Published online Jul 7, 2017. doi: 10.3748/wjg.v23.i25.4569
Peer-review started: February 9, 2017
First decision: March 16, 2017
Revised: March 27, 2017
Accepted: April 12, 2017
Article in press: April 12, 2017
Published online: July 7, 2017
Processing time: 149 Days and 18.6 Hours
To investigate the functional role and underlying molecular mechanism of miR-29a in hepatitis B virus (HBV) expression and replication.
The levels of miR-29a and SMARCE1 in HBV-infected HepG2.2.15 cells were measured by quantitative real-time PCR and western blot analysis. HBV DNA replication was measured by quantitative PCR and Southern blot analysis. The relative levels of hepatitis B surface antigen and hepatitis B e antigen were detected by enzyme-linked immunosorbent assay. The Cell Counting Kit-8 (CCK-8) was used to detect the viability of HepG2.2.15 cells. The relationship between miR-29a and SMARCE1 were identified by target prediction and luciferase reporter analysis.
miR-29a promoted HBV replication and expression, while SMARCE1 repressed HBV replication and expression. Cell viability detection indicated that miR-29a transfection had no adverse effect on the host cells. Moreover, SMARCE1 was identified and validated to be a functional target of miR-29a. Furthermore, restored expression of SMARCE1 could relieve the increased HBV replication and expression caused by miR-29a overexpression.
miR-29a promotes HBV replication and expression through regulating SMARCE1. As a potential regulator of HBV replication and expression, miR-29a could be a promising therapeutic target for patients with HBV infection.
Core tip: Although aberrant miR-29a expression has been found to be involved in the process of hepatitis B virus (HBV) infection, its underlying mechanism remains unclear. In this study, we analyzed the expression levels of miR-29a in HBV-infected HepG2.2.15 cells. Our results indicated that miR-29a expression was up-regulated in HBV-associated hepatocellular carcinoma cells. In addition, SMARCE1 was confirmed to be a functional target of miR-29a. miR-29a promoted HBV replication and expression through directly regulating SMARCE1. These findings provide potential therapeutic targets for patients with HBV infection.
- Citation: Wu HJ, Zhuo Y, Zhou YC, Wang XW, Wang YP, Si CY, Wang XH. miR-29a promotes hepatitis B virus replication and expression by targeting SMARCE1 in hepatoma carcinoma. World J Gastroenterol 2017; 23(25): 4569-4578
- URL: https://www.wjgnet.com/1007-9327/full/v23/i25/4569.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i25.4569
Hepatitis B virus (HBV) infection is a global public health problem[1]. HBV is a small encircled DNA virus belonging to the Hepadnaviridae family. HBV infection could cause either acute or chronic hepatitis, even leading to liver cirrhosis and hepatocellular carcinoma (HCC)[2]. A previous study reported that 2 billion people were infected with the HBV virus worldwide, and more than 350 million were chronic HBV carriers[3]. Additionally, over 600000 deaths have been triggered by HBV virus infection[4]. Although some clinical drugs, such as α-interferon and nucleoside analogs, could inhibit HBV replication to some extent for treatment of HBV infection, they could not induce persistent antivirus immune response and completely clear HBV in infected liver resulting from the existence of HBV covalently closed circular DNA and drug-resistant HBV mutants[5-7]. Due to the suboptimality and incompletion of the currently existing therapies for HBV infection[8], it is urgently necessary to elucidate the mechanisms underlying HBV replication and to identify novel molecular targets for HBV therapy.
microRNAs (miRNAs) are a class of small non-coding RNAs of 18 to 25 nucleotides in length, which play crucial regulatory roles in various physiological and pathological processes, especially in tumorigenesis and cancer progression[9]. These small RNAs function as post-transcriptional regulators by imperfect base pairing with the 3’-untranslated region (3’-UTR), coding region or 5’-untranslated region (5’-UTR) of target mRNAs, and usually lead to translational repression or RNA degradation[10,11]. In addition, miRNAs could also be used as a decoy interfering with the function of regulatory proteins[12]. For instance, miR-328 could interact with the RNA-binding protein hnRNP E2 independent of the miRNA’s seed sequence and lead to release of CEBPA mRNA from hnRNP E2-mediated translational inhibition[12]. Another study elucidated that miRNAs can also target non-coding RNA and regulate the expression of itself in a feedback control manner, a novel regulatory mechanism of miRNAs[13].
miRNAs are closely related to the occurrence of cancer and serve as oncogenes or tumor suppressor genes involved in rich disordered pathways in cancers[14]. Up to now, abnormal expression of miRNAs has been detected in almost all types of cancers[15]. Several studies have reported that miR-210, miR-199a-3p and miR-125a-5p were involved in regulative process of HBV replication, by either directly binding to HBV transcripts or targeting crucial transcription factors participating in HBV gene replication and expression[16-18]. Moreover, a recent study indicated that epigenetically regulated miR-449a promoted HBV replication and gene expression by targeting cAMP-responsive element binding protein 5 to increase the expression of FXRα, a transcription factor facilitating HBV replication[19].
The functional role of miR-29a varies in different cancers. For example, miR-29a was involved in tumorigenesis in acute myeloid leukemia and breast cancer as a tumor promotor[20,21]. On the contrary, miR-29a was implicated in lung cancer, cholangiocarcinoma and chronic lymphocytic leukemia acting as a tumor suppressor[22,23]. Additionally, miR-29a exerts important roles in liver diseases. For instance, a study stated that miR-29a could regulate hepatoma cell migration mediated by HBx through directly targeting PTEN in HBV-related HCC[24]. miR-29a was shown to have a protective role against acute liver injury in a mouse model of obstructive jaundice[25]. miR-29a has also been shown to induce apoptosis of LX-2 cells and lead to the reversion of liver fibrosis in mice[26]. Furthermore, miR-29a was found to be up-regulated in the HCC cell line HepG2.2.15 with HBV infection[27].
