Published online Mar 21, 2016. doi: 10.3748/wjg.v22.i11.3165
Peer-review started: September 28, 2015
First decision: October 14, 2015
Revised: November 4, 2015
Accepted: January 17, 2016
Article in press: January 17, 2016
Published online: March 21, 2016
Processing time: 170 Days and 14.8 Hours
AIM: To study the hepatitis B virus (HBV) and hepatitis D virus (HDV) replication interferences in patients with chronic hepatitis delta infected with different HBV genotypes.
METHODS: We conducted a transversal study including 68 chronic hepatitis delta (CHD) (37 HIV-positive) patients and a control group of 49 chronic hepatitis B (CHB) (22 HIV-positive) patients. In addition, a dynamic follow-up was performed in 16 CHD patients. In all the samples, the surface antigen of hepatitis B (HBsAg) serum titers were analyzed with the Monolisa HBsAg Ultra system (Bio-Rad), using as quantification standard a serial dilution curve of an international HBsAg standard. Serum HBV-DNA titers were analyzed using the Roche Cobas TaqMan (Roche, Barcelona, Spain), and the serum HDV-RNA using an in-house real-time qRT-PCR method, with TaqMan probes. HBV genotype was determined with the line immunoassay LiPA HBV genotyping system (Innogenetics, Ghent, Belgium). In those patients negative for LiPA assay, a nested PCR method of complete HBsAg coding region, followed by sequence analysis was applied.
RESULTS: No differences in the HBV-DNA levels were found in CHB patients infected with different HBV genotypes. However, in CHD patients the HBV-DNA levels were lower in those infected with HBV-A than in those with HBV-D, both in HIV negative [median (IQR): 1.25 (1.00-1.35) vs 2.95 (2.07-3.93) log10 (copies/mL), P = 0.013] and HIV positive patients [2.63 (1.24-2.69) vs 7.25 (4.61-7.55) log10 (copies/mL), P < 0.001]. This was confirmed in the dynamic study of the HBV/HDV patients. These differences induce an under-estimation of HBV-A incidence in patients with CHD analyzed with LiPA assay. Finally, the HBsAg titers reflected no significant differences in CHD patients infected with HBV-A or D.
CONCLUSION: Viral replication interference between HBV and HDV is HBV-genotype dependent, and more evident in patients infected with HBV-genotype A, than with HBV-D or E.
Core tip: Hepatitis B virus (HBV)-DNA titer is a predictive pattern of optimal response to treatment with interferon based regimens. Our results suggest that in chronic hepatitis delta patients the inhibition of HBV replication is genotype-dependent. Viral replication interference between HBV and hepatitis B virus (HDV) is more evident in patients infected with HBV-genotype A, than with HBV-D or E. Based on these results the analysis of HBV genotype could be taken into account in the algorithms for treatment indication for delta infected patients. For patients from geographical regions with a different distribution of HBV genotypes, the replication behaviour of HBV is warranted to clarify the HBV/HDV interference process.
- Citation: Madejón A, Romero M, Hernández &, García-Sánchez A, Sánchez-Carrillo M, Olveira A, García-Samaniego J. Hepatitis B and D viruses replication interference: Influence of hepatitis B genotype. World J Gastroenterol 2016; 22(11): 3165-3174
- URL: https://www.wjgnet.com/1007-9327/full/v22/i11/3165.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i11.3165
Hepatitis delta virus (HDV) is a defective infectious agent which only can infect patients previously (superinfection) or simultaneously (co-infection) infected with hepatitis B virus (HBV). Although during the last decades, in developed countries, effective vaccination programs for the prevention of HBV significantly decreased novel hepatitis delta virus HDV cases, chronic HDV infection continues to be an important public health problem, due to the lack of an effective therapy and the frequent severity of chronic hepatitis delta (CHD)[1]. Moreover, the lower than expected decrease of HDV infection in some Western countries, such as Italy, Germany, United Kingdom and Turkey, probably has been caused by persistent HDV reservoirs of immigrant populations[2-5].
The natural history of CHD is crucially influenced by HBV and HDV replication. Thus, HBV replication has been described as modulating HDV pathogenesis and the active replication of both viruses is associated with severe liver disease[6,7]. However, several factors can influence the dynamic replication of HBV and HDV. A potential inhibitory role of HDV over HBV replication has been shown in cross-sectional studies using low-sensitive HDV quantitative techniques[8,9]. Nevertheless, data from longitudinal studies, with high-sensitive HDV quantification assays, suggest more complex interactions between both viruses: patients show parallel replication patterns for HBV and HDV, contrasting with others in which both markers change in opposite ways, and these differences are not currently explained by molecular data. Thus, the role of the HBV genotype on the viral replication interference processes has not been extensively studied[10,11].
