo vs 4.7 mo (P = 0.843) for Beclin1 high expression and low expression, respectively; 4.70 mo vs 9.96 mo (P = 0.167) for LC3 high expression and low expression, respectively; and 5.32 mo vs 7.36 mo (P = 0.410) for 4E-BP1 high expression and low expression, respectively (Table 2)].
| Factor | DCR | ORR | PFS | |||||
| n (%) | n (%) | P-value | n (%) | P-value | Time (mo) | P-value | ||
| 25 | ||||||||
| Beclin1 | High | 12 (48.0) | 7 (58.3) | 0.325 | 5 (41.6) | 1.000 | 8.80 | 0.843 |
| Low | 8 (32.0) | 7 (87.5) | 4 (50.0) | 4.70 | ||||
| NA | 5 (20.0) | |||||||
| LC3 | High | 12 (48.0) | 7 (58.3) | 0.325 | 4 (33.3) | 0.362 | 4.70 | 0.167 |
| Low | 8 (32.0) | 7 (87.5) | 5 (62.5) | 9.96 | ||||
| NA | 5 (20.0) | |||||||
| 4E-BP1 | High | 15 (60.0) | 12 (80.0) | 0.560 | 9 (60.0) | 0.303 | 5.32 | 0.410 |
| Low | 5 (20.0) | 3 (60.0) | 1 (20.0) | 7.36 | ||||
| NA | 5 (20.0) | |||||||
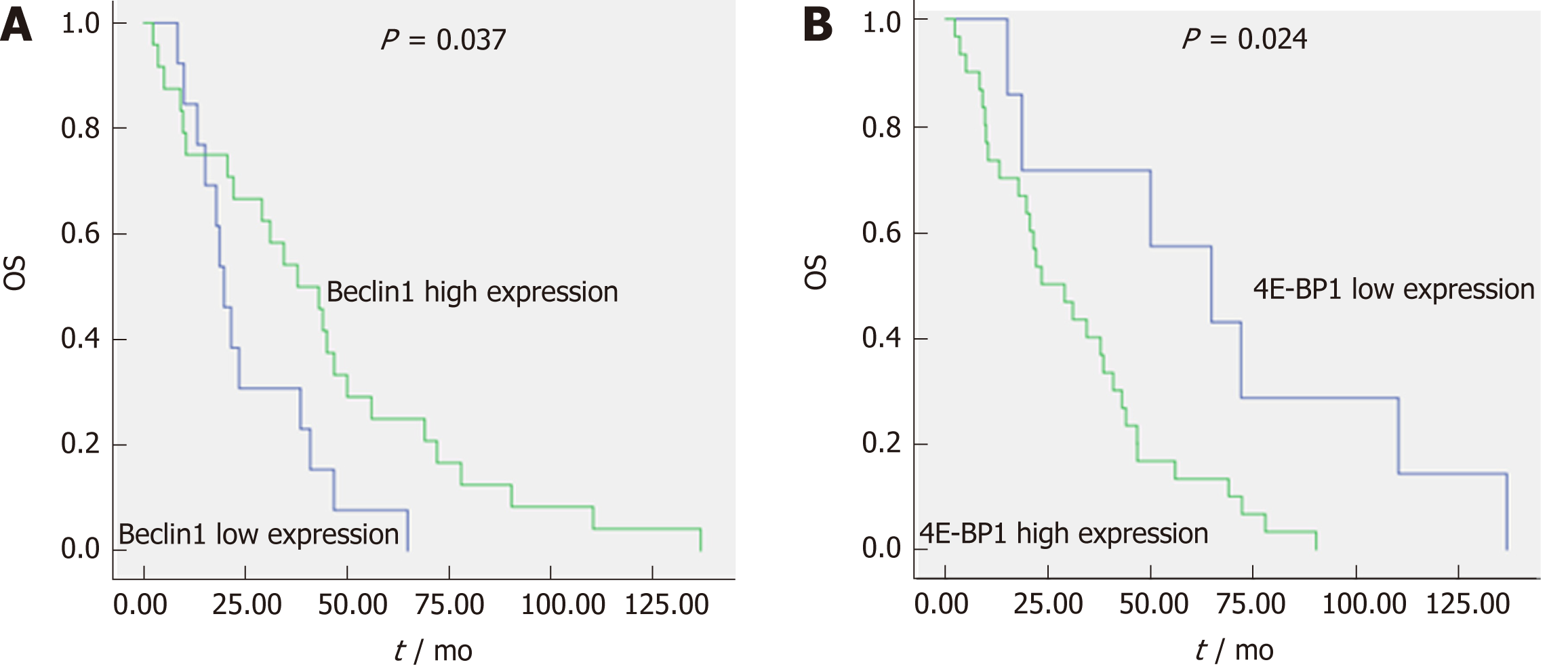
The median OS for the 45 patients with the wild-type KRAS gene who received cetuximab as either a first-line treatment or a subsequent treatment was 31 mo (2.2-137). The most common factors potentially affecting OS, such as gender, age, family history of cancer, tumor location, pathological grade, T stage, N stage, synchronous/metachronous metastasis, and the expression of Beclin1, LC3, and 4E-BP1, were analyzed, as shown in Table 3. According to univariate Cox regression analysis, pathological grade (43.00 mo vs 10.32 mo, P < 0.001), the expression level of Beclin1 (19.65 mo vs 37.82 mo, P = 0.037) (Figure 4A), and the expression level of 4E-BP1 (64.82 mo vs 23.66 mo, P = 0.024) (Figure 4B) were risk factors affecting OS. Multivariate Cox regression analysis revealed that pathological grade (P = 0.002), T stage (P = 0.004), the expression level of Beclin1 (P = 0.010), and the expression level of 4E-BP1 (P = 0.005) were independent prognostic factors for OS.
| OS | ||||
| Univariate analysis | Multivariate analysis | |||
| HR (95%CI) | P-value | HR (95%CI) | P-value | |
| Sex | 0.860 (0.452, 1.636) | 0.496 | ||