Pan et al[28] demonstrated that the cellular transcription modulator, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (SMARCE1), could repress HBV replication through binding to the core promoter of HBV. The expression of miR-29a was extensively investigated and well confirmed in many cancers, however, the functional role of miR-29a in HBV expression and replication regulation in HCC is still not clear. Here, we illustrate the underlying molecular mechanism of miR-29a in HBV replication and expression.
Human HCC cell line HepG2 without HBV expression, human HCC cell line HepG2.2.15 with HBV expression and HEK293T cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, United States). These cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Grand Island, NY, United States) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA, United States) at 37 °C in a humidified atmosphere containing 5% CO2. miR-control, miR-29a mimics, anti-miR-29a, pcDNA-control, pcDNA-SMARCE1, si-control or si-SMARCE1 was transfected into cells at the indicated concentrations using Lipofectamine 2000 (Invitrogen) in accordance with the manufacturer’s protocol.
Total RNA was isolated from HepG2 or HepG2.2.15 cells by using TRIzol Reagent (Invitrogen) following the manufacturer’s instructions. cDNA was synthesized by using a ImProm-II™ Reverse Transcription System (Promega, Madison, WI, United States) according to the manufacturer’s instructions. The expression of miR-29a was detected by TaqMan miRNA assay (Ambion, Austin, TX, United States), and U6 snRNA was used as the internal control. The expression of SMARCE1 mRNA was quantified by PrimeScript RT-PCR Kit (TaKaRa, Shiga, Japan), and β-actin served as the internal control. The primer sequences were as follows: for miR-29a 5’-ACAGGATATCGCATTGTTGG-3’ (forward) and 5’-TATACCACATGCAATTCAG-3’ (reverse); for SMARCE1 5’-CGGCTTATCTGGTGGCTTT-3’ (forward) and 5’-GGAGGGTCGGACATCAACAA-3’ (reverse); for U6 5’-CTCGCTTCGGCAGCACA-3’ (forward) and 5’-AACGCTTCACGAATTTGCGT-3’ (reverse); for β-actin 5’-TGAGAGGGAAATCGTGCGTGAC-3’ (forward) and 5’-AAGAAGGAAGGCTGGAAAAGAG-3’ (reverse). The relative expression of RNAs was calculated and normalized using the ∆∆Ct method, relative to the control gene. Each test was performed in triplicate.
Anti-SMARCE1 (Bethyl, Montgomery, TX, United States) and anti-β-actin (Sigma-Aldrich, St Louis, MO, United States) antibodies were used as monoclonal rabbit antibodies. Cells were harvested and total proteins were isolated with RIPA reagents (Thermo Scientific, Rockford, IL, United States) following the manufacturer’s instructions. Subsequently, protein concentrations were quantified by using the BCA protein assay kit (Thermo Scientific). Western blot analysis was then performed following the previously described protocol[29]. The protein bands were visualized and quantified using the ImageQuant software (Molecular Dynamics, Sunnyvale, CA, United States).
HBV replicative intermediates from intracellular core particles were extracted from hepatoma cell lines and detected by Southern blotting, according to the previously published protocols[30]. HBV DNA was extracted using the Column Viral DNAout kit (TIANDZ, Beijing, China) following the manufacturer’s protocol, and quantified by real-time PCR as described previously[30]. The levels of hepatitis B surface antigen (HBsAg) and hepatitis B e antigen (HBeAg) were determined using enzyme-linked immunosorbent assay (ELISA) kits (Kehua Biotech, Shanghai, China).
HepG2.2.15 cells transfected with miR-29a mimics or anti-miR-29a were cultured for 48 h, and then cell viability was determined using the Cell Counting Kit-8 (Dojindo, Kumamoto, Japan).
The SMARCE1 expression construct was generated by PCR amplification of SMARCE1 from HepG2 cDNA using the following primers: forward, 5’-GTACGAATTCCACCATGTCAAAAAGACCATCTTATGC-3’; reverse, 3’-GAATAAGTGTTGCCTTGTTTTGTGCTCGAGACTG-5’. The fragment was cloned into the pcDNA3.1 expression vector using the EcoR I and Xho I restriction sites in the multiple cloning site and sequence verified. For 3’-UTR reporter plasmids, the 3’-UTR of SMARCE1 containing the predicted miR-29a target site was amplified by the primers (forward, 5’-GTACGCTAGCGGCTCAGTCAGTCACCTTTC-3’; reverse, 3’-ATCTGGTCTCGGGTGGAAACTCGAGACTG-5’), and then cloned into a pGL3 reporter vector using the Nhe I and Xho I restriction sites in the multiple cloning site.
HEK293T cells were co-transfected with 50 nmol/L miR-29a mimics or control miRNA and 0.4 μg of pGL3 reporter vectors containing wild-type or mutant 3’-UTR of SMARCE1 using Lipofectamine 2000 (Invitrogen). pRL-CMV (Promega) was co-transfected as a normalization control. Luciferase activities were measured 48 h post-transfection using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocol.
All data were presented as mean ± SD and analyzed using SPSS 19.0 software (IBM Corp, Armonk, NY, United States). The statistical significance was calculated with the Student’s t-test or one-way ANOVA. A P value of less than 0.05 was considered statistically significant.