Previous reports suggest that HBV genotype may influence the natural history of chronic hepatitis B (CHB) at different levels. Studies performed in Asian patients have shown a higher percentage of spontaneous[12] and interferon treatment induced HBeAg seroconversion in patients with B than C genotypes[13,14]. In addition, a more severe evolution of the hepatic damage and a higher risk of developing hepatocelular carcinoma have been described for HBV-C compared with HBV-B infections[15]. There are no consistent data for A and D genotypes, but in some European studies a higher rate of HBe seroconversion in patients infected with genotype A treated with interferon or interferon plus lamivudine has beeb shown[16,17]. In addition, it has been suggested that HBV genotype may play a role in the natural history of CHD, and that CHD patients carrying the HBV-C present a worse outcome of liver disease than those infected with HBV-B[7-9]. The study of the role of HBV genotype on the evolution of CHD can be particularly important for countries with high immigration flows from HDV endemic regions and/or those with a varying prevalence of HBV genotypes.
The aim of this work was to study the HBV replication interference mediated by HDV in CHD patients infected with different HBV genotypes. Therefore, we analyzed, in both cross-sectional and dynamic follow-up approaches, the serum titers of HDV-RNA, HBV-DNA and HBsAg, a surrogate marker of HBV replication, in CHB and CHD patients infected with HBV genotypes A and D, the most prevalent genotypes in Western countries.
Retrospective, transversal study, in which 68 CHD (simultaneous CHB and CHD) patients (31 HIV-negative and 37 HIV-positive) were included. As a control group, 49 patients with CHB, negative for HDV superinfection (27 HIV-negative and 22 HIV-positive) were included (Table 1). No patient was under antiviral treatment at least 6 mo prior to inclusion in the study. In 16/31 (52%) CHD patients without HIV coinfection and with available sequential serum samples we carried out a longitudinal dynamic analysis of the viral replication markers (Table 2). Hepatitis C virus (HCV) infection was confirmed in 9 patients (1 with CHB, 2 with CHD, 2 with HBV + HIV and 4 with HBV + HDV + HIV).
| HBV | HBV + HIV | HBV + HDV | HBV + HDV + HIV | |
| (n = 27) | (n = 22) | (n = 31) | (n = 37) | |
| Age (yr)1 | 38.54 ± 12.22 | 47.33 ± 8.50 | 41.15 ± 9.33 | 42.32 ± 7.76 |
| Gender (M/F) | 21/6 | 12/10 | 22/9 | 17/20 |
| Caucasian | 18 (67) | 20 (91) | 27 (87) | 33 (89) |
| Subsaharian | 9 (33) | 2 (9) | 4 (13) | 4 (11) |
| ALT (IU/mL)1 | 66.58 ± 76.03c | 70.67 ± 18.58 | 112.92 ± 84.39d | 90.40 ± 21.92 |
| AST (IU/mL)1 | 42.65 ± 27.76 | 32.67 ± 19.40 | 89.77 ± 62.43 | 38.33 ± 20.12 |
| Anti-HCV | 4 (15) | 2 (9) | 2 (6) | 3 (8) |
| Anti-HBe | 23 (85) | 17 (77) | 29 (93) | 31 (84) |
| HBeAg | 4 (15) | 5 (23) | 2 (7) | 6 (16) |
| HBV genotype-A | 17 (63) | 12 (54) | 12 (42) | 16 (43) |
| HBV genotype-D | 10 (37) | 10 (46) | 13 (42) | 20 (54) |
| HBV genotype-E | 0 | 0 | 4 (9) | 1 (3) |
| HBV-DNA (Log10 IU/mL)2 | 3.90 (3.15-5.85)e | 6.97 (5.58-7.25)f | 1.58 (1.00-2.43)g | 2.80 (1.00-7.02) |
| HBsAg (Log10 IU/mL)2 | 4.00 (3.81-4.22)h | 5.46 (4.15-5.61)i | 3.83 (3.59-4.39)j | 3.83 (3.24-4.38)k |
| HDV-RNA (Log10 copies/mL)2 | NA | NA | 5.20 (4.37-5.74) | 6.89 (4.41-7.73) |
| Age1 | Gender | Precedence | Mode of | Duration of | HBV Genotype2 | Previous | Time without Treatment3 (mo) | Follow-up (mo) | Sampling | |
| transmission | infection (yr) | treatment | ||||||||
| Pat #1 | 43 | Male | Western Europe | Horizontal | 20 | Undetermined | IFN | 84 | 74 | 8 |
| Pat #2 | 30 | Male | Equatorial Ginnea | Unknown | Unknown | A | No | 5 | 4 | |
| Pat #3 | 41 | Male | Equatorial Ginnea | Unknown | Unknown | E | No | 2 | 3 | |
| Pat #4 | 43 | Male | Eastern Europe | Unknown | Unknown | A | No | 5 | 3 | |
| Pat #5 | 45 | Male | Western Europe | IVD | Unknown | A | No | 4 | 4 | |
| Pat #6 | 38 | Female | Western Europe | Horizontal | 20 | D | IFN | 36 | 94 | 10 |
| Pat #7 | 35 | Female | Western Europe | Horizontal | 25 | E | IFN | 60 | 89 | 13 |
| Pat #8 | 55 | Female | Western Europe | Unknown | Unknown | D | No | 17 | 3 | |
| Pat #9 | 37 | Male | Equatorial Ginnea | IVD | 22 | A | IFN/ADV | 8 | 77 | 12 |
| Pat #10 | 31 | Female | Western Europe | Horizontal | 20 | D | IFN | 12 | 78 | 12 |
| Pat #11 | 46 | Male | Western Europe | IVD | 25 | A | No | 53 | 4 | |
| Pat #12 | 42 | Male | Western Europe | Unknown | 26 | D | No | 93 | 13 | |
| Pat #13 | 45 | Male | Western Europe | Unknown | Unknown | A | No | 42 | 3 | |
| Pat #14 | 39 | Male | Western Europe | IVD | 20 | D | No | 41 | 6 | |
| Pat #15 | 53 | Male | Western Europe | IVD | 25 | D | No | 89 | 14 | |
| Pat #16 | 38 | Female | Equatorial Ginnea | Unknown | Unknown | E | No | 9 | 5 |
HBV serological markers were tested by commercial EIA assays: AxSym HBsAg (version 2), AxSym HBeAg (version 2.0) and AxSym anti-HBe (Abbott Laboratories, North Chicago, IL, United States). Serum HDAg, total anti-HDV antibodies and specific IgM anti-HDV antibodies were tested by commercial EIAs (Radim Iberica, Barcelona, Spain).