| Age | 2.050 (0.831, 5.056) | 0.111 | ||
| Family | 1.568 (0.786, 3.128) | 0.198 | ||
| Tumor location | 0.933 (0.463, 1.880) | 0.353 | ||
| Pathological grade | 2.273 (1.549, 3.335) | 0.000 | 2.421 (1.397, 4.197) | 0.002 |
| T stage | 1.240 (0.977, 1.573) | 0.072 | 1.749 (1.190, 2.569) | 0.004 |
| N stage | 1.159 (0.611, 2.200) | 0.409 | ||
| Synchronous/metachronous metastasis | 1.493 (0.793, 2.813) | 0.081 | ||
| Beclin1 expression | 0.466 (0.223, 0.971) | 0.037 | 0.209 (0.064, 0.685) | 0.010 |
| LC3 expression | 0.541 (0.262, 1.122) | 0.094 | ||
| 4E-BP1 expression | 2.926 (1.110, 7.713) | 0.024 | 6.385 (1.764, 23.112) | 0.005 |
Most studies on the roles of autophagy in cetuximab-mediated cancer therapy are limited to cancer cells, but results based on clinical patients are few. The present study investigated the influence of Beclin1, LC3, and 4E-BP1 expression in tumors, which are three key factors involved in the autophagic process, on the ORR, DCR, PFS, and OS of patients treated with cetuximab combined with chemotherapy. The efficacy found in this study population was the same as that in the previous stage III clinical trials[3], with an ORR of 45.5%, a DCR of 72.7%, and a median OS of 31 mo. First, we found that LC3 and 4E-BP1 were upregulated and P62 was downregulated in CACO-2 cells exposed to cetuximab, and autophagosome formation was observed by fluorescence microscopy, indicating that cetuximab induces autophagy in CRC cells. In addition, the effect of autophagy on the killing of cancer cells by cetuximab was confirmed by MTT assay. We further studied the influence of these autophagic factors on cetuximab efficacy in ACRC. Using IHC, we observed that there were significant correlations between the levels of Beclin1 and the levels of LC3 and 4E-BP1 in CRC tissue and that LC3 was significantly overexpressed in tumor tissues compared to normal tissues. The expression levels of Beclin1 and 4E-BP1 as well as pathological grade and T stage were independent prognostic factors for OS.