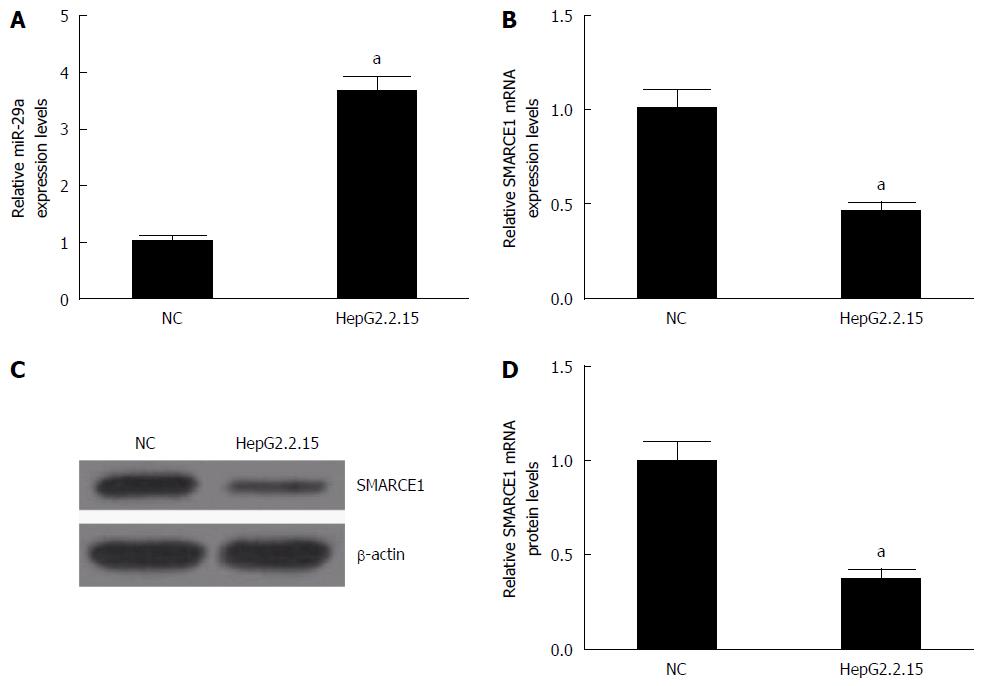
The levels of miR-29a and SMARCE1 in HepG2 (NC) or HepG2.2.15 cells were measured by qRT-PCR and western blotting. The results showed that miR-29a expression (Figure 1A) was significantly increased and SMARCE1 mRNA (Figure 1B) and protein (Figure 1C and D) were dramatically reduced in HepG2.2.15 cells compared with HepG2 cells. These results suggested that miR-29a and SMARCE1 may play some important roles in HepG2.2.15 cells.
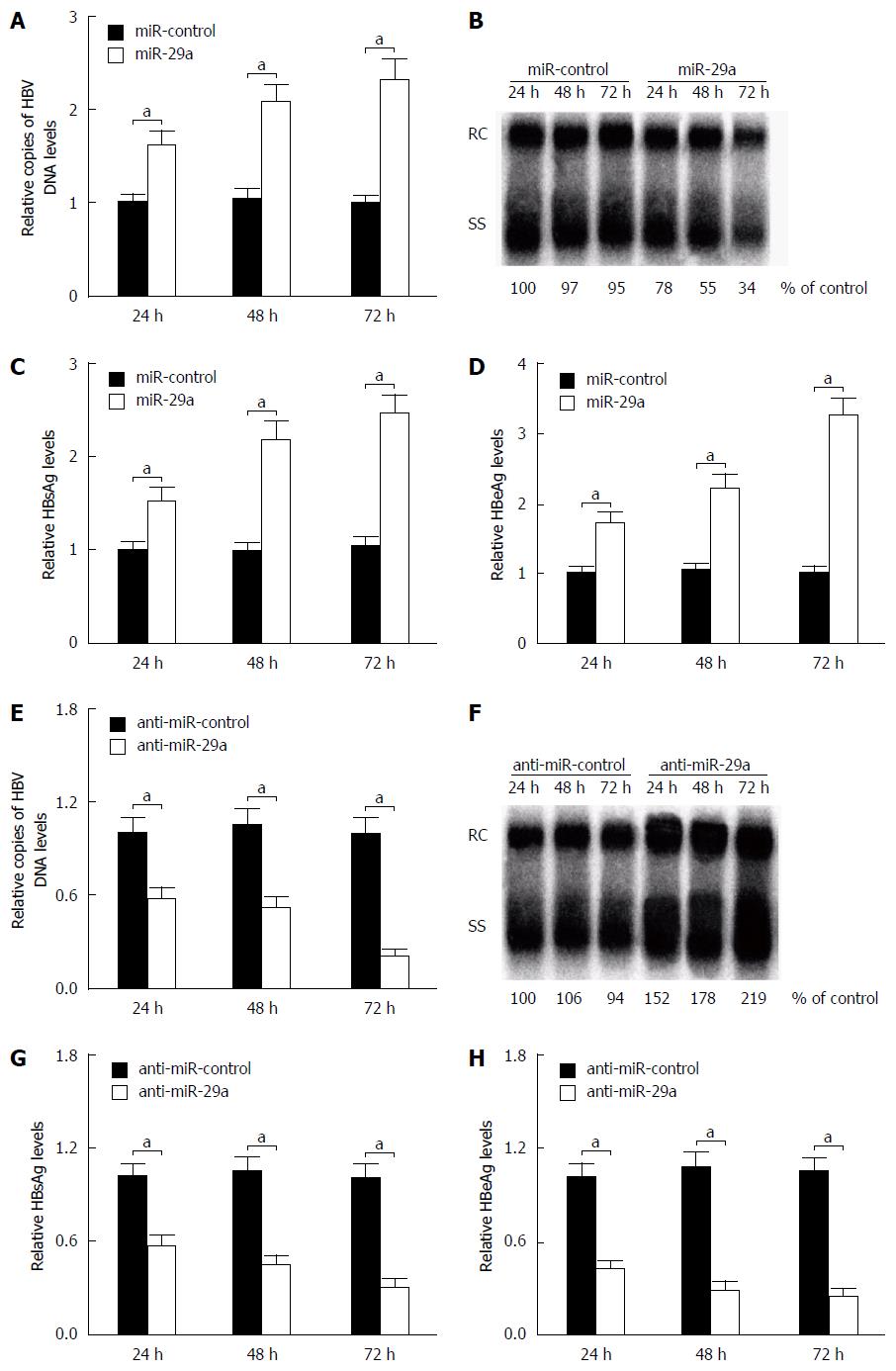
To examine whether miR-29a affected HBV replication and expression, HepG2.2.15 cells were transfected with miR-control, miR-29a mimics or anti-miR-29a, respectively. The influence of miR-29a on HBV DNA replication was measured by quantitative PCR (qPCR) and Southern blot, while the expression levels of HBsAg and HBeAg were detected by ELISA. The qPCR and Southern blot analysis indicated that the overexpression of miR-29a significantly increased the HBV DNA replication, while the endogenous miR-29a inhibition by anti-miR-29a obviously decreased its replication compared with the controls (Figure 2A and B). Moreover, ELISA assay demonstrated that miR-29a overexpression significantly elevated the expression levels of HBsAg and HBeAg, while the endogenous miR-29a inhibition by anti-miR-29a markedly reduced their expression levels compared with the controls (Figure 2C). Together, these results implied that miR-29a promoted HBV replication and expression in HepG2.2.15 cells.