The quantitative analysis of serum HBsAg levels was performed with the Monolisa HBsAg Ultra system (Bio-Rad), using as quantification standard a serial dilution curve of an international HBsAg standard of known concentration (NIBSC, Potters Bar, United Kingdom). HBsAg titers were expressed as log10 (IU/mL).
HBV genotype was determined with the line immunoassay LiPA HBV genotyping system (Innogenetics, Ghent, Belgium). In patients negative for LiPA assay, a high sensitivity nested PCR method for the complete HBsAg coding region was applied.
All the positive amplified products were subjected to direct sequencing and the sequences obtained analyzed using the Geno2pheno (HBV) software (Genafor, Max-Plank Institut Informatic, Saarbrücken, Germany).
Serum HBV-DNA levels were analyzed using the Roche Cobas TaqMan (Roche, Barcelona, Spain) with a lower detection limit of 12 IU/mL. The copies number of HDV-RNA, and HBV-DNA were tested using in-house real-time qRT-PCR methods, with TaqMan probes[18]. The lower detection limit of these assays was 10 and 20 copies/mL, respectively.
All parameters expressed as absolute number or percentages were analyzed using the Spearman’s ran-correlation, the Wilcoxon matched-pairs and the U Mann-Wiitney systems. The percentage of variability of each variable was expressed as the variant coefficient (VC). The mean comparisons were performed using the Student-t test. All the statistical analyses were performed using the SPSS v13 software (SPSS Inc. North Chicago, IL, United States).
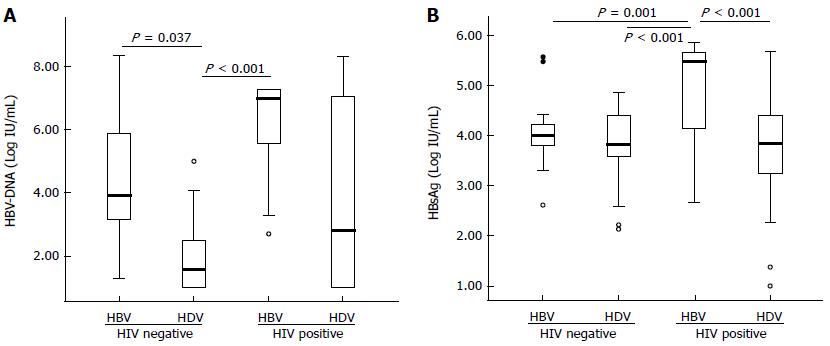
The analysis of the 117 serum samples showed that the HBV replication markers (serum HBV-DNA and HBsAg) tended to be higher in patients coinfected with HIV and lower in those with CHD. These differences were especially marked in HBV-DNA titers between CHB patients and those with CHD [median (IQR): 3.90 (3.15-5.85) vs 1.58 (1.00-2.43) log10 (copies/mL), respectively, P = 0.037] (Table 1 and Figure 1A). The HBsAg titers showed a similar behaviour except in the group of CHD patients, in which the decrease in the HBV-DNA levels was not accompanied by a similar decrease in the HBsAg (Table 1 and Figure 1B). Similarly, the HDV-RNA were higher, but without statistical significance, in HIV-patients than in HIV-negatives [median (IQR): 6.89 (4.41-7.73) vs 5.20 (4.37-5.74) log10 (copies/mL), respectively, P = NS).
