Published online Jun 21, 2016. doi: 10.3748/wjg.v22.i23.5393
Peer-review started: February 2, 2016
First decision: March 7, 2016
Revised: March 21, 2016
Accepted: April 7, 2016
Article in press: April 7, 2016
Published online: June 21, 2016
Processing time: 132 Days and 2.2 Hours
AIM: To determine the genomic changes in hepatitis B virus (HBV) and evaluate their role in the development of hepatocellular carcinoma (HCC) in patients chronically infected with genotype C HBV.
METHODS: Two hundred and forty chronic hepatitis B (CHB) patients were subjected and followed for a median of 105 mo. HCC was diagnosed in accordance with AASLD guidelines. The whole X, S, basal core promoter (BCP), and precore regions of HBV were sequenced using the direct sequencing method.
RESULTS: All of the subjects were infected with genotype C HBV. Out of 240 CHB patients, 25 (10%) had C1653T and 33 (14%) had T1753V mutation in X region; 157 (65%) had A1762T/G1764A mutations in BCP region, 50 (21%) had G1896A mutation in precore region and 67 (28%) had pre-S deletions. HCC occurred in 6 patients (3%). The prevalence of T1753V mutation was significantly higher in patients who developed HCC than in those without HCC. The cumulative occurrence rates of HCC were 5% and 19% at 10 and 15 years, respectively, in patients with T1753V mutant, which were significantly higher than 1% and 1% in those with wild type HBV (P < 0.001).
CONCLUSION: The presence of T1753V mutation in HBV X-gene significantly increases the risk of HCC development in patients chronically infected with genotype C HBV.
Core tip: In the present study, we determined the genomic changes in the X, S, basal core promoter (BCP), and precore regions of hepatitis B virus (HBV), and evaluate their role in the development of hepatocellular carcinoma (HCC) in chronic hepatitis B (CHB) patients with genotype C HBV. As the results, it was suggested that T1753V mutation in X region might significantly increase the risk of HCC development in CHB patients with genotype C HBV. Also, the BCP mutations might act in synergy with T1753V or G1896A mutation, and with pre-S deletion to promote the development of HCC in these patients.
- Citation: Lee D, Lyu H, Chung YH, Kim JA, Mathews P, Jaffee E, Zheng L, Yu E, Lee YJ, Ryu SH. Genomic change in hepatitis B virus associated with development of hepatocellular carcinoma. World J Gastroenterol 2016; 22(23): 5393-5399
- URL: https://www.wjgnet.com/1007-9327/full/v22/i23/5393.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i23.5393
Worldwide, hepatocellular carcinoma (HCC) is one of the most common malignant tumors[1]. One of the major risk factors closely associated with HCC development is chronic hepatitis B virus (HBV) infection, which in combination with the hepatitis C virus accounts for more than 80% of HCC worldwide[2]. The specific details of the hepatocarcinogenesis from the initial HBV infection is still unclear, although it has been suggested that many factors including but not limited to sex, age, viral genotype environmental factors, and genetic susceptibility all play an important role in the multistage process of HCC development[3].
Due to the high degree of genetic heterogeneity of the virus, there may be multiple mechanisms by which HBV causes HCC. Some studies have found that HBV can directly provoke hepatocarcinogenesis through chromosomal integration or transactivation of cellular genes[3,4]. Regardless, many of these molecular changes indirectly or directly leading to the development of HCC can be linked to genomic changes within the virus.
Various mutations in the HBV genome have been strongly associated with the development of HCC. A significant amount of evidence points to specific genetic mutations as essential viral factors contributing to the development of HCC that could serve as important prognostic biomarkers of the disease. The substitution of A by T at nucleotide 1762 (A1762T) and of G by A at nucleotide 1764 (G1764A) in the basal core promoter (BCP) region is the most common mutation in the HBV genome. This BCP mutation has been associated with higher occurrence of HCC[5,6]. In addition, it has been reported that other mutations of HBV such as G to A transition at nucleotide 1896 (G1896A) in the precore (PC) region, substitution from C to T at nucleotide 1653 (C1653T) and from T to either C/A/G at nucleotide 1753 (T1753V) in the X region, and the deletion in the pre-S region, may be associated with HCC, although their exact roles are still unknown[7-12].