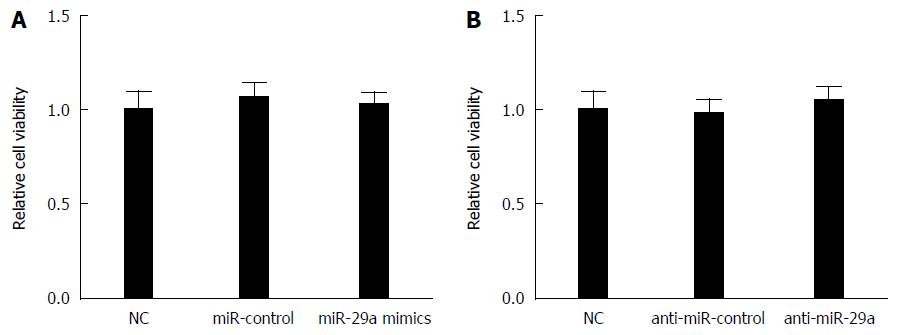
To further investigate whether miR-29a transfection had effects on host cells, CCK-8 was used to detect the cell viability of HepG2.2.15 cells transfected with miR-control, miR-29a mimics or anti-miR-29a. The CCK-8 assay showed that miR-29a mimics, as well as anti-miR-29a, had no significant effect on HepG2.2.15 cell viability compared to respective controls (Figure 3A and B). These results indicated that miR-29a transfection did not damage the host cells.
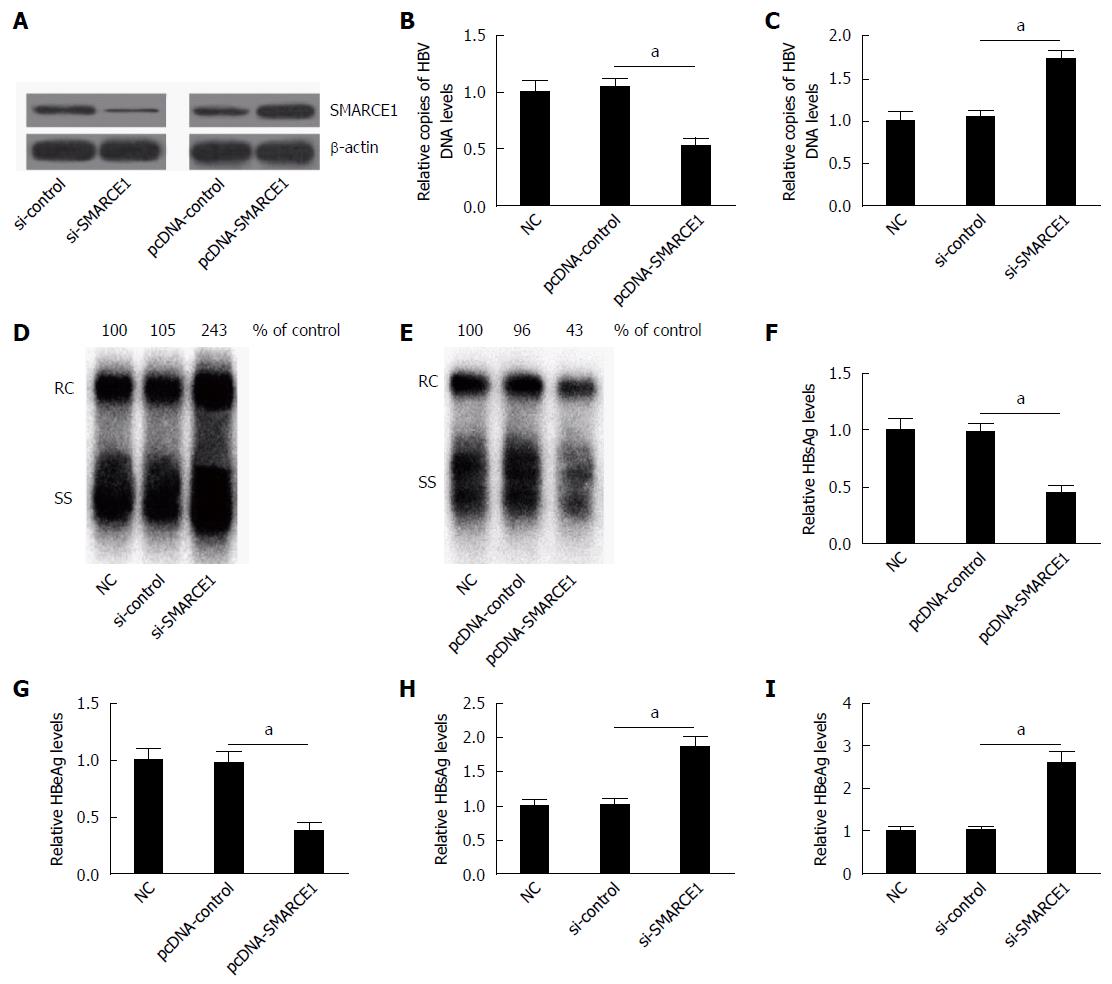
To confirm the effect of SMARCE1 on HBV replication and expression, HepG2.2.15 cells were transfected with pcDNA-control, pcDNA-SMARCE1, si-control or si-SMARCE1, respectively. Western blot analysis indicated that SMARCE1 was overexpressed or down-regulated after pcDNA-SMARCE1 or si-SMARCE1 transfection (Figure 4A). The effect of SMARCE1 on HBV DNA replication was measured by quantitative (q)PCR and Southern blot, while the expression levels of HBsAg and HBeAg were detected by ELISA.