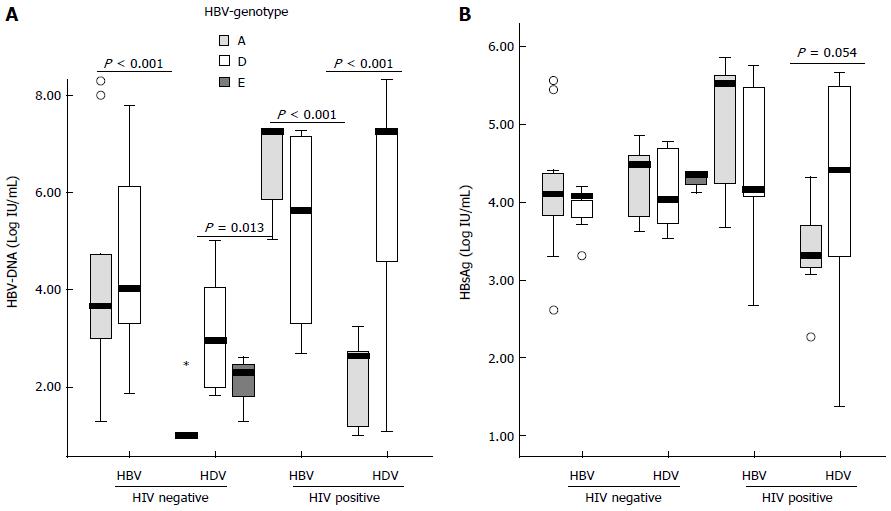
No differences were found in the HBV-DNA titers among patients infected with HBV genotype A or D [median (IQR): 3.90 (2.31-7.25) vs 4.81 (3.30-7.25) log10 (copies/mL), respectively, P = NS] or the HBsAg levels [median (IQR): 4.25 (3.71-5.45) vs 4.11 (3.72-4.72) log10(IU/mL), respectively, P = NS]. When the study population was stratified according to the viral coinfection status, a different HBV replication behaviour was observed between patients with different HBV genotypes (Figure 2A). In CHB patients no differences in the HBV-DNA levels were found between those infected with HBV-A or D irrespective of the HIV status. By contrast, significant higher levels of HBV DNA were found in CHD patients with HBV-D than HBV-A, both in HIV positive [median (IQR): 7.25 (4.61-7.55) vs 2.63 (1.24-2.69), P < 0.001] and negative patients [2.95 (2.07-3.93) vs 1.25 (1.00-1.35) log10 (copies/mL), P = 0.013]. No differences in the HBsAg titers were found in the different groups of patients (Figure 2B). No differences in the HCV prevalence was observed among patients with HVB-A and D [5/57 (9%) vs 4/53 (7%), respectively, P = NS]. Regarding to the treatment pressure, no differences were found in the number of patients with previous history of antiviral treatment in patients with CHD infected with HBV-A or D [9/28 (32%) vs 10/33 (30%), respectively, P = NS].
In order to confirm the data from the cross-sectional study regarding the HBV genotype dependent inhibition of HBV replication mediated by HDV superinfection, a longitudinal follow-up of 16 CHD patients without HIV infection was performed (see epidemiological features in Table 2). In all but one patient, HDV-RNA was higher than HBV-DNA (Table 3 and Table 4). HBV-DNA was persistently detected in only 6/16 patients (37.5%), negative in 4 (25%), and fluctuating in the remaining 6 patients (37.5%) (Table 3). From these 6 patients with fluctuations in HBV-DNA, 3 presented an alternating pattern with positive and negative samples; for the other 3 patients, HBV-DNA became negative during follow-up and remained negative until the end of the study. By contrast, serum HDV-RNA was detectable during the follow-up in 12/16 (75%) patients and alternating in 3 (Table 4).
| Serum HBV-DNA | HBsAg | ||||||
| Genotype1 | Anti-HBe | Status2 | Log10 (IU/mL) (range) | VC | Log10 (IU/mL) (range) | VC | |
| Pat #1 | Und | Positive | Negative | NA | NA | 4.40 ± 0.13 (4.20-4.48) | 0.03 |
| Pat #2 | A | Positive | Negative | NA | NA | 4.60 ± 0.09 (4.56-4.66) | 0.02 |
| Pat #3 | E | Positive | Positive | 2.80 ± 0.29 (2.60-3.01) | 0.10 | 4.46 ± 0.10 (4.39-4.53) | 0.02 |
| Pat #4 | A | Positive | Positive | 1.75 ± 1.20 (1.30-2.95) | 0.69 | 3.53 ± 0.14 (3.44-3.63) | 0.04 |
| Pat #5 | A | Positive | Positive | 3.25 ± 1.30 (2.13-4.35) | 0.40 | 4.87 ± 0.06 (4.86-4.93) | 0.01 |
| Pat #6 | D | Positive | Fluctuating | 2.32 ± 0.70 (1.00-3.42) | 0.30 | 4.52 ± 0.27 (4.01-4.78) | 0.06 |
| Pat #7 | E | Positive | Fluctuating | 1.36 ± 0.45 (1.00-2.48) | 0.33 | 3.94 ± 0.10 (3.80-4.12) | 0.03 |
| Pat #8 | D | Positive | Fluctuating | 1.65 ± 0.92 (1.00-2.30) | 0.56 | 4.64 ± 0.07 (4.59-4.69) | 0.02 |
| Pat #9 | A | Positive | Fluctuating | 1.62 ± 0.88 (1.00-2.48) | 0.54 | 4.17 ± 0.68 (3.68-4.64) | 0.16 |
