Published online Jun 21, 2018. doi: 10.3748/wjg.v24.i23.2468
Peer-review started: March 1, 2018
First decision: March 15, 2018
Revised: March 27, 2018
Accepted: May 11, 2018
Article in press: May 11, 2018
Published online: June 21, 2018
Processing time: 106 Days and 20.3 Hours
To investigate changes in gut microbiota and metabolism during nonalcoholic steatohepatitis (NASH) development in mice fed a methionine-choline-deficient (MCD) diet.
Twenty-four male C57BL/6J mice were equally divided into four groups and fed a methionine-choline-sufficient diet for 2 wk (Control 2w group, n = 6) or 4 wk (Control 4w group, n = 6) or the MCD diet for 2 wk (MCD 2w group, n = 6) or 4 wk (MCD 4w group, n = 6). Liver injury, fibrosis, and intestinal barrier function were evaluated after 2 and 4 wk of feeding. The fecal microbiome and metabolome were studied using 16s rRNA deep sequencing and gas chromatography-mass spectrometry.
The mice fed the MCD diet presented with simple hepatic steatosis and slight intestinal barrier deterioration after 2 wk. After 4 wk of feeding with the MCD diet, however, the mice developed prominent NASH with liver fibrosis, and the intestinal barrier was more impaired. Compared with the control diet, the MCD diet induced gradual gut microbiota dysbiosis, as evidenced by a marked decrease in the abundance of Alistipes and the (Eubacterium) coprostanoligenes group (P < 0.001 and P < 0.05, respectively) and a significant increase in Ruminococcaceae UCG 014 abundance (P < 0.05) after 2 wk. At 4 wk, the MCD diet significantly reduced the promising probiotic Bifidobacterium levels and markedly promoted Bacteroides abundance (P < 0.05, and P < 0.01, respectively). The fecal metabolomic profile was also substantially altered by the MCD diet: At 2 wk, arachidic acid, hexadecane, palmitic acid, and tetracosane were selected as potential biomarkers that were significantly different in the corresponding control group, and at 4 wk, cholic acid, cholesterol, arachidic acid, tetracosane, and stearic acid were selected.
The MCD diet induced persistent alterations in the gut microbiota and metabolome.
Core tip: Nonalcoholic steatohepatitis (NASH) is increasingly prevalent as a remarkable problem worldwide. Increased evidence indicates the critical role of gut microbiota in NASH progression. We aimed to investigate the dynamic alterations in the gut microbiota and the related metabolites during NASH development in mice fed a methionine-choline-deficient (MCD) diet. We for the first time find that the MCD diet may induce persistent gut microbiota and metabolome deterioration.
- Citation: Ye JZ, Li YT, Wu WR, Shi D, Fang DQ, Yang LY, Bian XY, Wu JJ, Wang Q, Jiang XW, Peng CG, Ye WC, Xia PC, Li LJ. Dynamic alterations in the gut microbiota and metabolome during the development of methionine-choline-deficient diet-induced nonalcoholic steatohepatitis. World J Gastroenterol 2018; 24(23): 2468-2481
- URL: https://www.wjgnet.com/1007-9327/full/v24/i23/2468.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i23.2468
The prevalence of nonalcoholic fatty liver disease (NAFLD), the most common chronic liver disease worldwide, is increasing, encompassing a pathological spectrum from simple steatosis to nonalcoholic steatohepatitis (NASH)[1]. NASH is a liver characteristic of metabolic syndrome, accounting for lipid accumulation and hepatic inflammation[2] and is projected to be the main indication for liver transplantation in the next decade[3]. NASH is commonly associated with type 2 diabetes, cardiovascular disease, and end-stage kidney disease[4,5], and no approved pharmacological therapies currently exist for NASH[6].
Recently, compelling evidence has indicated the critical role of the gut microbiota in the pathogenesis and progression of NAFLD to NASH[7]. Normally, the commensal gut microbiota and the host maintain a beneficial symbiotic relationship. The liver may function as a vascular firewall, mediating mutualism between the host and the intestinal commensal bacteria[8]. This intimate connection between the gastrointestinal tract and liver defines the term gut-liver axis. However, gut microbiota dysbiosis increases the influx of harmful substances, such as lipopolysaccharide (LPS), ethanol, and bacterial DNA, into the liver through portal vein circulation and promotes NASH development[9,10]. Currently, early and noninvasive diagnosis of NASH with high sensitivity and specificity remains challenging. Metabolomics, along with other “omics” technologies, helps provide a detailed understanding of biochemical events using the systems biology approach and facilitates early diagnosis and exploitation of treatment strategies[11].
Despite the importance of the gut microbiota and metabolome in NASH, detailed information regarding their role during NASH development is limited. The purpose of the present study was to investigate dynamic alterations in the gut microbiota and metabolome during the development of methionine-choline-deficient (MCD) diet-induced NASH in a mouse model.
The animal protocol was designed to minimize the animals’ pain or discomfort and was approved by the Animal Care Committee of Zhejiang University School of Medicine (Permit number: 2017-591). Male C57BL/6J mice (8 wk old) were purchased from SLAC Laboratory (Shanghai, China). After their arrival, the mice were acclimatized for 1 wk in a specific pathogen-free environment (23 °C, 12 h/12 h light/dark, 50% humidity, and ad libitum access to food and water) prior to experimentation. The mice were fed a methionine-choline-sufficient diet (Research Diet, New Brunswick, United States) for 2 wk (the Control 2w group, n = 6) or 4 wk (the Control 4w group, n = 6). Alternatively, the mice were fed an MCD diet (Research Diet) for 2 wk (the MCD 2w group, n = 6) or 4 wk (the MCD 4w group, n = 6). The diet specifications are summarized in Supplementary Table 1. After 2 or 4 wk on the diet, the mice were euthanized by an intraperitoneal injection of 4% chloral hydrate (with 1 mg/100 mL of atropine, to inhibit respiratory secretions) for tissue collection.
Fecal samples were collected from all mice upon defecation and were stored at -80 °C for further procedures. Blood samples were centrifuged (3000 rpm, 15 min) for serum separation, and all serum aliquots were stored in a -80 °C freezer. Liver and colon samples were fixed in either neutral-buffered formalin or optimal cutting temperature (OCT)-embedding media for histological staining or were snap-frozen in liquid nitrogen and kept in a -80 °C freezer for further procedures.
Paraffin-embedded liver sections were stained with hematoxylin-eosin (HE) or Masson’s trichrome to detect liver injury or fibrosis. The stained sections were scanned using a NanoZoomer Digital Pathology system (Hamamatsu Photonics, KK, Japan), which digitally scanned the sections into a particular image format for further assessment. The HE-stained sections were scored in accordance with the NAFLD activity score (NAS) system (score 0-2: Not NASH, 3-4: Borderline NASH, and 5-8: NASH)[12]. Six fields from each section were selected and analyzed at 200 × magnification. The Masson’s trichrome-stained sections were analyzed to quantify fibrosis using Image-Pro Plus software (version 6.0, Media Cybernetics, Rockville, United States) as previously detailed[13]. For each section, the blue area (collagen) was normalized to the red area (hepatocyte). The fibrosis index (%) was calculated as a percent of the total tissue region and represented the average of six randomly selected fields from each section.