The results of qPCR and Southern blot analysis indicated that SMARCE1 overexpression significantly reduced the HBV DNA replication (Figure 4B and E), while SMARCE1 knockdown dramatically increased its replication (Figure 4C and D). Moreover, ELISA assay revealed that SMARCE1 overexpression significantly decreased the expression levels of HBsAg and HBeAg (Figure 4F and G), while the si-SMARCE1 knockdown notably enhanced their expression levels (Figure 4H and I). These results indicated that SMARCE1 suppressed HBV replication and expression.
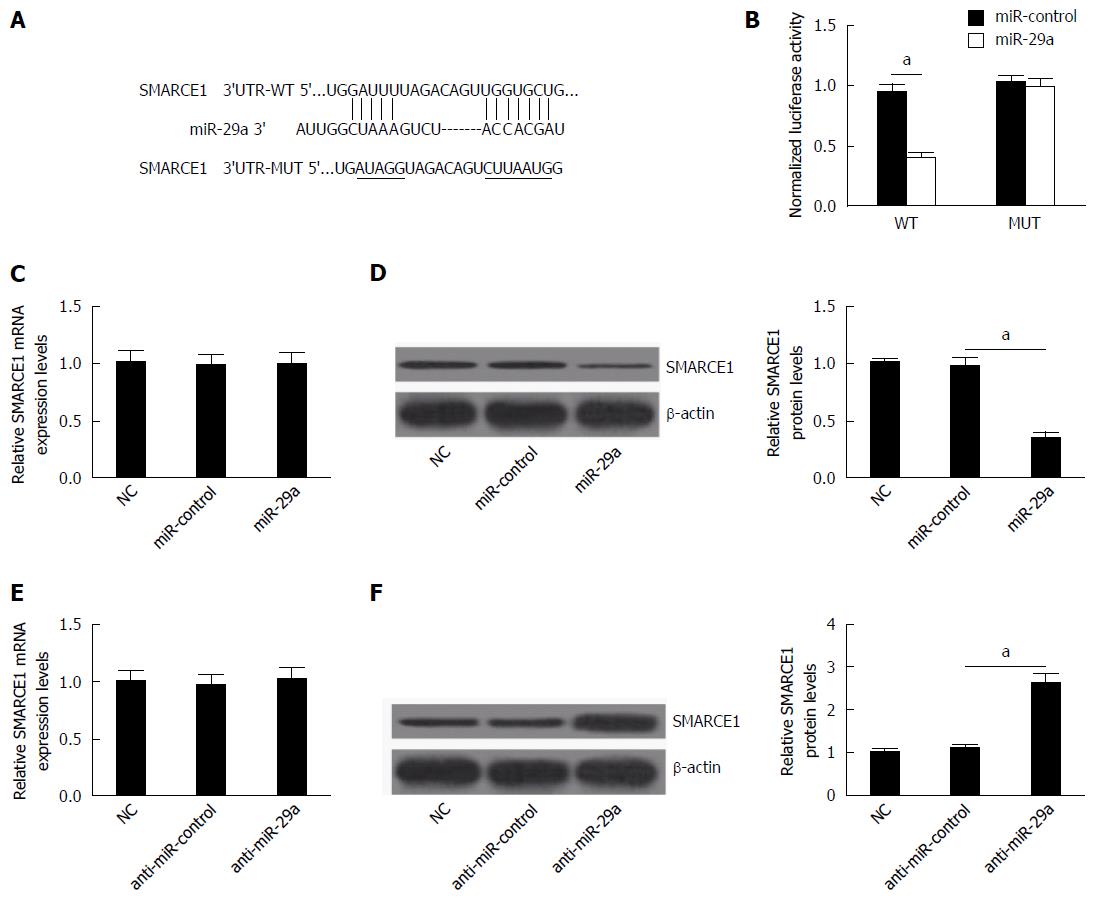
Considering that the expression and function of miR-29a and SMARCE1 were inverse and that miRNAs play their roles by targeting specific target, we speculated that miR-29a might promote HBV replication and expression by regulating the expression of SMARCE1. Therefore, miRNA target analysis tools TargetScan and PicTar were used to predict the potential targets of miR-29a. The results showed that 3’-UTR of SMARCE1 contained the binding sequence of miR-29a (Figure 5A). Then, dual-luciferase reporter assay was used to confirm that SMARCE1 was directly targeted by miR-29a. Results showed that miR-29a overexpression significantly decreased the luciferase activity of the wild-type reporter gene (WT), but not the mutant reporter gene (MUT) (Figure 5B).