Our study found that cetuximab, which targets EGFR, induced autophagy in colon cancer cells. The mechanism of the downstream signaling effect of EGFR on autophagy and the role of autophagy in EGFR-driven cancer cell proliferation remain unclear. One study of EGFR and autophagy in cancer cells showed that the mechanisms underlying the enhancement of autophagy involve anti-EGFR agent-stimulated downregulation of HIF1 as well as Beclin2, which in turn releases Beclin1 from Beclin2 suppression; this demonstrated that autophagy is a protective cellular response to cetuximab treatment[17]. Another study found that autophagy played different roles in the cetuximab-mediated treatment of different cancer cells, including vulvar squamous carcinoma cells, colorectal adenocarcinoma cells, and head and neck cancer cells. Autophagy protected cancer cells from death when apoptosis was strongly induced by cetuximab and enhanced cetuximab-induced apoptosis when only a minimal level of apoptosis was induced after cetuximab treatment[18]. We also found that autophagy was activated by cetuximab in colon cancer cells, and cetuximab increased cell death through autophagy. Autophagy is involved in the process by which cetuximab inhibits cell proliferation[8], but the negative or positive effects of cetuximab treatment on autophagy are controversial, and further studies are needed.
In our study, patients with metachronous metastasis showed significantly higher expression levels of Beclin1 than those with synchronous metastasis. Beclin1 overexpression might indicate late metastasis of CRC, which is consistent with the previous results observed in cells. Koneri et al[19] transfected the Beclin1 gene into HT29 colon cancer cells and found that ectopic Beclin1 overexpression resulted in slow cell growth and G1 arrest with downregulated expression levels of cyclin E and phosphorylated Rb. This finding suggested that Beclin1 overexpression may reverse aggressive phenotypes and act as a tumor suppressor in CRC. Additionally, we found that patients with mutant KRAS had higher expression levels of the autophagic markers Beclin1 and LC3. Several studies have shown that autophagy is upregulated in cancers with KRAS mutations. Alves et al[20] explained that KRAS effector pathways are involved in the regulation of autophagy through the MEK/ERK and PI3K/AKT/mTOR signaling pathways. Guo et al[21] observed that cells with mutated KRAS require autophagy to maintain oxidative metabolism and tumorigenesis when nutrients are limiting.
The present study demonstrated that CRC tissues overexpressed LC3 compared with normal colorectal tissues, with a sensitivity and specificity of 71.4% and 91.3%, respectively, indicating that LC3 might be a means of distinguishing between normal and cancer tissues. This finding was supported by the results of a study involving 163 gastrointestinal cancer patients in which LC3 was differentially expressed in the cytoplasm of cancer cells and normal epithelial cells[22]. The overexpression of LC3 could be interpreted in several ways. During the development and progression of cancers, cancer cells are often exposed to metabolic stress, such as insufficient nutrients or oxygen, because of their high proliferation rate and insufficient vascularization[23]. It is thought that normal cells exhibit a basal level of autophagy to maintain cellular homeostasis. Moreover, increased autophagy in CRC cells may also play a crucial role in tumor metastasis and survival[24]. We observed that Beclin1 and LC3 (0.657, P < 0.001) were significantly correlated in CRC tissue, and Beclin1 and 4E-BP1 (0.211, P = 0.042) were also significantly correlated. These results are the same as the conclusions drawn in a previous study[25] and our experimental results in vitro.
As a type of programmed cell death, autophagy is closely related to chemotherapy resistance[26]. Studies have shown that autophagy occurs abnormally in the chemotherapy courses for liver cancer, leukemia, lung cancer, CRC, and other tumors[27-29]. In the present study, we attempted to identify the proteins involved in autophagy that could predict the efficacy of cetuximab. We observed that Beclin1, LC3, and 4E-BP1 expression levels were not related to the ORR, DCR, or PFS. Interestingly, the prognosis of patients with ACRC who received cetuximab plus chemotherapy was not only correlated with clinical and pathological factors, such as the degree of pathological grade and T stage but also with Beclin1 and 4E-BP1 expression levels. A high expression level of Beclin1 and low expression level of 4E-BP1 independently predicted longer OS in ACRC patients, which might indicate a positive relationship between autophagy and cetuximab efficacy. This finding is consistent with our results in colon cancer cells, which showed that autophagy was enhanced in cells treated with cetuximab, and autophagy increased the effect of cetuximab on inhibiting cell proliferation. It is necessary to design a more rigorous clinical trial to confirm our primary results. We believe that both Beclin1 and 4E-BP1 could be used as independent predictors of cetuximab efficacy.