Frozen liver sections fixed in OCT (10 μm) were stained with Oil red O (Sigma-Aldrich, St. Louis, MO, United States). Images were captured using the abovementioned NanoZoomer Digital Pathology system. To quantify intrahepatic lipid accumulation, the mean optical density of the red intensity was assessed using Image-Pro Plus software.
The liver triglyceride (TG) content was determined using a commercial kit from Applygen Technologies Inc. (Beijing, China) according to the manufacturer’s protocols, and the final TG concentrations were normalized to the corresponding protein content.
Paraffin-embedded liver sections were stained for F4/80 (anti-active macrophage) (Abcam, Cambridge, United Kingdom) and α-SMA (fibrosis hallmark) with immunohistochemistry (IHC) staining procedures as previously detailed[14]. Briefly, liver sections were incubated with a specific primary antibody, followed by incubation with horseradish peroxidase (HRP)-linked secondary antibody (Dako, Glostrup, Denmark) and 3,3’-diaminobenzidine; the sections were then scanned with the NanoZoomer Digital Pathology system. Image-Pro Plus software was used to count F4/80+ cells and quantitatively analyze the staining intensity of α-SMA as previously described[13]. Six fields of view were randomly selected in each section.
Likewise, paraffin-embedded colon sections were stained for Zonula occludens-1 (ZO-1) (intestinal barrier hallmark) (Proteintech, Rosemont, IL, United States) with standard immunofluorescence staining procedures as previously detailed[15]. Briefly, sections were incubated with the rabbit polyclonal ZO-1 antibody, followed by incubation with Texas Red-conjugated goat anti-rabbit antibody (Jackson ImmunoResearch, West Grove, PA, United States) and 4’,6-diamino-2-phenyl indole (DAPI), and images were captured using a Zeiss LSM T-PMT confocal microscope (Zeiss, Jena, Germany).
Total DNA was extracted from fecal samples with a QIAamp fast DNA stool mini kit (Qiagen, Valencia, CA, United States) following the manufacturer’s handbook. After determining the DNA concentration and integrity, an amplicon sequencing library was constructed based on the PCR-amplified V3-V4 variable regions of 16s rDNA. Then, the qualified libraries were paired-end sequenced on an Illumina MiSeq platform according to manufacturer’s procedures. Raw sequencing data were subjected to filtration using Trimmomatic, FLASH, and QIIME software. Then, clean reads were clustered into operational taxonomic units (OTUs) using UPARSE software with a 97% threshold. The representative read from each OTU was selected using the QIIME package[16]. Representative OTU sequences were annotated and taxonomically classified using Ribosomal Database Project (RDP) Classifier v.2.2, trained on the Silva database version 123[17]. The linear discriminant analysis (LDA) effect size (LEfSe) method (http://huttenhower.sph.harvard.edu/galaxy/) was applied to differentiate taxa with statistical significance and biological relevance[18].
Metabolomic profiling analysis was performed as previously described[19] with slight modification. Briefly, fecal metabolites were extracted by mixing 15 mg of feces with 800 μL of ice-cold methanol. After homogenization and centrifugation, the supernatant was transferred into an Eppendorf tube containing 20 μL of 1 mg/mL heptadecanoic acid as the internal standard. Then, the sample was dried using a nitrogen stream (Aosheng, Hangzhou, China). The residue was reconstituted in 50 μL of 15 mg/mL methoxylamine hydrochloride in anhydrous pyridine and was incubated at 37 °C for 24 h. Then, 50 μL of N,O-bistrifluoroacetamide (BSTFA) [with 1% trimethylsilyl chloride (TMCS)] (Sigma-Aldrich, St. Louis, MO, United States) was added to the mixture, and the sample was incubated at 70 °C for 120 min. Metabolomic analysis was performed with a gas chromatography-mass spectrometry (GC-MS) on an Agilent 7890A GC system coupled to an Agilent 5975C inert mass selective detector (MSD) system (Agilent Technologies, Santa Clara, CA, United States). For data analysis, ChemStation software (version E.02.02.1431), and ChromaTOF software (version 4.34, LECO, St. Joseph, MI, United States) were used. Metabolites were identified by the National Institute of Standards and Technology (NIST) and Fiehn databases. Principal component analysis (PCA) and orthogonal partial least-squares-discriminant analysis (OPLS-DA) were performed to visualize metabolic differences among the experimental groups. Differential metabolites were selected according to the statistically significant VIP values obtained from the OPLS-DA model, and the P values from two-tailed Student’s t-tests on the normalized peak areas; metabolites with VIP values > 1 and P values < 0.05 were included.
The data are shown as the means ± SEM, and the Kolmogorov-Smirnov test was performed to assess data normality. For most data, one-way ANOVA with Tukey’s post hoc test was used to determine the significance between the groups. The Wilcoxon rank sum test was used to evaluate alpha diversity and principal coordinates between the different cohorts in the 16s sequencing analysis. Analysis of similarities (ANOSIM) was performed to test for microbial community clustering according to unweighted UniFrac distance matrices. P values < 0.05 were considered significant. The data were analyzed using SPSS version 17.0 for Windows (SPSS Inc., Chicago, IL, United States).