| Pat #10 | D | Positive | Positive | 3.17 ± 0.56 (2.22-4.08) | 0.18 | 3.99 ± 0.19 (3.64-4.13) | 0.05 |
| Pat #11 | A | Positive | Negative | NA | NA | 4.55 ± 0.07 (4.48-4.63) | 0.02 |
| Pat #12 | D | Negative | Fluctuating | 3.22 ± 1.40 (1.00-4.99) | 0.43 | 4.45 ± 0.09 (4.30-4.58) | 0.02 |
| Pat #13 | A | Positive | Negative | NA | NA | 3.85 ± 0.17 (3.73-3.97) | 0.04 |
| Pat #14 | D | Positive | Fluctuating | 2.11 ± 0.73 (1.00-2.93) | 0.35 | 3.75 ± 0.10 (3.68-3.82) | 0.03 |
| Pat #15 | D | Positive | Positive | 2.76 ± 0.95 (1.97-4.76) | 0.34 | 3.55 ± 0.20 (3.20-3.73) | 0.06 |
| Pat #16 | E | Positive | Positive | 2.36 ± 0.10 (2.30-2.48) | 0.04 | 4.28 ± 0.10 (4.21-4.35) | 0.02 |
| HDV markers | HCV markers | ||||||
| Serum HDV-RNA | Serum HCV-RNA | ||||||
| Genotype | Anti-HD IgM | Status1 | Log10 (copies/mL) (range) | VC | Anti-HCV | HCV-RNA | |
| Pat #1 | Und | Negative | Positive | 5.90 ± 0.31 (5.53-6.11) | 0.05 | Negative | NA |
| Pat #2 | A | Negative | Positive | 5.69 ± 0.05 (5.64-5.72) | 0.01 | Negative | NA |
| Pat #3 | E | Negative | Positive | 5.11 ± 0.12 (5.02-5.20) | 0.02 | Negative | NA |
| Pat #4 | A | Negative | Positive | 4.38 ± 1.64 (2.60-5.84) | 0.37 | Negative | NA |
| Pat #5 | A | Negative | Positive | 6.47 ± 0.64 (5.70-7.18) | 0.10 | Negative | NA |
| Pat #6 | D | Positive | Positive | 4.55 ± 2.26 (1.08-5.75) | 0.50 | Negative | NA |
| Pat #7 | E | Positive | Positive | 4.01 ± 0.31 (3.57-4.43) | 0.08 | Negative | NA |
| Pat #8 | D | Positive | Positive | 4.13 ± 0.35 (3.89-4.38) | 0.08 | Negative | NA |
| Pat #9 | A | Positive | Fluctuating | 2.14 ± 0.99 (1.00-3.31) | 0.46 | Negative | NA |
| Pat #10 | D | Positive | Fluctuating | 4.07 ± 1.72 (1.00-5.07) | 0.42 | Negative | NA |
| Pat #11 | A | Negative | Positive | 5.47 ± 1.33 (4.53-5.47) | 0.24 | Negative | NA |
| Pat #12 | D | Positive | Positive | 4.76 ± 1.46 (1.70-6.22) | 0.31 | Negative | NA |
| Pat #13 | A | Positive | Positive | 4.53 ± 0.16 (4.41-4.64) | 0.04 | Negative | NA |
| Pat #14 | D | Negative | Fluctuating | 3.08 ± 2.95 (1.00-5.17) | 0.96 | Positive | Negative |
| Pat #15 | D | Negative | Negative | NA | NA | Positive | Negative |
| Pat #16 | E | Positive | Positive | 6.26 ± 0.74 (5.74-6.78) | 0.12 | Negative | NA |
HBV genotyping by commercial LiPA assay was available for only 7 patients: 4 (57%) HBV-D, 1 (14%) HBV-A and 2 (28%) HBV-E. For the remaining 9 patients, with HBV-DNA titers lower than 1000 IU/mL, the LiPA assay yielded negative results. The application of a sensitive nested-PCR, allowed the genotyping of 8/9 of LiPA negative patients. The HBV genotype composition for this group with low HBV replication was: 2 (22%) HBV-D, 5 (56%) HBV-A and 1 (14%) HBV-E; quite different from that found in patients with higher levels of HBV replication. The global results (LiPA plus sequence analysis) presented the following genotype distribution: 6 (37%) HBV-A, 6 (37%) HBV-D, 3 (19%) HBV-E, and only one patient who remained HBV-DNA negative for all follow-up samples independently of the amplification technique used (Table 3).
Although the rate of patients having received previous antiviral treatment was higher in the group infected with the genotype HBV-D (2/6: 33%), the differences did not reach a statistical significance compared to those infected with HBV-A (1/6: 17%, P = 0.416) or HBV-E (1/3: 33%, P = 0.635) (Table 2).