Moreover, it has been demonstrated that genotype C HBV is strongly associated with mutations in the core promoter region and these particular mutations have been shown to be independent risk factors for HCC development[6,13,14]. Consequently, those infected with genotype C HBV may have a poorer prognosis with more aggressive liver disease[11].
Individual mutations within the HBV genome have been studied in order to prove their role in HCC development in chronic hepatitis B patients. Nevertheless, combined mutations in the HBV genome must also be recognized to have the potential to serve as predictive markers for HCC due to the potential ability of the virus to acquire mutations over time. Although there are few reports about the effects of combined mutations of HBV on HCC development, these results are still controversial.
Therefore, in this study, we intended to investigate the genomic changes in the X, S, BCP, and PC regions of HBV. And also we aimed to evaluate their roles in the development of HCC in patients chronically infected with genotype C HBV.
This study included 240 patients with chronic hepatitis B (CHB) diagnosed at Asan Medical Center, Seoul, Korea, between 1991 and 1998. All patients fulfilled the following requirements: (1) seropositivity for hepatitis B surface antigen for at least 6 mo; (2) age > 20 years; and (3) diagnosis with genotype C HBV. The exclusion criteria were as follows: (1) co-infection with hepatitis C virus, hepatitis D virus or human immunodeficiency virus; or (2) diagnosis with HCC at the time of screening.
The patients were followed up regularly at 3-6 month intervals. Serum biochemistry, hematology, HBV DNA titers, hepatitis B e antigen (HBeAg), and anti-HBe, were obtained at each visit. Serum alpha-fetoprotein (AFP), and imaging studies such as ultrasonography or dynamic computed tomographic scan were also performed for the surveillance of HCC development during follow-up. The diagnosis of HCC was determined using AASLD guidelines requiring typical imaging patterns on dynamic CT scan or MRI or pathologic findings[15]. This study was approved by the Institutional Review Board of Asan Medical Center.
HBV DNA was extracted from patient’s serum or liver tissue samples using QIAmp DNA extraction kit (Qiagen K.K., Tokyo, Japan). The HBV genotypes were determined by polymerase chain reaction-restriction fragment length polymorphism of the surface gene of the HBV genome. False positives were avoided by preventing cross-contamination and duplicating the tests in order to validate the results.
The sequence was analyzed by amplifying the entire X, S, PC/core, and BCP regions of the HBV genomes. These regions of HBV genome were then sequenced using a direct sequencing method. The methods used for the genotyping and sequencing of the HBV genome were detailed in our previous study[6]. The sequencing conditions, including A1762T or G1764A in the BCP region, G1896A in the PC region, C1653T or T1753V in the region encoding HBx, and the pre-S deletion mutation were specified in the protocol for the Taq DyeDeoxy Terminator Cycle Sequencing Kit (ABI).
All variables are expressed as the median (range) or number (percentage). Follow-up duration was calculated from the date of enrollment to the last visit and time to recurrence was defined as the duration from enrollment to HCC diagnosis. To identify factors predisposing the development of HCC, the Kaplan-Meier method and log-rank test were used. And also multivariate analysis was performed using the Cox proportional hazards model. All significance tests were two-tailed, and P < 0.05 was considered to be statistically significant. All statistical analyses were performed using the SPSS statistical software package (version 20; SPSS Inc., Chicago, IL).