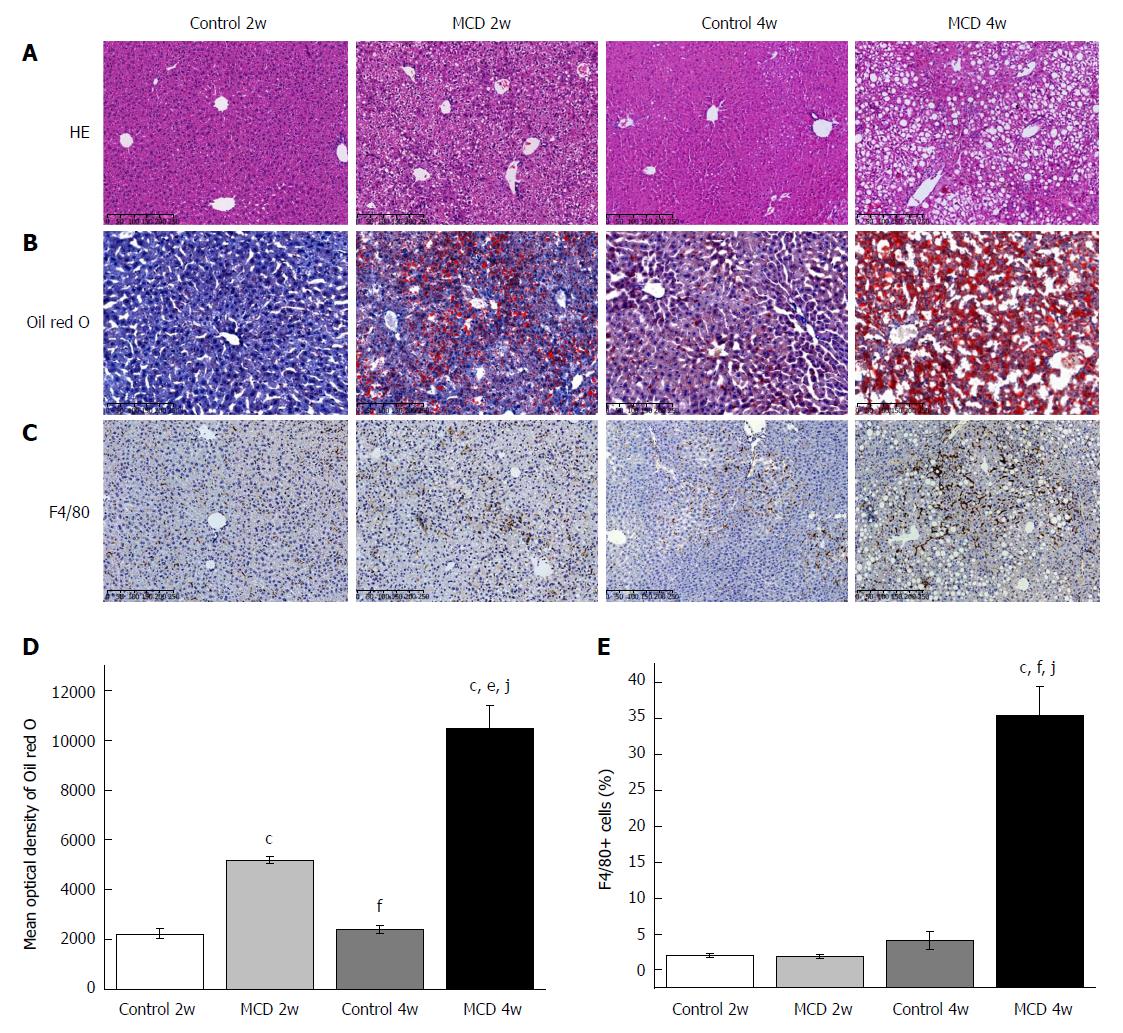
After 2 wk, the mice fed the MCD diet developed simple hepatic steatosis: the liver showed predominantly microvesicular fat accumulation without lobular inflammation and ballooning (Figure 1A, Table 1). Hepatic steatosis was significantly increased in the MCD 2w group compared with that in the Control 2w group, as further indicated by Oil red O staining and hepatic TG quantification (Figure 1B and D, Table 1). No significant difference was found in the number of F4/80+ cells between the MCD 2w group and the Control 2w group (Figure 1C and E). Compared with the Control 2w group, the MCD 2w group presented significantly increased serum alanine aminotransferase (ALT) (P < 0.01) and aspartate aminotransferase (AST) (P < 0.01) levels (Table 1). Evidence of liver fibrosis was not found, as shown by Masson’s trichrome and α-SMA immunohistochemical staining (Figure 2). Intestinal barrier destruction is associated with NAFLD progression[10]; therefore, we detected tight junction protein expression in the colon, where the most abundant gut microbiota reside[20]. ZO-1 immunostaining revealed that the colon tissues of the mice in the MCD 2w group exhibited increased disruption and disorganization on the apical surface and in the crypts (Figure 3).
| Control 2w | MCD 2w | Control 4w | MCD 4w | |
| NAS | 0.17 ± 0.17 | 1.17 ± 0.17a | 0.33 ± 0.21 | 4.83 ± 0.31cfj |
| Steatosis | 0.00 ± 0.00 | 1.17 ± 0.17b | 0.33 ± 0.21 | 2.50 ± 0.22cej |
| Inflammation | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 1.17 ± 0.17bei |
| Ballooning | 0.17 ± 0.17 | 0.00 ± 0.00 | 0.00 ± 0.00 | 1.17 ± 0.31 |
| TG (nmol/mg protein) | 179.41 ± 21.72 | 592.88 ± 55.84b | 204.45 ± 6.76e | 731.60 ± 113.46ah |
| ALT (U/L) | 19.33 ± 0.88 | 413.83 ± 81.77a | 34.83 ± 2.64bd | 974.17 ± 163.00ah |
| AST (U/L) | 105.50 ± 10.55 | 327.33 ± 39.92a | 109.83 ± 11.91d | 749.677 ± 92.34bdi |
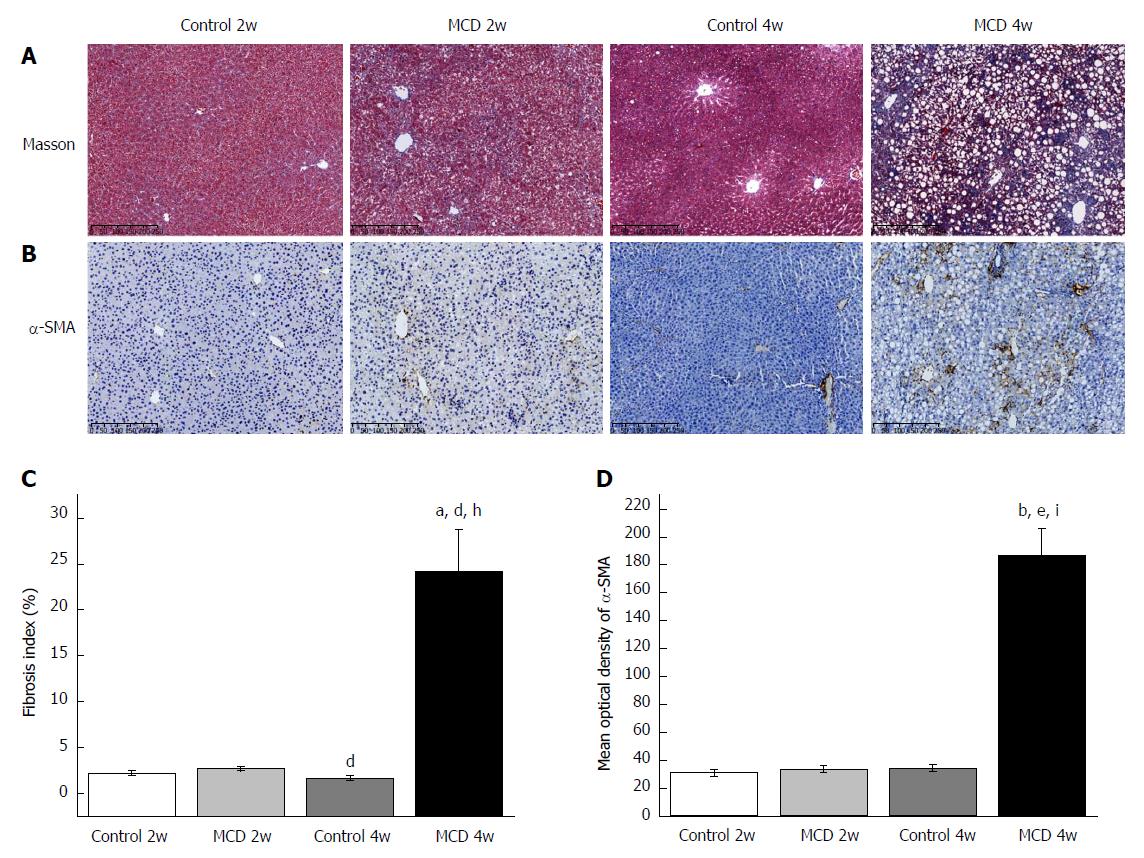
As expected, the mice in the MCD 4w group developed prominent NASH, as evidenced by major hepatic steatosis with lobular inflammation and ballooning hepatocytes in the liver (Figure 1A, Table 1). Oil red O staining and hepatic TG quantification also revealed that fat accumulation was significantly increased in the livers of the mice fed the MCD diet for 4 wk (the MCD 4w group) compared with that in the mice fed the control diet for 4 wk (the Control 4w group) (Figure 1B and D, Table 1). F4/80+ cell infiltration was significantly increased in the MCD 4w group compared with that in the Control 4w group (Figure 1C and E). In addition, we found that the mice in the MCD 4w group exhibited indications of liver fibrosis, including periportal and interstitial collagen deposition (Figure 2A and C). Immunohistochemical analysis further confirmed our result: α-SMA protein expression was significantly up-regulated in the MCD 4w group compared with that in the Control 4w group (Figure 2B and D). Intestinal barrier function was further impaired in the mice fed the MCD diet for 4 wk compared with that in the mice fed the control diet, as revealed by ZO-1 immunostaining (Figure 3).