To further determine whether miR-29a can indeed repress the expression of SMARCE1, HepG2.2.15 cells were transfected with miR-control, miR-29a mimics or anti-miR-29a, and then the mRNA and protein levels of SMARCE1 were detected by qRT-PCR and western blot, respectively. Results displayed that miR-29a overexpression significantly lowered the protein level of SMARCE1 and miR-29a inhibition significantly promoted the protein level of SMARCE1 (Figure 5D and E), and that miR-29a overexpression or inhibition had no obvious effects on the mRNA level of SMARCE1, which indicated that miR-29a suppressed SMARCE1 expression at the post-transcriptional level. Taken together, these results illuminated that miR-29a directly targeted 3’-UTR of SMARCE1 to regulate its expression in HepG2.2.15 cells.
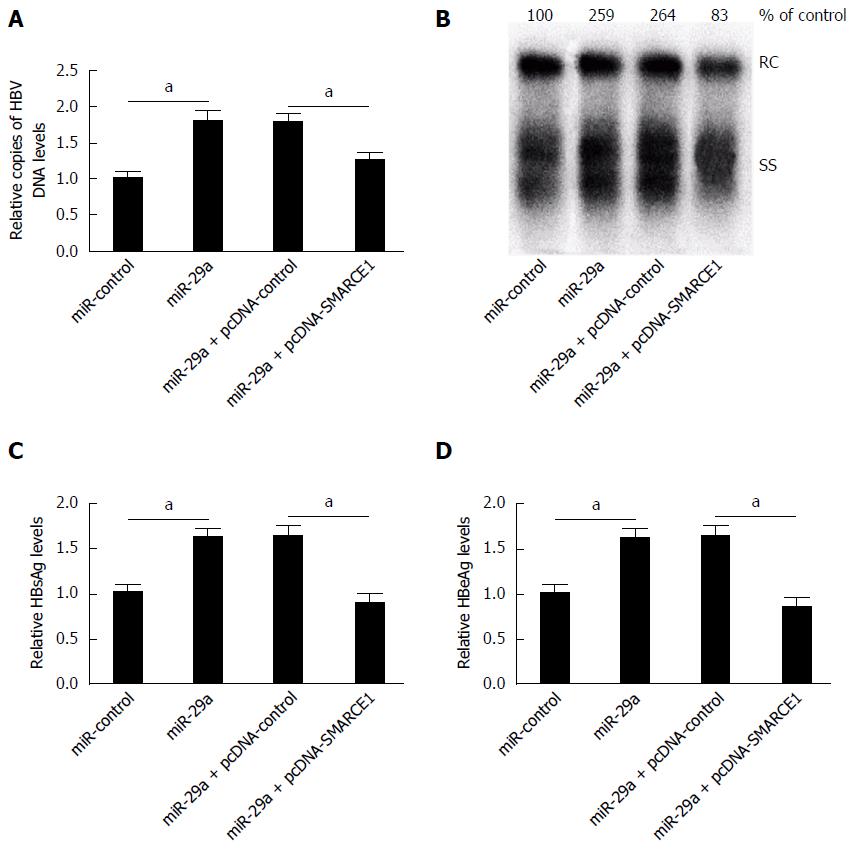
To clarify whether miR-29a promoted HBV replication and expression through regulating SMARCE1, HepG2.2.15 cells were transfected with miR-control or miR-29a mimics or co-transfected miR-29a mimics with pcDNA-control or pcDNA-SMARCE1. The results showed that miR-29a overexpression strikingly augmented HBV replication, whereas co-transfection of pcDNA-SMARCE1 attenuated the effect of miR-29a on HepG2.2.15 cells (Figure 6A and B). As shown in Figure 6C and D, miR-29a overexpression evidently enhanced HBsAg and HBeAg expression, whereas co-transfection of pcDNA-SMARCE1 significantly restored the effect of miR-29a on HBsAg and HBeAg expression in HepG2.2.15 cells. These results demonstrated that miR-29a promoted HBV replication and expression through targeting SMARCE1 in HepG2.2.15 cells.
To improve the treatment of HBV infection, new therapeutic targets and strategies must be identified and developed. Recently, studies on miRNAs have provided novel insights into the potential cure of HBV infection. Moreover, a growing number of works have revealed that miRNAs play critical roles in the HBV replication and expression processes[16-18]. However, their underlying molecular mechanisms remain to be further elucidated.
In our study, we explored the role of miR-29a in the regulation of HBV replication and expression. We found that the expression of miR-29a was significantly up-regulated in the HBV-infected HCC cell line HepG2.2.15. Overexpression of miR-29a increased the HBV replication and expression, while miR-29a knockdown decreased the replication and expression of HBV. These results suggested that miR-29a could be a promising diagnostic biomarker and therapeutic target for HBV infection.
Recent studies indicated that the host cellular miRNAs could regulate the HBV replication and expression by directly targeting HBV transcripts or indirectly targeting HBV transcription regulatory factors needed for HBV transcription[31]. For instance, Zhang et al[17] showed that miR-199a-3p targeted the coding regions of HBsAg and miR-210 specifically bound to the preS1 region of HBV, and they were further confirmed to inhibit HBV replication and expression through directly targeting HBV RNA. Potenza et al[16] also found that miR-125a-5p interacted with the RNA encoded by HBV surface antigen, eventually reducing the secretion of HBsAg. Wu et al[32] reported that miR-7, miR-196, miR-433 and miR-511 might directly target HBV DNA polymerase and surface antigen genes, and miR-205 and miR-345 could target HBx and the pre-core gene, respectively. In addition, Wang et al[33] demonstrated that miR-155 could promote HBV enhancer II activity in a dose-dependent manner by directly targeting CEBP-β. Zhang et al[30] confirmed that miR-1 improved farnesoid X receptor alpha (FXRA) expression and enhanced HBV core promoter transcription activity. Furthermore, HCC cell proliferation and cell cycle arrest modulated by miR-29a may also influence HBV replication and gene expression[34]. In the present study, we found that miR-29a was up-regulated in the HCC cell line HepG2.2.15, consistent with findings from a previous study[27]. However, the possible influencing mechanism of miR-29a on HBV replication and expression was not illustrated. Our study enriches the miRNAs and HBV-host interaction theory.