The demographic data of the 240 patients with CHB are summarized in Table 1. All of the patients had a genotype C HBV infection. About 84% of the patients were male and the median age was 48 years (range, 27-86 years). More than 90% of the patients had relatively well-preserved liver function with Child-Pugh Class A and the median serum alanine aminotransferase (ALT) level was 149 IU/L (range, 10-2170 IU/L). Over 80% of the study subjects were positive for HBeAg. The median follow-up period was 105 mo (range, 1-237 mo).
| Variables | n = 240 |
| Age, yr1 | 48 (27-86) |
| Gender, M/F | 201/39 (84/16) |
| 1Platelet, × 103/mm3 | 176 (62-556) |
| Prothrombin time, %1 | 87 (30-147) |
| ALT, IU/L1 | 149 (10-2170) |
| Total bilirubin, mg/dL1 | 0.8 (0.3-15.4) |
| Albumin, g/dL1 | 4.1 (2.2-5.2) |
| Serum AFP, ng/mL1 | 5.9 (1-2150) |
| Child-Pugh class, A/B/C | 216/15/0 (94/6/0) |
| HBeAg positivity | 195 (81) |
| Serum HBV DNA, copies/mL1 | 4.2 × 106 (ND-1.1 × 109) |
| Presence of HBV genotype C | 240 (100) |
| Follow-up periods, mo1 | 105 (1-237) |
The frequencies of genomic mutations in HBV are shown in Figure 1. The C1653T mutation in the X region was found in 10% of the study cases (n = 25). Another mutation in the X region, T1753V, was seen in 14% (n = 33). The BCP double mutation, A1762T/G1764A was detected in 65% (n = 157). The G1896A mutation in the PC region was seen in 21% (n = 50) and the pre-S deletion was found in 28% (n = 67).
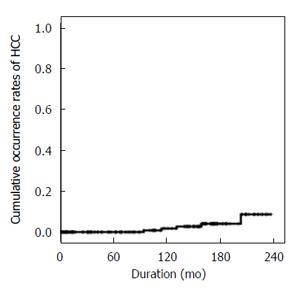
During follow-up, 6 of 240 patients with CHB (3%) newly developed HCC. The 5-, 10-, and 15-year overall cumulative occurrence rates of HCC were 0%, 1.8% and 4.1%, respectively (Figure 2).
The presence of the T1753V mutation in the X region was significantly associated with the development of HCC (Figure 3). The 10- and 15-year cumulative occurrence rates of HCC were 5% and 19%, respectively, in patients with the T1753V mutation, which were significantly higher than the 1% 10- and 15-year rates seen in those with wild-type HBV (P < 0.001). However, other point mutations, such as C1653T in the X region, the BCP double mutation A1762T/G1764A, G1896A in the PC region, and the pre-S deletion, did not affect HCC occurrence (Table 2).
| Variables | RR (95%CI) | P value |
| Genomic changes | ||
| C1653T in X region | 1.583 (0.176-14.210) | NS |
| T1753V in X region | 17.565 (1.956-157.737) | < 0.01 |
| A1762T/G1764A in BCP | 30.512 (0.004-230706) | NS |
| G1896A in PC | 5.765 (0.519-64.076) | NS |
| Pre-S deletion | 2.491 (0.502-12.370) | NS |
| Clinical characteristics | ||
| Age > 50 yr | 57.5 (0.035-93428) | NS |
| Male | 27.3 (0.001-547718) | NS |
| ALT > 80 IU/L | 0.55 (0.091-3.29) | NS |
| PT ≤ 70 % | 4.12 (0.682-24.845) | NS |
| Serum AFP > 200 ng/mL | 0.044 (0.00-209046) | NS |
| HBeAg positive | 0.034 (0.00-360.724) | NS |
| HBV DNA > 105 copies/mL | 0.741 (0.123-4.452) | NS |
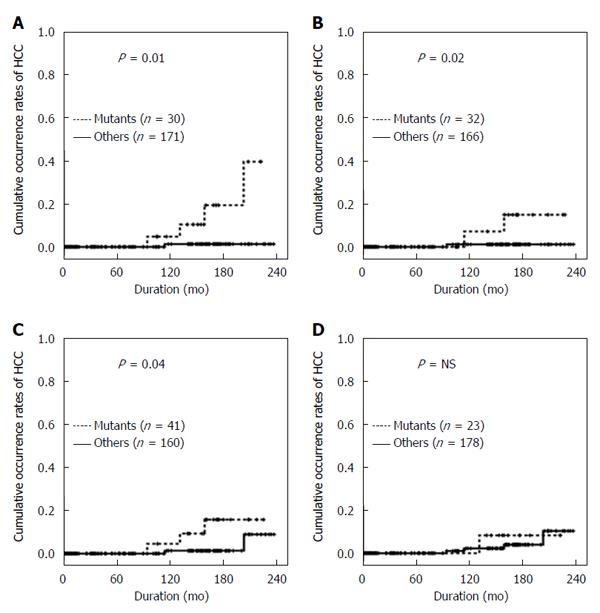
The occurrence rates of HCC were assessed in relation to the presence of combined mutations to evaluate the synergic effects of genomic changes in HBV. The combination of the BCP double mutations A1762T/G1764A and the T1753V mutation had a close association with HCC development (P < 0.01) (Figure 4A). Patients with both BCP A1762T/G1764A mutations and the G1896A mutation also had a significantly higher occurrence rate of HCC (P < 0.05) (Figure 4B). In addition, coexistence of BCP double mutations and pre-S deletion showed the higher HCC occurrence rates (P < 0.05) (Figure 4C). However, the combination of BCP double mutations and C1653T in the X region was not significantly associated with HCC development (Figure 4D).