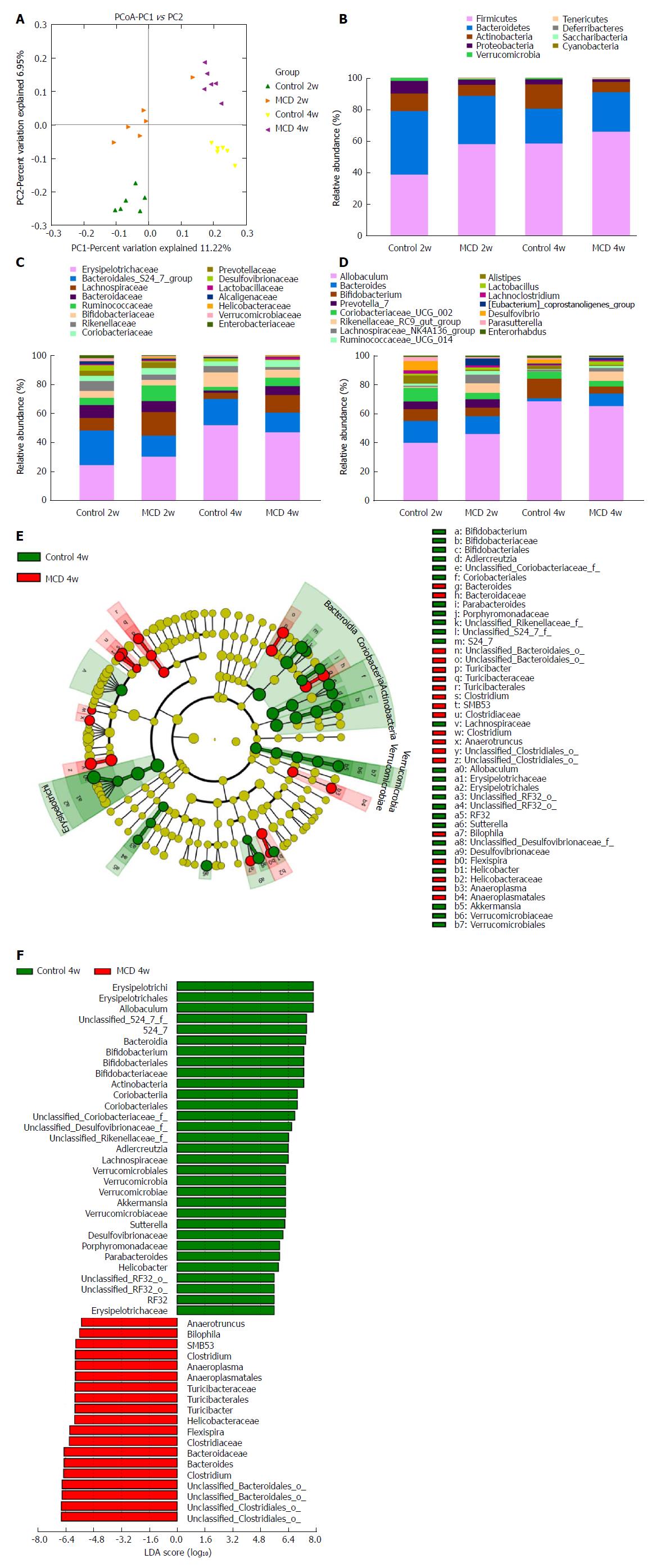
The MCD diet clearly altered the gut microbiota configuration. No significant differences were observed in alpha diversity between the control and MCD groups after 2 and 4 wk of treatment, as estimated by the Chao1, Shannon, and Simpson indices (data not shown). However, these two groups were clearly separated into different clusters at 2 wk (ANOSIM, P < 0.01, r = 0.6352) and at 4 wk (ANOSIM, P < 0.01, r = 0.9074) in the unweighted UniFrac principal coordinate analysis (PCoA), which was performed to calculate the beta-diversity values (Figure 4A). The most abundant taxa at the phylum, family, and genus levels are shown in Figure 4B-D. At the phylum level, Tenericutes was increasingly more abundant in the fecal microbiota of the MCD group compared with that of the control group at 2 and 4 wk (P < 0.05, and P < 0.01, respectively), while Verrucomicrobia was consistently less abundant in the MCD group at 2 and 4 wk (P < 0.05, and P < 0.05, respectively). Compared with the control group, the MCD group had a significantly higher abundance of Firmicutes and a significantly reduced abundance of Proteobacteria at 2 wk (P < 0.001, and P < 0.05, respectively). At 4 wk, the abundance of Actinobacteria was significantly lower in the MCD group than that in the corresponding control group (P < 0.01). At the family level, the relative abundance levels of Rikenellaceae, Desulfovibrionaceae, and Verrucomicrobiaceae were persistently reduced in the MCD group compared with those in the control group at 2 wk (P < 0.001, P < 0.05, and P < 0.05 respectively) and at 4 wk (P < 0.05, P < 0.05, and P < 0.05 respectively). The relative abundance of the Bacteroidales S24-7 group was significantly lower in the MCD group than that in the control group at 2 wk (P < 0.01), while Ruminococcaceae was significantly higher in the MCD group (P < 0.05). Compared with the control group, the MCD group had significantly higher abundance levels of Bacteroidaceae and Enterobacteriaceae at 4 wk (P < 0.01, and P < 0.05 respectively) and a significantly reduced abundance of Bifidobacteriaceae (P < 0.05). At the genus level, the abundance of the Rikenellaceae RC9 gut group was significantly reduced in the MCD group compared with that in the control group at 2 and 4 wk (P < 0.001, and P < 0.05, respectively). Compared with the control group, the MCD group presented with marked decreases in the abundance levels of Alistipes and (Eubacterium) coprostanoligenes at 2 wk (P < 0.001, and P < 0.05, respectively) and a significant increase in Ruminococcaceae UCG 014 abundance (P < 0.05). However, at 4 wk, the MCD diet significantly reduced the abundance of the promising probiotic Bifidobacterium and markedly promoted Bacteroides abundance (P < 0.05, and P < 0.01, respectively).