Our study had several shortcomings. First, it was retrospective, and the number of cases was small. Second, our study focused on the associations between the levels of expression of Beclin-1, LC3, and 4E-BP1 and the efficacy of cetuximab in patients with ACRC with wild-type KRAS; the statuses of NRAS and HRAS were not available. However, we identified two new independent predictors of the efficacy of cetuximab treatment in ACRC patients with wild-type KRAS, Beclin1 and 4E-BP1, and, to the best of our knowledge, our report of the association between 4E-BP1 and cetuximab is novel. Furthermore, our data were obtained from the clinical setting; therefore, the findings are important despite the small sample size.
Great effects have been made in exploring the treatment of colorectal cancer (CRC), but the effectiveness is still limited due to the limited drug selection. Cetuximab, a monoclonal antibody targeting epidermal growth factor receptor (EGFR), significantly increased OS (overall survival) of advanced CRC patients (ACRC). But the response rate (RR) is only 40%-60% for patients with wild-type KRAS. Autophagy, showing a key role in the cancer progression and induced by EGFR siRNA, may allow to develop effective strategies for improving cetuximab effect in CRC, while there have been few studies regarding the predictive role of autophagy in ACRC patients with wild-type KRAS.
Autophagy-related factors were investigated in ACRC with wild-type KRAS to find new biomarkers for cetuximab efficacy, which may offer the potential for developing novel therapeutic strategies for these patients.
The role of Beclin1, microtubule-associated protein 1A/B-light chain 3 (LC3), and 4E-binding protein 1 (4E-BP1), the key factors in autophagy, in predicting the efficacy of cetuximab in ACRC with wild-type KRAS was explored. It was found that Beclin1 and 4E-BP1 were independent prognostic factors for overall survival (OS). In the future research, elucidating the molecular mechanism of association of these factors with cetuximab may help us find more new biomarkers and make cetuximab treatment more accurate.
We detected the expression of autophagy-related proteins 4E-BP1, Beclin-1, and LC3 in CRC samples and adjacent non-tumor tissues by immunohistochemistry. And the three proteins were examined by Western blot in CRC cells. The effect of autophagy on cetuximab-treated cancer cells was confirmed by MTT assay. The associations of Beclin1, LC3, and 4E-BP1 expression in tumor tissue with the efficacy of cetuximab-based therapy were analyzed.
Autophagosome formation was observed in colon cancer cells, with LC3 and 4E-BP1 upregulated, and autophagy increased the efficacy of cetuximab. Beclin1 expression was significantly correlated with LC3 and 4E-BP1 expression in ACRC tissues. LC3 was significantly overexpressed in tumor tissues compared to normal tissues. In patients with wild-type KRAS, the expression levels of Beclin1 and 4E-BP1 were independent prognostic factors for OS. The molecular mechanism of association of these factors with cetuximab effect and their weight coefficients in prognostic models need further study.
LC3 is overexpressed in tumor tissues, and Beclin1 and 4E-BP1 are significant predictors of OS in wild-type KRAS ACRC treated with cetuximab. Autophagy has a role in improving the effects of cetuximab on colon cancer cell.