The occurrence rates of HCC in relation to baseline characteristics were also evaluated (Table 2). Age and gender did not showed significant association with the occurrence of HCC in univariate analysis. In addition, other clinical characteristics, such as serum ALT level, prothrombin time, serum AFP level, HBeAg positivity, and HBV DNA titers, did not affect HCC development. In multivariate analysis, the presence of the T1753V mutation in the X region was an independent risk factor for the development of HCC (relative risk 14.03, P < 0.05) (Table 3).
| Variables | RR (95%CI) | P value |
| Age > 50 yr | 102903 (0.000-1838) | NS |
| Male | 196796 (0.000-2834) | NS |
| T1753V in X region | 14.029 (1.568-125.550) | < 0.05 |
The impact of genomic changes in HBV on the clinical course of the infection-from the initial viral infection to HCC development-has been of great interest due to the unclear nature of the relationship between the two components. Our work clarifies some of the uncertainty because our results suggest that patients with the T1753V mutation in the X region of the HBV genome are more likely to develop HCC as a result of their chronic HBV infection, especially that of genotype C. The HBx, a nonstructural regulatory protein of HBV, may play an essential role in hepatocarcinogenesis in the context of HBV. Studies of HBx transgenic mice have provided limited results regarding the role of the X-gene in live tumor formation[3,7,11]. Moreover, the exact role of the genomic mutations in the X region is still unclear. However, the present findings indicated that the T1753V mutation in the X region is an independent risk factor for the development of HCC in patients with CHB. Our results thus suggest that an amino acid change at nucleotide 1753 in the X region can affect the function of the HBx protein and thereby contribute to the process of hepatocarcinogenesis[7,11,16]. Therefore, it seems likely that mutations in the X region resulting in the translation of a truncated HBx protein contribute to the initiation of tumor formation in the liver[3,17].
The BCP double mutation A1762T/G1764A is one of the most common mutations in the HBV genome. The current study also showed high prevalence (65%) of A1762T/G1764A in patients with CHB of genotype C. However, the BCP double mutation was not significantly associated with HCC development. Nevertheless, we found that A1762T/G1764A tended to have high occurrence rates of HCC, although the results were not statistically significant. Interestingly, the BCP double mutation A1762T/G1764A significantly contributed to HCC development when it was combined with other individual mutations such as T1753V in the X region, G1896A in the PC region, or the pre-S deletion. Thus, the BCP mutations may also be associated with hepatocarcinogenesis[13,18]. The BCP mutations may change the viral pre-genomic secondary structure or increase the transcription of the pre-genomic RNA, consequently, the BCP mutations can increase viral replication[14,19].