To characterize further the distinguishing phylotypes in the gut microbiota of the two groups, LEfSe analysis was performed. No significant differences were found between the MCD 2w group and Control 2w group. At 4 wk, however, we found that the MCD diet increased the abundance levels of Anaerotruncus, Bilophila, SMB53, Clostridium, Anaeroplasma, Turicibacter, Helicobacteraceae, Flexispira, and Bacteroides [LDA score (-log10) > 4.8] and decreased the abundance levels of Allobaculum, S24-7, Bifidobacterium, Adlercreutzia, Lachnospiraceae, Akkermansia, Sutterella, Desulfovibrionaceae, Porphyromonadaceae, Parabacteroides, and Erysipelotrichaceae [LDA score (log10) > 4.8] (Figure 4E and F).
Using an untargeted strategy, we studied the fecal metabolome associated with functional characteristics of the gut microbiome, and a total of 322 metabolites were ultimately identified and quantified.
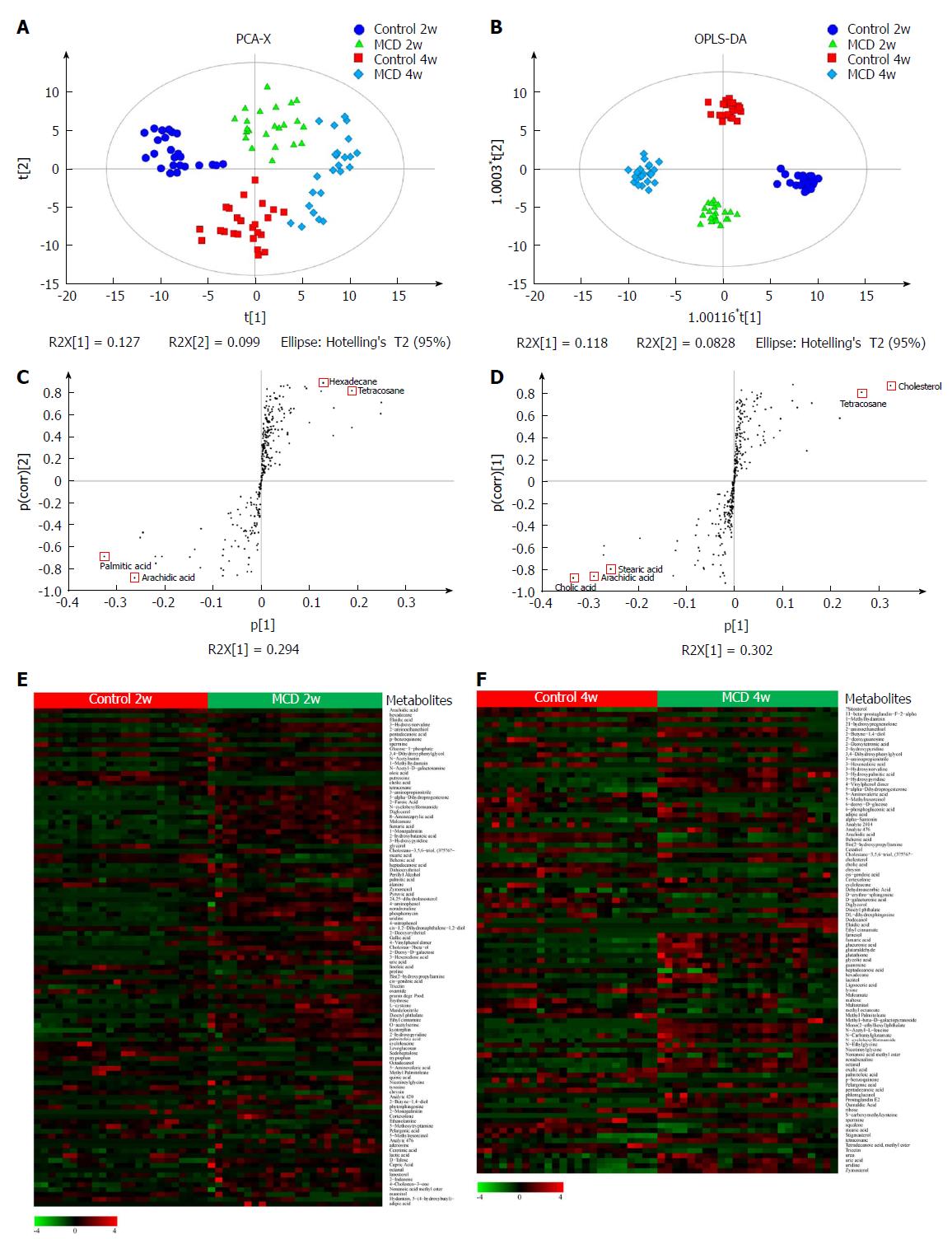
The PCA model was established (R2X = 0.526, Q2 = 0.223), corresponding to the four groups (the Control 2w group, MCD 2w group, Control 4w group, and MCD 4w group) (Figure 5A), and the score plot showed a clustering tendency in the first predictive principal component (X axis) and second predictive principal component (Y axis). Then, an OPLS-DA model was constructed. As depicted in Figure 5B, the score plot in the direction of the X axis (the first predictive principal component) and Y axis (the first orthogonal principal component) showed a significant separation in the metabolomics datasets among the four groups. The explained variance, R2, was 0.968. The cross-validated predictive ability Q2 was 0.914, indicating that a random fecal GC-MS spectrum discriminates among the four groups 91.4% of the time. Therefore, these results indicate the distinct clustering of the fecal metabolomic profiles induced by the MCD diet over time. Characteristic metabolites that had been significantly modified by the MCD diet were further identified according to the OPLS-DA model, with VIP values > 1 and P values < 0.05 (Figure 5E and F, and Supplementary Tables 2 and 3). Ultimately, 103 and 93 metabolites were selected at 2 and 4 wk, respectively, most of which were mainly associated with pathways involved in lipid, amino acid, carbohydrate, nucleotide, cofactors, and vitamin metabolism. S-plots were used to identify further potential biomarkers among the metabolites. As shown in Figure 5C and D, arachidic acid, hexadecane, palmitic acid, and tetracosane at 2 wk and cholic acid, cholesterol, arachidic acid, tetracosane, and stearic acid at 4 wk were the farthest from the origin and were selected as potential biomarkers due to their marked contribution to the separation between the control and MCD groups.