Autophagy might play a new role to predict the effects of cetuximab in ACRC with wild-type KRAS in the future, and inducing or blocking autophagy may be a new way to improve cetuximab treating ACRC.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Budai B, Mohamed SY, Perse M, Sung WWS-Editor: Ma RY L-Editor: Wang TQ E-Editor: Ma YJ
| 1. | Giacchetti S, Perpoint B, Zidani R, Le Bail N, Faggiuolo R, Focan C, Chollet P, Llory JF, Letourneau Y, Coudert B, Bertheaut-Cvitkovic F, Larregain-Fournier D, Le Rol A, Walter S, Adam R, Misset JL, Lévi F. Phase III multicenter randomized trial of oxaliplatin added to chronomodulated fluorouracil-leucovorin as first-line treatment of metastatic colorectal cancer. J Clin Oncol. 2000;18:136-147. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1017] [Cited by in F6Publishing: 998] [Article Influence: 41.6] [Reference Citation Analysis (0)] |
| 2. | Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, McAllister PK, Morton RF, Schilsky RL. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091-2096. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 893] [Cited by in F6Publishing: 885] [Article Influence: 59.0] [Reference Citation Analysis (0)] |
| 3. | Venook AP, Niedzwiecki D, Lenz HJ, Innocenti F, Fruth B, Meyerhardt JA, Schrag D, Greene C, O'Neil BH, Atkins JN, Berry S, Polite BN, O'Reilly EM, Goldberg RM, Hochster HS, Schilsky RL, Bertagnolli MM, El-Khoueiry AB, Watson P, Benson AB, Mulkerin DL, Mayer RJ, Blanke C. Effect of First-Line Chemotherapy Combined With Cetuximab or Bevacizumab on Overall Survival in Patients With KRAS Wild-Type Advanced or Metastatic Colorectal Cancer: A Randomized Clinical Trial. JAMA. 2017;317:2392-2401. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 507] [Cited by in F6Publishing: 614] [Article Influence: 87.7] [Reference Citation Analysis (0)] |
| 4. | Heinemann V, von Weikersthal LF, Decker T, Kiani A, Vehling-Kaiser U, Al-Batran SE, Heintges T, Lerchenmüller C, Kahl C, Seipelt G, Kullmann F, Stauch M, Scheithauer W, Hielscher J, Scholz M, Müller S, Link H, Niederle N, Rost A, Höffkes HG, Moehler M, Lindig RU, Modest DP, Rossius L, Kirchner T, Jung A, Stintzing S. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): a randomised, open-label, phase 3 trial. Lancet Oncol. 2014;15:1065-1075. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1117] [Cited by in F6Publishing: 1227] [Article Influence: 122.7] [Reference Citation Analysis (0)] |
| 5. | Alan P, Venook DN. Impact of primary (1°) tumor location on Overall Survival (OS) and Progression Free Survival (PFS) in patients (pts) with metastatic colorectal cancer (mCRC): Analysis of All RAS wt patients on CALGB/SWOG 80405 (Alliance) [Abstract]. ESMO Congress, 2016. . [Cited in This Article: ] |
| 6. | Lièvre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Côté JF, Tomasic G, Penna C, Ducreux M, Rougier P, Penault-Llorca F, Laurent-Puig P. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992-3995. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1669] [Cited by in F6Publishing: 1670] [Article Influence: 92.8] [Reference Citation Analysis (0)] |
| 7. | Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol. 2013;15:713-720. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 767] [Cited by in F6Publishing: 912] [Article Influence: 82.9] [Reference Citation Analysis (0)] |
| 8. | Kenific CM, Thorburn A, Debnath J. Autophagy and metastasis: another double-edged sword. Curr Opin Cell Biol. 2010;22:241-245. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 218] [Cited by in F6Publishing: 235] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 9. | Zhang J, Yang Z, Xie L, Xu L, Xu D, Liu X. Statins, autophagy and cancer metastasis. Int J Biochem Cell Biol. 2013;45:745-752. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 67] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 10. | Johnson SM, Gulhati P, Rampy BA, Han Y, Rychahou PG, Doan HQ, Weiss HL, Evers BM. Novel expression patterns of PI3K/Akt/mTOR signaling pathway components in colorectal cancer. J Am Coll Surg. 2010;210:767-776, 776-778. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 148] [Cited by in F6Publishing: 189] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 11. | Ekstrand AI, Jönsson M, Lindblom A, Borg A, Nilbert M. Frequent alterations of the PI3K/AKT/mTOR pathways in hereditary nonpolyposis colorectal cancer. Fam Cancer. 2010;9:125-129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 44] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 12. | Pause A, Belsham GJ, Gingras AC, Donzé O, Lin TA, Lawrence JC, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5'-cap function. Nature. 1994;371:762-767. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 955] [Cited by in F6Publishing: 963] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 13. | Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927-939. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2607] [Cited by in F6Publishing: 2722] [Article Influence: 143.3] [Reference Citation Analysis (0)] |