SMARCE1 is a member of the SWI/SNF family of chromatin-remodeling complexes, playing key roles in transcriptional control. A previous study reported that SWI/SNF was recruited to neuronal genes by the CoREST corepressor, interacting with the DNA binding repressor[35]. In another study, the potential tumor suppressor, prohibitin, was implied to recruit SWI/SNF to a particular E2F-dependent promoter and inhibit E2F-mediated transcription[36]. However, the role of SMARCE1 in HBV replication and expression was largely unknown. In the present study, we found that overexpression of SMARCE1 decreased the HBV replication and expression, while inhibition of SMARCE1 increased the HBV replication and expression. SMARCE1 repressed HBV replication through binding to the mutant core promoter of HBV in HepG2 cells[28], which also revealed that SMARCE1 could repress HBV replication; however, the functional mechanism of SMARCE1 involved in HBV replication may be different from that observed in our study. Furthermore, a dual-luciferase assay was performed and SMARCE1 was verified to be a target of miR-29a and was regulated by miR-29a at the protein levels. Restoration of SMARCE1 expression by pcDNA-SMARCE1 recuperated the promoting effect of miR-29a on the HBV replication and expression. These findings suggested that miR-29a promoted HBV replication and expression by targeted inhibition of SMARCE1 expression, providing a theoretical foundation for the clinical application of miRNAs in therapy of HBV infection.
In summary, the expression of miR-29 was up-regulated in the HCC cell line HepG2.2.15 infected with HBV. Aberrantly expressed miR-29a affected the HBV replication and expression at least partially by targeted inhibition of SMARCE1 expression. Our findings suggest that the inhibition of miR-29a could be a promising therapeutic strategy for the treatment of patients with HBV infection.
MicroRNAs (miRNAs) are involved in the progression of numerous types of cancers. Chronic hepatitis B virus (HBV) infection is one of the major risks for hepatocellular carcinoma (HCC), and through the regulation of HBV replication and expression, miRNAs promote or suppress carcinogenesis in HCC.
HBV infection not only induces liver inflammation but also produces viral oncoproteins to influence HCC progression. Previous studies have reported that many miRNAs are dysregulated in the HBV-associated HCC. The dysregulated miRNAs can regulate HBV replication by either directly binding to HBV transcripts or targeting crucial transcription factors participating in HBV gene replication and expression. However, the role of miRNA in regulation of HBV replication and expression remains largely unclear. In the present study, the authors report that miR-29a promotes HBV replication and expression through targeting SMARCE1.
Previous studies regarding the role of miR-29a were limited in the regulation of HBV-associated HCC carcinogenesis. In addition, the function of miR-29a in the regulation of HBV replication is unclear. This work demonstrated that miR-29a is overexpressed in HBV-associated HCC. The results also suggest that miR-29a promotes HBV replication and expression through targeting SMARCE1.
Because miR-29a is up-regulated in patients with HBV-associated HCC and promotes HBV replication and expression, these finding could provide a novel therapeutic target for HBV-associated HCC.
This study is well designed and the manuscript is well written.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Aizawa Y S- Editor: Ma YJ L- Editor: Filipodia E- Editor: Wang CH
| 1. | Belongia EA, Costa J, Gareen IF, Grem JL, Inadomi JM, Kern ER, McHugh JA, Petersen GM, Rein MF, Sorrell MF. NIH consensus development statement on management of hepatitis B. NIH Consens State Sci Statements. 2008;25:1-29. [PubMed] |
| 2. | McMahon BJ. The natural history of chronic hepatitis B virus infection. Hepatology. 2009;49:S45-S55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 519] [Cited by in RCA: 556] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 3. | Lavanchy D. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J Viral Hepat. 2004;11:97-107. [PubMed] |
| 4. | Hoofnagle JH, Doo E, Liang TJ, Fleischer R, Lok AS. Management of hepatitis B: summary of a clinical research workshop. Hepatology. 2007;45:1056-1075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 438] [Cited by in RCA: 440] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 5. | Billioud G, Pichoud C, Puerstinger G, Neyts J, Zoulim F. The main hepatitis B virus (HBV) mutants resistant to nucleoside analogs are susceptible in vitro to non-nucleoside inhibitors of HBV replication. Antiviral Res. 2011;92:271-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 64] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 6. | Daniels HM, Meager A, Eddleston AL, Alexander GJ, Williams R. Spontaneous production of tumour necrosis factor alpha and interleukin-1 beta during interferon-alpha treatment of chronic HBV infection. Lancet. 1990;335:875-877. [PubMed] |
| 7. | Lam A, Espiritu C, Flores O, Hartmanet G, Klumpp K. P0640: Effect of the combination of the HBV core inhibitor NVR 3-778 with nucleoside analogs or other HBV core inhibitors on the inhibition of HBV DNA replication in HepG2. 2.15 cells. J Hepatol. 2015;62:S559. [RCA] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 8. | Mailliard ME, Gollan JL. Emerging therapeutics for chronic hepatitis B. Annu Rev Med. 2006;57:155-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 9. | Liu M, Lang N, Qiu M, Xu F, Li Q, Tang Q, Chen J, Chen X, Zhang S, Liu Z. miR-137 targets Cdc42 expression, induces cell cycle G1 arrest and inhibits invasion in colorectal cancer cells. Int J Cancer. 2011;128:1269-1279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 137] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 10. | Ambros V. The functions of animal microRNAs. Nature. 2004;431:350-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7919] [Cited by in RCA: 8589] [Article Influence: 409.0] [Reference Citation Analysis (0)] |
| 11. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [PubMed] |
| 12. | Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, Liu S, Schwind S, Santhanam R, Hickey CJ. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652-665. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 441] [Cited by in RCA: 394] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 13. | Zisoulis DG, Kai ZS, Chang RK, Pasquinelli AE. Autoregulation of microRNA biogenesis by let-7 and Argonaute. Nature. 2012;486:541-544. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 173] [Cited by in RCA: 177] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 14. | Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1354] [Cited by in RCA: 1476] [Article Influence: 92.3] [Reference Citation Analysis (0)] |