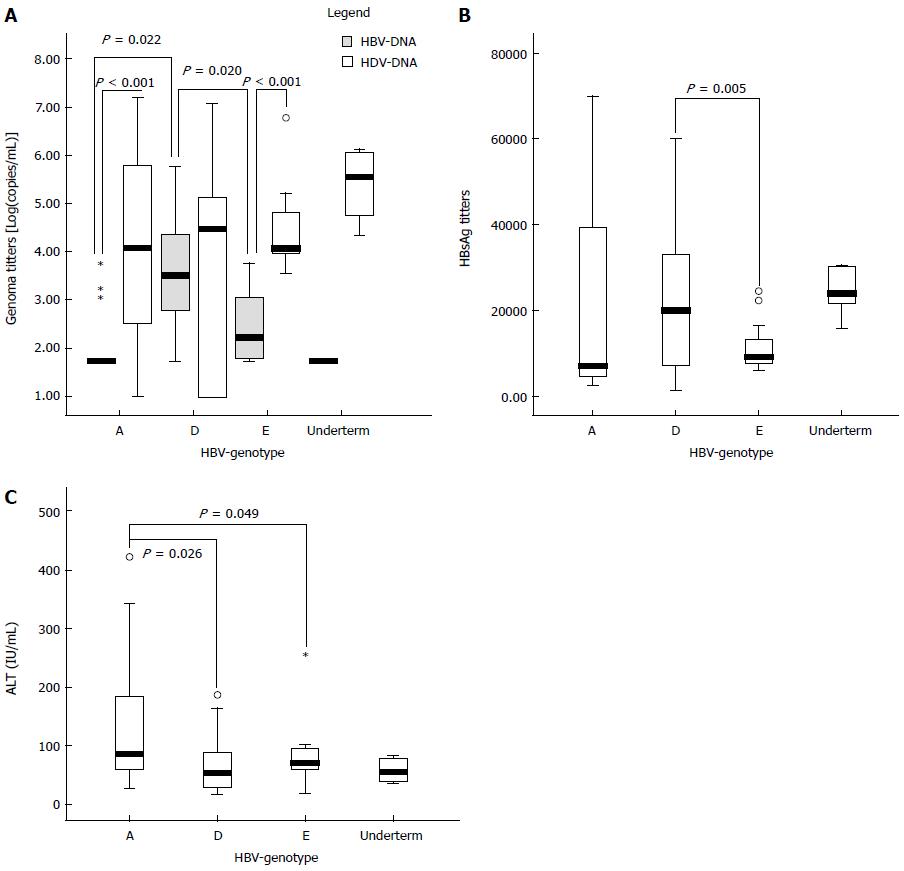
Quantitative analysis of serum HBV-DNA levels also showed a different replication level according to the HBV genotype (Figure 3A): HBV-DNA levels were significantly higher in patients with HBV-D [median (IQR): 3.49 (3.82-4.31) log10 (copies/mL)] than in those infected with HBV-A or E [median (IQR): 1.79 (1.61-1.83), P = 0.022; and 1.75 (1.62-1.80), P = 0.02; respectively]. In contrast, HDV-RNA titers were similar in the three groups of patients [median (IQR): 4.8 (2.54-5.78); 4.48 (1.00-5.10) and 4.06 (3.98-4.82) log10 (copies/mL), for genotypes A, D and E, respectively]. No differences in HBV-DNA and HDV-RNA levels were found in patients infected with genotype HBV-D. By contrast, in patients carrying HBV genotypes A and E, HDV-RNA levels were significantly higher.
Serum HBsAg was the most conserved HBV marker for all patients, with lower fluctuations in the follow-up period than that observed for HBV-DNA or HDV-RNA. Moreover, the variation coefficient of HBsAg titers was lower than the values observed for the viral genomes for 12/16 (75%) of patients (Tables 2 and 3). However, differences were found once more in the profile of this marker by HBV genotype. HBsAg tended to be higher in patients with HBV-D [median (IQR): 2.01 × 104 (7.32 × 103-3.32 × 104) IU/mL] than in those with HBV-A or E [7.13 × 103 (4.89 × 103-3.81 × 104); and 9.17 × 103 (7.85 × 103-1.32 × 104) IU/mL, respectively]. These differences reached statistical significance in HBV-E and HBV-D patients (See Figure 3B). Neither direct correlation between HBsAg and HDV-RNA levels in the study population (P = 0.252; r2 = 0.034) nor in the different groups of patients according to the HBV genotype were observed in the logistic regression analysis.
ALT levels also showed differences by HBV genotype. As shown in Figure 3C, the mean ALT value was significantly higher in patients with genotype A (mean ± SD: 1.44 × 102± 35.59 IU) than in those with genotype D (mean ± SD: 58.59 ± 5.16, P = 0.026) or E (mean ± SD: 69.25 ± 5.93, P = 0.049). In several patients, the longitudinal analysis of ALT fluctuations indicated a good correlation with the changes in HDV-RNA levels. However, no direct correlation among ALT levels and HBV or HDV titers was found in the regression analysis of the total samples.
The analysis of HBV and HDV replication markers in CHD patients showed that HDV was pre-eminent over HBV replication. Thus, the mean serum HDV-RNA titers were significantly higher than those for HBV-DNA. In addition, in the longitudinal analysis, HDV-RNA levels were persistently higher during follow-up in 15/16 (94%) patients. These results agree with previously described cross-sectional and longitudinal studies[8,9,11]. However, the percentage of patients with predominantly HBV replication was lower for our series of patients (6%) than those reported (up to 30%), probably due to the different ratio of HBeAg positive patients included in these studies[11].