The PC mutation terminates the translation of HBeAg by creating a premature stop codon in the PC gene[6,7,20]. However, the exact role of this mutation is unclear because, although the mutation has been demonstrated to cause more severe liver disease, it was frequently detected in HBeAg-negative asymptomatic carriers[9,21,22]. In particular, one study has shown that the PC 1896 mutation alone appears to have no direct pathogenic role in HBV genotype C[21]. Our results also showed that the PC G1896A mutation alone was not significantly associated with HCC development, but a combination of the PC mutation and the BCP double mutation significantly influenced HCC occurrence.
Truncated pre-S2/S sequences can often be found in HBV DNA integration sites in HCC[3,23]. As in our present results, a recent study in Taiwan showed that the combination of the pre-S deletion mutation and the BCP double mutation was significantly associated with the development of HCC[6,24]. It is possible that hepatocytes expressing altered large and middle surface proteins encoded by the mutated S gene have a potential growth advantage[10,25]. Moreover, the pre-S mutant has been found to upregulate cyclin A expression and induce nodular proliferation of hepatocytes[10,17]. The modified surface antigen may induce oxidative DNA damage and mutations in hepatocytes in the late stages of a chronic HBV infection[10,26].
One limitation of our present study was the small number of newly developed HCC cases during follow-up. Out of 240 patients with CHB, only 6 patients developed HCC. However, most subjects had CHB, not liver cirrhosis. In addition, the median follow-up duration was about 9 years, which may be insufficient duration for HCC development, especially in patients with CHB. Nevertheless, our current study is one of the largest of its kind to date and had a longer follow-up period than other similar reports.
In conclusion, the presence of the T1753V mutation in the X region significantly increases the risk of HCC development in patients with genotype C CHB. The BCP mutations may act in synergy with the T1753V mutation in the X region, G1896A in the PC region, and with the pre-S deletion to significantly increase the occurrence of HCC in CHB patients with genotype C HBV.
Hepatitis B virus (HBV) is the most common risk factor for hepatocellular carcinoma development. Especially, among the various characteristics of HBV, genomic variation of the virus may effect on hepatocellular carcinoma (HCC) development directly. However, the exact role of the genomic mutation of HBV is still controversial for hepatocarcinogenesis. Thus, it is very important to clarify obvious function of the genomic mutation for the development of HCC in patients with HBV infection.
The authors are part of the leading group in the studies of carcinogenesis for HBV associated HCC. The authors believe that some mutations in the basal core promoter, the precore, and the X regions may be associated with HCC development. We provide reasonable evidence to support the hypothesis which we have made from this paper.
Generally agreed by the most researchers on genomic changes of HBV are able to effect on the development of HCC. However, their opinions are far from being unanimous as to obvious role of each mutation for the development of HCC. This paper shows that the T1753V mutation in X region plays an important role as the risk factor for HCC development. Moreover, combined mutations of the basal core promoter mutation and other mutations such as T1753V, G1896A, and the pre-S deletion are also the risk factors for the development of HCC in patients with genotype C HBV infection.
High risk group for HCC development can be identified among the patients infected with genotype C HBV. Furthermore, the authors can establish efficient process to detect HCC earlier and also the authors may prevent the development of HCC ultimately.
HBV is a member of hepadnaviridae family, and it can cause acute hepatitis, fulminant hepatitis, asymptomatic carrier state, chronic hepatitis, cirrhosis, and HCC. Especially, HBV is the most common risk factor for HCC development. Because HBV has the high degree of genetic diversity, it is considered that the genomic changes of HBV can influence on the carcinogenesis of HCC, although the exact role of each genomic mutation is not obvious. HCC is a primary cancer of the liver and usually occurs in patients with chronic liver disease, especially chronic hepatitis B. Multiple mechanisms impact on the development of HCC and some of mechanisms has not been fully verified as yet.