To the best of our knowledge, this is the first study to demonstrate dynamic alterations in the gut microbiota and metabolome in an experimental model of MCD diet-induced steatohepatitis: We sought to determine the key microbiota and metabolites involved in NASH progression over time.
Mice fed the MCD diet developed simple hepatic steatosis at 2 wk and prominent NASH at 4 wk. Among the most abundant taxa, the prevalence of the Rikenellaceae RC9 gut group in the family Rikenellaceae, family Desulfovibrionaceae, and family Verrucomicrobiaceae in the phylum Verrucomicrobia was consistently decreased at 2 and 4 wk in the MCD-supplemented group compared with that in the control group, while the phylum Tenericutes was consistently increased in the MCD-supplemented group. The phylum Tenericutes was also increased in the mice fed a high-fat diet and is correlated with obesity-associated metabolic characteristics[21]. The Rikenellaceae RC9 gut group was previously shown to be decreased in rats with hypertriglyceridemia-related acute necrotizing pancreatitis and in cows receiving oil supplementation[22,23], but the specific function of this taxa remains poorly understood. High Rikenellaceae abundance is reportedly associated with healthy metabolic states[24], and one study reported the ability of some of the bacteria in the family Rikenellaceae to produce butyrate[25]. Desulfovibrionaceae are known as sulfate-reducing bacteria that produce hydrogen sulfide (H2S)[26], which has its pros and cons in relation to gut health. Some studies have demonstrated that H2S functions as a mucosal barrier breaker and therefore leads to inflammation[27,28]. Regarding the advantages of H2S, previous studies have reported that H2S is a crucial mediator in gastrointestinal mucosal defense and repair and in reducing systemic inflammation[26,29]. Verrucomicrobiaceae abundance is positively correlated with gastrointestinal health and negatively correlated with gut inflammation[30]. Therefore, disrupted barrier homeostasis, as evidenced by the ZO-1 immunostaining results in this study, may be associated with a decrease in MCD-sensitive microbiota.
In this study, we found that Bifidobacterium in the family Bifidobacteriaceae, Bacteroides in the family Bacteroidaceae, and the family Enterobacteriaceae respond to the MCD diet in a time-dependent manner. The first of these taxa tended to decrease, while the latter two tended to increase after only 4 wk of feeding. Consistently, Bifidobacterium was significantly decreased in NASH subjects compared with that in healthy and obese subjects[10]. Various members of the genus Bifidobacterium have recently attracted substantial interest due to their multiple beneficial effects on the host, such as protection against enteropathogenic infection through acetate production[31], promotion of antitumor immunity, and facilitation of programmed cell death protein 1 ligand 1 (PD-L1)-specific antibody therapy[32]. Although Bacteroides spp. are overall regarded as beneficial microorganisms[33], they can also cause gut-related bacteremia in conditions of high portal venous pressure[34]. Enterobacteriaceae include many potential pathogens and LPS-producing bacteria and are significantly increased in NASH patients[10]. Furthermore, as evidenced by LEfSe analysis, the MCD diet favored LPS-producing bacteria (Bilophila, and Anaeroplasma[35,36]), pro-inflammatory bacteria (Anaerotruncus, and Turicibacter[37,38]), and other opportunistic pathogens (SMB53, Clostridium, Helicobacteraceae and Flexispira[39-42]) after 4 wk of feeding and inhibited the promising probiotics Sutterella and Akkermansia[43,44]. Therefore, these data indicate an important role for the microbiota in NAFLD progression to NASH.
To link the gut microbiota with their functional states, an untargeted metabolomics analysis was integrated into this study. A core of the MCD diet-responsive signaling pathways were involved in lipid, nucleotide, cofactor, vitamin, carbohydrate, and amino acid metabolism. S-plots further identified potential biomarkers among the metabolites during NASH development. Arachidic acid under-representation and tetracosane over-representation were prominent features in the group with MCD diet-induced steatosis and NASH compared with the corresponding control groups at 2 and 4 wk. However, functional exploration of the association between arachidic acid and tetracosane with non-alcoholic fatty liver disease has not been well documented and warrants further studies. Kuroda et al[45] reported that hexadecane causes nonspecific inflammation. Therefore, we speculated in this study that the increased hexadecane levels after the MCD diet treatment for 2 wk may contribute to disease progression, although we have not yet obtained direct evidence for this effect. Jiao et al[46] found that the cholic acid level is increased in NASH patients. JianHua et al[47] found that cholic acid is significantly decreased in humans with NASH, which is consistent with our study results in the MCD 4w group. Stearic acid is a potent anti-inflammatory lipid and may accelerate hepatic dysfunction recovery in a rat model of liver injury[48]. Our study found that the stearic acid level was significantly decreased in the NASH mice after the MCD diet treatment for 4 wk. Although our study found an association between altered gut microbiota and metabolism and NASH, a causative contribution of the gut microbiota and metabolism to NASH progression has not been sufficiently documented. Establishing a better understanding of the gut microbiota and metabolomics in this disease state will provide beneficial information for the treatment and prevention of NAFLD.
In conclusion, the MCD diet induced gut microbiota and metabolome deterioration. Fundamental observations of these alterations will provide new insight into NASH-associated intestinal disorder and gut-targeted therapies for NASH.
The contributing role of the gut microbiota in the pathogenesis of nonalcoholic steatohepatitis (NASH) has been extensively studied.
Gut microbiota dysbiosis in NASH is mainly depicted as an endpoint, and little is known regarding the microbiota disturbances during NASH progression.
Our goal was to investigate dynamic changes in the gut microbiota and its metabolism during the progression from simple hepatic steatosis to NASH in mice fed a methionine-choline-deficient (MCD) diet.
C57BL/6J mice were equally divided into four groups and fed either a methionine-choline-sufficient diet for 2 or 4 wk (the Control 2w group and Control 4w group, respectively) or the MCD diet for 2 or 4 wk (the MCD 2w group and MCD 4w group, respectively) (n = 6 per group). Liver injury, fibrosis, intestinal barrier function, and the fecal microbiome and metabolome were studied.
The mice fed with the MCD diet for 2 wk developed simple hepatic steatosis, which progressed to prominent NASH with liver fibrosis after 4 wk. Compared with the control diet, the MCD diet induced gradual intestinal barrier impairment and gut microbiota dysbiosis; the fecal metabolomic profile was also substantially altered by the MCD diet.
The MCD diet induced persistent alteration of the gut microbiota and metabolome.