| 14. | Wild P, McEwan DG, Dikic I. The LC3 interactome at a glance. J Cell Sci. 2014;127:3-9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 183] [Cited by in F6Publishing: 193] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 15. | Weihua Z, Tsan R, Huang WC, Wu Q, Chiu CH, Fidler IJ, Hung MC. Survival of cancer cells is maintained by EGFR independent of its kinase activity. Cancer Cell. 2008;13:385-393. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 355] [Cited by in F6Publishing: 373] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 16. | Guo GF, Jiang WQ, Zhang B, Cai YC, Xu RH, Chen XX, Wang F, Xia LP. Autophagy-related proteins Beclin-1 and LC3 predict cetuximab efficacy in advanced colorectal cancer. World J Gastroenterol. 2011;17:4779-4786. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 55] [Cited by in F6Publishing: 57] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 17. | Li X, Fan Z. The epidermal growth factor receptor antibody cetuximab induces autophagy in cancer cells by downregulating HIF-1alpha and Bcl-2 and activating the beclin 1/hVps34 complex. Cancer Res. 2010;70:5942-5952. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 147] [Cited by in F6Publishing: 151] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 18. | Li X, Lu Y, Pan T, Fan Z. Roles of autophagy in cetuximab-mediated cancer therapy against EGFR. Autophagy. 2010;6:1066-1077. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 78] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 19. | Koneri K, Goi T, Hirono Y, Katayama K, Yamaguchi A. Beclin 1 gene inhibits tumor growth in colon cancer cell lines. Anticancer Res. 2007;27:1453-1457. [PubMed] [Cited in This Article: ] |
| 20. | Alves S, Castro L, Fernandes MS, Francisco R, Castro P, Priault M, Chaves SR, Moyer MP, Oliveira C, Seruca R, Côrte-Real M, Sousa MJ, Preto A. Colorectal cancer-related mutant KRAS alleles function as positive regulators of autophagy. Oncotarget. 2015;6:30787-30802. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 21. | Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, Coller HA, Dipaola RS, Gelinas C, Rabinowitz JD, White E. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460-470. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 921] [Cited by in F6Publishing: 1008] [Article Influence: 77.5] [Reference Citation Analysis (0)] |
| 22. | Yoshioka A, Miyata H, Doki Y, Yamasaki M, Sohma I, Gotoh K, Takiguchi S, Fujiwara Y, Uchiyama Y, Monden M. LC3, an autophagosome marker, is highly expressed in gastrointestinal cancers. Int J Oncol. 2008;33:461-468. [PubMed] [Cited in This Article: ] |
| 23. | Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:1845-1846. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 260] [Cited by in F6Publishing: 453] [Article Influence: 41.2] [Reference Citation Analysis (0)] |
| 24. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39812] [Cited by in F6Publishing: 44348] [Article Influence: 3411.4] [Reference Citation Analysis (4)] |
| 25. | Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753-766. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1625] [Cited by in F6Publishing: 1697] [Article Influence: 84.9] [Reference Citation Analysis (0)] |
| 26. | Zou Z, Yuan Z, Zhang Q, Long Z, Chen J, Tang Z, Zhu Y, Chen S, Xu J, Yan M, Wang J, Liu Q. Aurora kinase A inhibition-induced autophagy triggers drug resistance in breast cancer cells. Autophagy. 2012;8:1798-1810. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 124] [Cited by in F6Publishing: 144] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 27. | Xu N, Zhang J, Shen C, Luo Y, Xia L, Xue F, Xia Q. Cisplatin-induced downregulation of miR-199a-5p increases drug resistance by activating autophagy in HCC cell. Biochem Biophys Res Commun. 2012;423:826-831. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 146] [Cited by in F6Publishing: 153] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 28. | Li N, Li X, Li S, Zhou S, Zhou Q. Cisplatin-induced downregulation of SOX1 increases drug resistance by activating autophagy in non-small cell lung cancer cell. Biochem Biophys Res Commun. 2013;439:187-190. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 29. | Yang M, Zeng P, Kang R, Yu Y, Yang L, Tang D, Cao L. S100A8 contributes to drug resistance by promoting autophagy in leukemia cells. PLoS One. 2014;9:e97242. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 58] [Article Influence: 5.8] [Reference Citation Analysis (0)] |