Excellent study has been performed by Danbi Lee et al reporting about the association between genomic changes of genotype C HBV and HCC development. This study definitely help to verify mechanism of HCC development. Congratulation to authors for the valuable results for role of genetic mutation of HBV for the hepatocarcinogenesis.
P- Reviewer: Liu EQ, Qin JM S- Editor: Qi Y L- Editor: A E- Editor: Wang CH
| 1. | van Zonneveld M, Honkoop P, Hansen BE, Niesters HG, Darwish Murad S, de Man RA, Schalm SW, Janssen HL. Long-term follow-up of alpha-interferon treatment of patients with chronic hepatitis B. Hepatology. 2004;39:804-810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 252] [Article Influence: 12.0] [Reference Citation Analysis (1)] |
| 2. | Nguyen VT, Law MG, Dore GJ. Hepatitis B-related hepatocellular carcinoma: epidemiological characteristics and disease burden. J Viral Hepat. 2009;16:453-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 232] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 3. | Park NH, Song IH, Chung YH. Chronic hepatitis B in hepatocarcinogenesis. Postgrad Med J. 2006;82:507-515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 58] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 4. | Pang R, Tse E, Poon RT. Molecular pathways in hepatocellular carcinoma. Cancer Lett. 2006;240:157-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 69] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 5. | Kao JH, Chen PJ, Lai MY, Chen DS. Basal core promoter mutations of hepatitis B virus increase the risk of hepatocellular carcinoma in hepatitis B carriers. Gastroenterology. 2003;124:327-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 414] [Cited by in RCA: 428] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 6. | Lyu H, Lee D, Chung YH, Kim JA, Lee JH, Jin YJ, Park W, Mathews P, Jaffee E, Zheng L. Synergistic effects of A1896, T1653 and T1762/A1764 mutations in genotype c2 hepatitis B virus on development of hepatocellular carcinoma. J Viral Hepat. 2013;20:219-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 7. | Asim M, Malik A, Sarma MP, Polipalli SK, Begum N, Ahmad I, Khan LA, Husain SA, Akhtar N, Husain S. Hepatitis B virus BCP, Precore/core, X gene mutations/genotypes and the risk of hepatocellular carcinoma in India. J Med Virol. 2010;82:1115-1125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 41] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 8. | Omata M, Ehata T, Yokosuka O, Hosoda K, Ohto M. Mutations in the precore region of hepatitis B virus DNA in patients with fulminant and severe hepatitis. N Engl J Med. 1991;324:1699-1704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 377] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 9. | Okamoto H, Yotsumoto S, Akahane Y, Yamanaka T, Miyazaki Y, Sugai Y, Tsuda F, Tanaka T, Miyakawa Y, Mayumi M. Hepatitis B viruses with precore region defects prevail in persistently infected hosts along with seroconversion to the antibody against e antigen. J Virol. 1990;64:1298-1303. [PubMed] |
| 10. | Fang ZL, Sabin CA, Dong BQ, Wei SC, Chen QY, Fang KX, Yang JY, Huang J, Wang XY, Harrison TJ. Hepatitis B virus pre-S deletion mutations are a risk factor for hepatocellular carcinoma: a matched nested case-control study. J Gen Virol. 2008;89:2882-2890. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 87] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 11. | Shinkai N, Tanaka Y, Ito K, Mukaide M, Hasegawa I, Asahina Y, Izumi N, Yatsuhashi H, Orito E, Joh T. Influence of hepatitis B virus X and core promoter mutations on hepatocellular carcinoma among patients infected with subgenotype C2. J Clin Microbiol. 2007;45:3191-3197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 55] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 12. | Chen BF, Liu CJ, Jow GM, Chen PJ, Kao JH, Chen DS. High prevalence and mapping of pre-S deletion in hepatitis B virus carriers with progressive liver diseases. Gastroenterology. 