We may have for the first time shown that an MCD diet induced persistent gut microbiota and metabolome deterioration. Fundamental observations of these alterations will provide new insight into NASH-associated intestinal disorder and gut-targeted therapies for NASH.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P- Reviewer: Das UN, Shimizu Y, Trovato GMM S- Editor: Wang XJ L- Editor: Filipodia E- Editor: Huang Y
| 1. | Musso G, Cassader M, Gambino R. Non-alcoholic steatohepatitis: emerging molecular targets and therapeutic strategies. Nat Rev Drug Discov. 2016;15:249-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 342] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 2. | Ishioka M, Miura K, Minami S, Shimura Y, Ohnishi H. Altered Gut Microbiota Composition and Immune Response in Experimental Steatohepatitis Mouse Models. Dig Dis Sci. 2017;62:396-406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 123] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 3. | Wong RJ, Aguilar M, Cheung R, Perumpail RB, Harrison SA, Younossi ZM, Ahmed A. Nonalcoholic steatohepatitis is the second leading etiology of liver disease among adults awaiting liver transplantation in the United States. Gastroenterology. 2015;148:547-555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1211] [Cited by in RCA: 1384] [Article Influence: 138.4] [Reference Citation Analysis (1)] |
| 4. | Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, Charlton M, Sanyal AJ. The diagnosis and management of non-alcoholic fatty liver disease: practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology. 2012;55:2005-2023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2413] [Cited by in RCA: 2613] [Article Influence: 201.0] [Reference Citation Analysis (1)] |
| 5. | Musso G, Gambino R, Tabibian JH, Ekstedt M, Kechagias S, Hamaguchi M, Hultcrantz R, Hagström H, Yoon SK, Charatcharoenwitthaya P. Association of non-alcoholic fatty liver disease with chronic kidney disease: a systematic review and meta-analysis. PLoS Med. 2014;11:e1001680. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 545] [Cited by in RCA: 525] [Article Influence: 47.7] [Reference Citation Analysis (0)] |
| 6. | Armstrong MJ, Gaunt P, Aithal GP, Barton D, Hull D, Parker R, Hazlehurst JM, Guo K; LEAN trial team, Abouda G, Aldersley MA, Stocken D, Gough SC, Tomlinson JW, Brown RM, Hübscher SG, Newsome PN. Liraglutide safety and efficacy in patients with non-alcoholic steatohepatitis (LEAN): a multicentre, double-blind, randomised, placebo-controlled phase 2 study. Lancet. 2016;387:679-690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1100] [Cited by in RCA: 1470] [Article Influence: 163.3] [Reference Citation Analysis (1)] |
| 7. | Kirpich IA, Marsano LS, McClain CJ. Gut-liver axis, nutrition, and non-alcoholic fatty liver disease. Clin Biochem. 2015;48:923-930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 233] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 8. | Balmer ML, Slack E, de Gottardi A, Lawson MA, Hapfelmeier S, Miele L, Grieco A, Van Vlierberghe H, Fahrner R, Patuto N. The liver may act as a firewall mediating mutualism between the host and its gut commensal microbiota. Sci Transl Med. 2014;6:237ra66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 374] [Cited by in RCA: 350] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 9. | Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, Thaiss CA, Kau AL, Eisenbarth SC, Jurczak MJ. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179-185. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1620] [Cited by in RCA: 1881] [Article Influence: 144.7] [Reference Citation Analysis (0)] |
| 10. | Zhu L, Baker SS, Gill C, Liu W, Alkhouri R, Baker RD, Gill SR. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013;57:601-609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 1282] [Article Influence: 106.8] [Reference Citation Analysis (1)] |
| 11. | Safaei A, Arefi Oskouie A, Mohebbi SR, Rezaei-Tavirani M, Mahboubi M, Peyvandi M, Okhovatian F, Zamanian-Azodi M. Metabolomic analysis of human cirrhosis, hepatocellular carcinoma, non-alcoholic fatty liver disease and non-alcoholic steatohepatitis diseases. Gastroenterol Hepatol Bed Bench. 2016;9:158-173. [PubMed] |
| 12. | Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, Ferrell LD, Liu YC, Torbenson MS, Unalp-Arida A. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313-1321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6807] [Cited by in RCA: 8244] [Article Influence: 412.2] [Reference Citation Analysis (5)] |
| 13. | Julio Junior HR, Costa SF, Costa WS, Barcellos Sampaio FJ, Favorito LA. Structural study of the bladder in fetuses with prune belly syndrome. Neurourol Urodyn. 2018;37:148-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 14. | Wu YS, Tseng JK, Chou CH, Chiu CH, Lin YL, Chen YC. Preventive effects of Ophiocordyceps sinensis mycelium on the liver fibrosis induced by thioacetamide. Environ Toxicol. 2017;32:1792-1800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 15. | Chung CY, Alden SL, Funderburg NT, Fu P, Levine AD. Progressive proximal-to-distal reduction in expression of the tight junction complex in colonic epithelium of virally-suppressed HIV+ individuals. PLoS Pathog. 2014;10:e1004198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 16. | Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335-336. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29318] [Cited by in RCA: 23745] [Article Influence: 1583.0] [Reference Citation Analysis (0)] |
| 17. | Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141-D145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3794] [Cited by in RCA: 3483] [Article Influence: 217.7] [Reference Citation Analysis (0)] |
| 18. | Lu H, Ren Z, Li A, Zhang H, Jiang J, Xu S, Luo Q, Zhou K, Sun X, Zheng S. Deep sequencing reveals microbiota dysbiosis of tongue coat in patients with liver carcinoma. Sci Rep. 2016;6:33142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 85] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 19. | Liu R, Hong J, Xu X, Feng Q, Zhang D, Gu Y, Shi J, Zhao S, Liu W, Wang X. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat Med. 2017;23:859-868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1164] [Cited by in RCA: 1032] [Article Influence: 129.0] [Reference Citation Analysis (0)] |
| 20. | Wu S, Yi J, Zhang YG, Zhou J, Sun J. Leaky intestine and impaired microbiome in an amyotrophic lateral sclerosis mouse model. Physiol Rep. 2015;3:pii: e12356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 127] [Cited by in RCA: 188] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 21. | Robertson RC, Seira Oriach C, Murphy K, Moloney GM, Cryan JF, Dinan TG, Ross RP, Stanton C. Deficiency of essential dietary n-3 PUFA disrupts the caecal microbiome and metabolome in mice. Br J Nutr. 2017;118:959-970. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 22. | Huang C, Chen J, Wang J, Zhou H, Lu Y, Lou L, Zheng J, Tian L, Wang X, Cao Z. Dysbiosis of Intestinal Microbiota and Decreased Antimicrobial Peptide Level in Paneth Cells during Hypertriglyceridemia-Related Acute Necrotizing Pancreatitis in Rats. Front Microbiol. 2017;8:776. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 86] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 23. | Zened A, Combes S, Cauquil L, Mariette J, Klopp C, Bouchez O, Troegeler-Meynadier A, Enjalbert F. Microbial ecology of the rumen evaluated by 454 GS FLX pyrosequencing is affected by starch and oil supplementation of diets. FEMS Microbiol Ecol. 2013;83:504-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 188] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 24. | Lippert K, Kedenko L, Antonielli L, Kedenko I, Gemeier C, Leitner M, Kautzky-Willer A, Paulweber B, Hackl E. Gut microbiota dysbiosis associated with glucose metabolism disorders and the metabolic syndrome in older adults. Benef Microbes. 2017;8:545-556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 246] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 25. | Su XL, Tian Q, Zhang J, Yuan XZ, Shi XS, Guo RB, Qiu YL. Acetobacteroides hydrogenigenes gen. nov., sp. nov., an anaerobic hydrogen-producing bacterium in the family Rikenellaceae isolated from a reed swamp. Int J Syst Evol Microbiol. 2014;64:2986-2991. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 148] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 26. | Konturek PC, Koziel J, Dieterich W, Haziri D, Wirtz S, Glowczyk I, Konturek K, Neurath MF, Zopf Y. Successful therapy of Clostridium difficile infection with fecal microbiota transplantation. J Physiol Pharmacol. 2016;67:859-866. [PubMed] |