| 15. | Li C, Feng Y, Coukos G, Zhang L. Therapeutic microRNA strategies in human cancer. AAPS J. 2009;11:747-757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 140] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 16. | Potenza N, Papa U, Mosca N, Zerbini F, Nobile V, Russo A. Human microRNA hsa-miR-125a-5p interferes with expression of hepatitis B virus surface antigen. Nucleic Acids Res. 2011;39:5157-5163. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 136] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 17. | Zhang GL, Li YX, Zheng SQ, Liu M, Li X, Tang H. Suppression of hepatitis B virus replication by microRNA-199a-3p and microRNA-210. Antiviral Res. 2010;88:169-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 182] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 18. | Zhang X, Hou J, Lu M. Regulation of hepatitis B virus replication by epigenetic mechanisms and microRNAs. Front Genet. 2013;4:202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 59] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 19. | Zhang X, Liu H, Xie Z, Deng W, Wu C, Qin B, Hou J, Lu M. Epigenetically regulated miR-449a enhances hepatitis B virus replication by targeting cAMP-responsive element binding protein 5 and modulating hepatocytes phenotype. Sci Rep. 2016;6:25389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 20. | Gebeshuber CA, Zatloukal K, Martinez J. miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep. 2009;10:400-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 297] [Cited by in RCA: 319] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 21. | Han YC, Park CY, Bhagat G, Zhang J, Wang Y, Fan JB, Liu M, Zou Y, Weissman IL, Gu H. microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J Exp Med. 2010;207:475-489. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 228] [Cited by in RCA: 240] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 22. | Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805-15810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1300] [Cited by in RCA: 1279] [Article Influence: 71.1] [Reference Citation Analysis (0)] |
| 23. | Pekarsky Y, Santanam U, Cimmino A, Palamarchuk A, Efanov A, Maximov V, Volinia S, Alder H, Liu CG, Rassenti L. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66:11590-11593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 463] [Cited by in RCA: 461] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 24. | Kong G, Zhang J, Zhang S, Shan C, Ye L, Zhang X. Upregulated microRNA-29a by hepatitis B virus X protein enhances hepatoma cell migration by targeting PTEN in cell culture model. PLoS One. 2011;6:e19518. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 149] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 25. | Tiao MM, Wang FS, Huang LT, Chuang JH, Kuo HC, Yang YL, Huang YH. MicroRNA-29a protects against acute liver injury in a mouse model of obstructive jaundice via inhibition of the extrinsic apoptosis pathway. Apoptosis. 2014;19:30-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 47] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 26. | Matsumoto Y, Itami S, Kuroda M, Yoshizato K, Kawada N, Murakami Y. MiR-29a Assists in Preventing the Activation of Human Stellate Cells and Promotes Recovery From Liver Fibrosis in Mice. Mol Ther. 2016;24:1848-1859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 67] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 27. | Zhang ZZ, Liu X, Wang DQ, Teng MK, Niu LW, Huang AL, Liang Z. Hepatitis B virus and hepatocellular carcinoma at the miRNA level. World J Gastroenterol. 2011;17:3353-3358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 50] [Cited by in RCA: 58] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 28. | Pan H, Niu DD, Feng H, Ng LF, Ren EC, Chen WN. Cellular transcription modulator SMARCE1 binds to HBV core promoter containing naturally occurring deletions and represses viral replication. Biochim Biophys Acta. 2007;1772:1075-1084. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 29. | Hung PS, Kao SY, Shih YH, Chiou SH, Liu CJ, Chang KW, Lin SC. Insulin-like growth factor binding protein-5 (IGFBP-5) suppresses the tumourigenesis of head and neck squamous cell carcinoma. J Pathol. 2008;214:368-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 42] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 30. | Zhang X, Zhang E, Ma Z, Pei R, Jiang M, Schlaak JF, Roggendorf M, Lu M. Modulation of hepatitis B virus replication and hepatocyte differentiation by MicroRNA-1. Hepatology. 2011;53:1476-1485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 160] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 31. | Liu WH, Yeh SH, Chen PJ. Role of microRNAs in hepatitis B virus replication and pathogenesis. Biochim Biophys Acta. 2011;1809:678-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 74] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 32. | Wu FL, Jin WB, Li JH, Guo AG. Targets for human encoded microRNAs in HBV genes. Virus Genes. 2011;42:157-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 33. | Wang B, Majumder S, Nuovo G, Kutay H, Volinia S, Patel T, Schmittgen TD, Croce C, Ghoshal K, Jacob ST. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. 2009;50:1152-1161. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 255] [Cited by in RCA: 249] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 34. | Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y, Jia WH, Zhuang SM. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology. 2010;51:836-845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 296] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 35. | Battaglioli E, Andrés ME, Rose DW, Chenoweth JG, Rosenfeld MG, Anderson ME, Mandel G. REST repression of neuronal genes requires components of the hSWI.SNF complex. J Biol Chem. 2002;277:41038-41045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 159] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 36. | Wang S, Zhang B, Faller DV. Prohibitin requires Brg-1 and Brm for the repression of E2F and cell growth. EMBO J. 2002;21:3019-3028. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 108] [Article Influence: 4.7] [Reference Citation Analysis (0)] |