2006;130:1153-1168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 244] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 13. | Yuen MF, Sablon E, Tanaka Y, Kato T, Mizokami M, Doutreloigne J, Yuan HJ, Wong DK, Sum SM, Lai CL. Epidemiological study of hepatitis B virus genotypes, core promoter and precore mutations of chronic hepatitis B infection in Hong Kong. J Hepatol. 2004;41:119-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 55] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 14. | Yuen MF, Tanaka Y, Shinkai N, Poon RT, But DY, Fong DY, Fung J, Wong DK, Yuen JC, Mizokami M. Risk for hepatocellular carcinoma with respect to hepatitis B virus genotypes B/C, specific mutations of enhancer II/core promoter/precore regions and HBV DNA levels. Gut. 2008;57:98-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 135] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 15. | Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020-1022. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5972] [Cited by in RCA: 6573] [Article Influence: 469.5] [Reference Citation Analysis (1)] |
| 16. | Takahashi K, Ohta Y, Kanai K, Akahane Y, Iwasa Y, Hino K, Ohno N, Yoshizawa H, Mishiro S. Clinical implications of mutations C-to-T1653 and T-to-C/A/G1753 of hepatitis B virus genotype C genome in chronic liver disease. Arch Virol. 1999;144:1299-1308. [PubMed] |
| 17. | Wang Y, Lau SH, Sham JS, Wu MC, Wang T, Guan XY. Characterization of HBV integrants in 14 hepatocellular carcinomas: association of truncated X gene and hepatocellular carcinogenesis. Oncogene. 2004;23:142-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 89] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 18. | Chu CM, Lin CC, Lin SM, Lin DY, Liaw YF. Viral load, genotypes, and mutants in hepatitis B virus-related hepatocellular carcinoma: special emphasis on patients with early hepatocellular carcinoma. Dig Dis Sci. 2012;57:232-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 19. | Kidd AH, Kidd-Ljunggren K. A revised secondary structure model for the 3’-end of hepatitis B virus pregenomic RNA. Nucleic Acids Res. 1996;24:3295-3301. [PubMed] |
| 20. | Yotsuyanagi H, Hino K, Tomita E, Toyoda J, Yasuda K, Iino S. Precore and core promoter mutations, hepatitis B virus DNA levels and progressive liver injury in chronic hepatitis B. J Hepatol. 2002;37:355-363. [PubMed] |
| 21. | Yoo BC, Park JW, Kim HJ, Lee DH, Cha YJ, Park SM. Precore and core promoter mutations of hepatitis B virus and hepatitis B e antigen-negative chronic hepatitis B in Korea. J Hepatol. 2003;38:98-103. [PubMed] |
| 22. | Baptista M, Kramvis A, Kew MC. High prevalence of 1762(T) 1764(A) mutations in the basic core promoter of hepatitis B virus isolated from black Africans with hepatocellular carcinoma compared with asymptomatic carriers. Hepatology. 1999;29:946-953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 195] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 23. | Bréchot C. Pathogenesis of hepatitis B virus-related hepatocellular carcinoma: old and new paradigms. Gastroenterology. 2004;127:S56-S61. [PubMed] |
| 24. | Chen CH, Hung CH, Lee CM, Hu TH, Wang JH, Wang JC, Lu SN, Changchien CS. Pre-S deletion and complex mutations of hepatitis B virus related to advanced liver disease in HBeAg-negative patients. Gastroenterology. 2007;133:1466-1474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 198] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 25. | Fan YF, Lu CC, Chang YC, Chang TT, Lin PW, Lei HY, Su IJ. Identification of a pre-S2 mutant in hepatocytes expressing a novel marginal pattern of surface antigen in advanced diseases of chronic hepatitis B virus infection. J Gastroenterol Hepatol. 2000;15:519-528. [PubMed] |
| 26. | Hsieh YH, Su IJ, Wang HC, Chang WW, Lei HY, Lai MD, Chang WT, Huang W. Pre-S mutant surface antigens in chronic hepatitis B virus infection induce oxidative stress and DNA damage. Carcinogenesis. 2004;25:2023-2032. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 256] [Article Influence: 12.2] [Reference Citation Analysis (0)] |