A more detailed analysis of the virological data has shown that in CHD patients with CHD the extent of inhibition of HBV replication could be HBV genotype dependent: HBV-DNA titers were significantly higher in HBV-D patients than in HBV-A and E patients. Differences observed in the HBV-DNA titers were due to a significant decrease of HBV-DNA in those patients infected with genotype A, rather than to an increase in patients infected with HBV-D, as confirmed by the lack of significant differences in the HBV titers among patients infected with genotype D from the four groups of patients. This finding is in contrast with data obtained in the group CHB patients, and with previously reported data[14], in which no differences in HBV-DNA levels were found between patients infected with genotypes A or D.
No significant differences in HDV-RNA levels have been found. Comparative analysis of serum HBV/HDV titers showed that HDV-RNA remained significantly higher than HBV-DNA for patients infected with HBV-A and E, but no differences for the titers of both replication markers were found for those infected with HBV-D. The HBV and HDV replication data suggest that for these patients there is a specific inhibition of HBV replication, but not for HDV replication. The extent of this inhibitory effect seems to be genotype dependent, with a higher HDV inhibitory effect for HBV genotypes A and E than for HBV-D.
A practical consequence of the correlation between HBV genotype and viral replication level in the study population is the potential misrepresentation of some HBV genotypes. Only 44% of these patients had “high-enough” HBV-DNA levels (higher than 1000 IU/mL) for LiPA testing of at least one sample during follow-up. The genotypic composition of this group of patients showed a predominance of HBV genotype D over genotypes A or E. These results agree with other reports finding differences in the transmission route associated with HBV genotype for HBV/HIV coinfected patients[19,20]. However, HBV genotyping with a highly-sensitive “in-house” nested PCR method showed HBV-A to be the predominant genotype in the group of patients with low HBV replication levels, showing that no difference existed in the prevalence of HBV-A or D (37%). These results suggest that the prevalence of HBV-A may be underestimated CHD patients due to its association with the lowest HBV replication.
In chronic HBV patients, serum HBsAg levels are considered to be a surrogate marker of HBV replication[21]. The HBsAg titers were the most conserved HBV marker compared with HBV or HDV genome levels. The dynamic analysis, therefore, showed that HBsAg titers showed no significant fluctuations, even in the presence of variability in the quantitative analysis of HBV and HDV viremia. These data suggest that HBsAg dynamics may not be a good indicator of HBV fluctuations in the natural history of untreated patients with CHD. This lack of correlation can be explained by the high prevalence of HBeAg negative patients in the study population. In this group of patients, HBsAg correlates poorly with serum HBV-DNA and cccDNA titers when compared with HBe positive patients[22]. On the other hand, in patients with CHD, HBsAg kinetics is influenced by the simultaneous replication of both HBV and HDV, since both agents carried HBsAg in their envelopes[23]. In this context, the dynamic evolution of HBsAg is a complex result in the number of circulating HBV and HDV particles and the circulating HBV uninfectious subviral particles.
Noteworthy in this study the correlation of HBsAg levels and HBV-DNA in patients with HBV-A or D was absent. Recent data suggest that HBV replication is inhibited by HDV infection at the intrahepatic level, as shown by the decrease of cccDNA and pgRNA[24]. In contrast, the transcription activity of HBsAg remained conserved, showing no difference between patients with or without HDV superinfection. According to these data, the transcription level of HBsAg is conserved for both HBV-A or D irrespective of their differences in replication levels[24].
Differences in HBV-DNA and HBsAg titers among patients infected with different HBV genotypes do not seem to be related with simultaneous infection with HCV, or with the previous treatment pressure. Thus, no difference in the incidence of HCV infection or previous treatment history was found among the different groups of patients.
The evolution of ALT levels showed an indirect relation with serum HBV-DNA titers. HBV-A patients, with the lowest replication level, showed significantly higher ALT values than those with HBV-D or E. In patients infected with HBV-A, HDV-RNA tended to be higher than in other patients, probably indicating that HDV replication plays a greater role than HBV in the pathogenic process. However, recent studies on the histopathological status of CHD patients found a lack of correlation for the extent of pathological damage and viral replication levels, suggesting that liver damage for these patients may no longer be attributed to a direct cytopathic effect of either HDV or HBV[25].
Differences in HBV-DNA and ALT levels between patients infected with HBV-A or D can have therapeutical consequences. Therefore, those patients infected with HBV-A show, in these series of patients, simultaneous low HBV-DNA levels and high ALT titers. This is a predictive pattern of optimal response to treatment with interferon based regimens. Based on these results, the analysis of HBV genotype could be taken into account in the algorithms for treatment indication for delta infected patients.
The main limitations of our work were: (1) the relative small sample number due to the fact that this study was unicentric; and (2) we did not analyze viral replication markers in liver tissue as most patients did not have liver biopsy (for diagnostic proposal these patients underwent Fibroscan). Thus, we have no data on delta antigen and/or HDV-RNA levels in liver tissue.