| 27. | Sheng L, Jena PK, Liu HX, Kalanetra KM, Gonzalez FJ, French SW, Krishnan VV, Mills DA, Wan YY. Gender Differences in Bile Acids and Microbiota in Relationship with Gender Dissimilarity in Steatosis Induced by Diet and FXR Inactivation. Sci Rep. 2017;7:1748. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 109] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 28. | Figliuolo VR, Dos Santos LM, Abalo A, Nanini H, Santos A, Brittes NM, Bernardazzi C, de Souza HSP, Vieira LQ, Coutinho-Silva R. Sulfate-reducing bacteria stimulate gut immune responses and contribute to inflammation in experimental colitis. Life Sci. 2017;189:29-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 104] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 29. | Wallace JL, Motta JP, Buret AG. Hydrogen sulfide: an agent of stability at the microbiome-mucosa interface. Am J Physiol Gastrointest Liver Physiol. 2018;314:G143-G149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 77] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 30. | Rodriguez C, Taminiau B, Korsak N, Avesani V, Van Broeck J, Brach P, Delmée M, Daube G. Longitudinal survey of Clostridium difficile presence and gut microbiota composition in a Belgian nursing home. BMC Microbiol. 2016;16:229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 31. | Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1444] [Cited by in RCA: 1693] [Article Influence: 120.9] [Reference Citation Analysis (0)] |
| 32. | Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084-1089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1979] [Cited by in RCA: 2843] [Article Influence: 284.3] [Reference Citation Analysis (2)] |
| 33. | Pferschy-Wenzig EM, Koskinen K, Moissl-Eichinger C, Bauer R. A Combined LC-MS Metabolomics- and 16S rRNA Sequencing Platform to Assess Interactions between Herbal Medicinal Products and Human Gut Bacteria in Vitro: a Pilot Study on Willow Bark Extract. Front Pharmacol. 2017;8:893. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 34. | Yao S, Yagi S, Uozumi R, Iida T, Nagao M, Okamura Y, Anazawa T, Okajima H, Kaido T, Uemoto S. A High Portal Venous Pressure Gradient Increases Gut-Related Bacteremia and Consequent Early Mortality After Living Donor Liver Transplantation. Transplantation. 2018;102:623-631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 35. | Song JJ, Tian WJ, Kwok LY, Wang YL, Shang YN, Menghe B, Wang JG. Effects of microencapsulated Lactobacillus plantarum LIP-1 on the gut microbiota of hyperlipidaemic rats. Br J Nutr. 2017;118:481-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 77] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 36. | Smith PF, Langworthy TA, Mayberry WR. Distribution and composition of lipopolysaccharides from mycoplasmas. J Bacteriol. 1976;125:916-922. [PubMed] |
| 37. | Zinkernagel MS, Zysset-Burri DC, Keller I, Berger LE, Leichtle AB, Largiadèr CR, Fiedler GM, Wolf S. Association of the Intestinal Microbiome with the Development of Neovascular Age-Related Macular Degeneration. Sci Rep. 2017;7:40826. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 151] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 38. | Suriano F, Bindels LB, Verspreet J, Courtin CM, Verbeke K, Cani PD, Neyrinck AM, Delzenne NM. Fat binding capacity and modulation of the gut microbiota both determine the effect of wheat bran fractions on adiposity. Sci Rep. 2017;7:5621. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 39. | Horie M, Miura T, Hirakata S, Hosoyama A, Sugino S, Umeno A, Murotomi K, Yoshida Y, Koike T. Comparative analysis of the intestinal flora in type 2 diabetes and nondiabetic mice. Exp Anim. 2017;66:405-416. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 87] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 40. | Collins J, Robinson C, Danhof H, Knetsch CW, van Leeuwen HC, Lawley TD, Auchtung JM, Britton RA. Dietary trehalose enhances virulence of epidemic Clostridium difficile. Nature. 2018;553:291-294. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 278] [Cited by in RCA: 269] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 41. | Miller WG, Parker CT, Rubenfield M, Mendz GL, Wösten MM, Ussery DW, Stolz JF, Binnewies TT, Hallin PF, Wang G. The complete genome sequence and analysis of the epsilonproteobacterium Arcobacter butzleri. PLoS One. 2007;2:e1358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 166] [Cited by in RCA: 169] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 42. | Freeman AF, Holland SM. Persistent bacterial infections and primary immune disorders. Curr Opin Microbiol. 2007;10:70-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 43. | Berer K, Gerdes LA, Cekanaviciute E, Jia X, Xiao L, Xia Z, Liu C, Klotz L, Stauffer U, Baranzini SE. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc Natl Acad Sci. 2017;114:10719-10724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 470] [Cited by in RCA: 646] [Article Influence: 80.8] [Reference Citation Analysis (0)] |
| 44. | Wu W, Lv L, Shi D, Ye J, Fang D, Guo F, Li Y, He X, Li L. Protective Effect of Akkermansia muciniphila against Immune-Mediated Liver Injury in a Mouse Model. Front Microbiol. 2017;8:1804. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 189] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 45. | Kuroda Y, Ono N, Akaogi J, Nacionales DC, Yamasaki Y, Barker TT, Reeves WH, Satoh M. Induction of lupus-related specific autoantibodies by non-specific inflammation caused by an intraperitoneal injection of n-hexadecane in BALB/c mice. Toxicology. 2006;218:186-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 46. | Jiao N, Baker SS, Chapa-Rodriguez A, Liu W, Nugent CA, Tsompana M, Mastrandrea L, Buck MJ, Baker RD, Genco RJ. Suppressed hepatic bile acid signalling despite elevated production of primary and secondary bile acids in NAFLD. Gut. 2017;pii:gutjnl-2017-314307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 389] [Cited by in RCA: 516] [Article Influence: 73.7] [Reference Citation Analysis (1)] |
| 47. | Han J, Dzierlenga AL, Lu Z, Billheimer DD, Torabzadeh E, Lake AD, Li H, Novak P, Shipkova P, Aranibar N. Metabolomic profiling distinction of human nonalcoholic fatty liver disease progression from a common rat model. Obesity (Silver Spring). 2017;25:1069-1076. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 48. | Hashemi Goradel N, Eghbal MA, Darabi M, Roshangar L, Asadi M, Zarghami N, Nouri M. Improvement of Liver Cell Therapy in Rats by Dietary Stearic Acid. Iran Biomed J. 2016;20:217-222. [PubMed] |