In conclusion, HBV-DNA titer is a predictive pattern of optimal response to treatment with interferon based regimens. Our results suggest that in patients with CHD the inhibition of HBV replication is genotype-dependent. Viral replication interference between HBV and HDV is more evident in patients infected with HBV-genotype A, than with HBV-D or E. Based on these results, the analysis of HBV genotype, could be taken into account in the algorithms for treatment indication for delta infected patients. Therefore, in CHD patients from geographical regions with a different distribution of HBV genotypes, the replication behaviour of the other HBV genotypes in large studies including a high number of patients is warranted to clarify the HBV/HDV interference process and its influence in the disease outcome. In addition, the development of effective HBV/HDV in vitro replication systems could help to elucidate the molecular mechanism involved in the replicative interference process in future works. Finally, the analysis of other viral genetic variants, such as HDV genotype and HBV pre-core/core variants should be further studied to better understand the HBV/HDV interference.
We thank Brenda Ashley Morris for English revision.
Chronic hepatitis delta virus (HDV) infection continues to be an important public health problem, due to both the lack of an effective therapy and the frequent severity of chronic hepatitis delta. Hepatitis B virus (HBV) replication has been described as modulating HDV pathogenesis and the active replication of both viruses is associated with more severe liver disease.
The most important topics in the treatment of chronic hepatitis delta (CHD) are: (1) the identification of factors related with the evolution of liver fibrosis, like HDV/HBV interactions; and (2) the identification of prognostic factors of liver disease evolution.
Results obtained in this work suggest that in CHD patients the inhibition of HBV replication is genotype-dependent. Viral replication interference between HBV and HDV is more evident in patients infected with HBV-genotype A, than with HBV-D or E.
Since HBV-DNA titer is a predictive pattern of optimal response to treatment with interferon based regimens, results obtained in this work suggest that the analysis of HBV genotype, could be taken into account in the algorithms for treatment indication for delta infected patients.
Authors of this article described that HBV-A genotype infection interfere viral replication than genotype D or E in HDV co-infected patients. The study is really interesting and well conducted.
P- Reviewer: De Paschale M, Kanda T, Kay A, Kim K, Romeo R S- Editor: Gong ZM L- Editor: A E- Editor: Liu XM
| 1. | Mele A, Mariano A, Tosti ME, Stroffolini T, Pizzuti R, Gallo G, Ragni P, Zotti C, Lopalco P, Curtale F. Acute hepatitis delta virus infection in Italy: incidence and risk factors after the introduction of the universal anti-hepatitis B vaccination campaign. Clin Infect Dis. 2007;44:e17-e24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 39] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 2. | Cross TJ, Rizzi P, Horner M, Jolly A, Hussain MJ, Smith HM, Vergani D, Harrison PM. The increasing prevalence of hepatitis delta virus (HDV) infection in South London. J Med Virol. 2008;80:277-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 3. | Wedemeyer H, Heidrich B, Manns MP. Hepatitis D virus infection--not a vanishing disease in Europe! Hepatology. 2007;45:1331-1332; author reply 1332-1333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 4. | Gaeta GB, Stroffolini T, Smedile A, Niro G, Mele A. Hepatitis delta in Europe: vanishing or refreshing? Hepatology. 2007;46:1312-1313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 5. | Niro GA, Smedile A, Ippolito AM, Ciancio A, Fontana R, Olivero A, Valvano MR, Abate ML, Gioffreda D, Caviglia GP. Outcome of chronic delta hepatitis in Italy: a long-term cohort study. J Hepatol. 2010;53:834-840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 122] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 6. | Smedile A, Rosina F, Saracco G, Chiaberge E, Lattore V, Fabiano A, Brunetto MR, Verme G, Rizzetto M, Bonino F. Hepatitis B virus replication modulates pathogenesis of hepatitis D virus in chronic hepatitis D. Hepatology. 1991;13:413-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 72] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 7. | Su CW, Huang YH, Huo TI, Shih HH, Sheen IJ, Chen SW, Lee PC, Lee SD, Wu JC. Genotypes and viremia of hepatitis B and D viruses are associated with outcomes of chronic hepatitis D patients. Gastroenterology. 2006;130:1625-1635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 161] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 8. | Kiesslich D, Crispim MA, Santos C, Ferreira Fde L, Fraiji NA, Komninakis SV, Diaz RS. Influence of hepatitis B virus (HBV) genotype on the clinical course of disease in patients coinfected with HBV and hepatitis delta virus. J Infect Dis. 2009;199:1608-1611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 43] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 9. | Jardi R, Rodriguez F, Buti M, Costa X, Cotrina M, Galimany R, Esteban R, Guardia J. Role of hepatitis B, C, and D viruses in dual and triple infection: influence of viral genotypes and hepatitis B precore and basal core promoter mutations on viral replicative interference. Hepatology. 2001;34:404-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 181] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 10. | Madejón A, Castillo I, Cotonat T, Carreño V. Significance of low HDV replication levels during the natural history of HDV infection. Long-term follow-up. J Hepatol. 1993;17 Suppl 3:S157-S160. [PubMed] |
| 11. | Schaper M, Rodriguez-Frias F, Jardi R, Tabernero D, Homs M, Ruiz G, Quer J, Esteban R, Buti M. Quantitative longitudinal evaluations of hepatitis delta virus RNA and hepatitis B virus DNA shows a dynamic, complex replicative profile in chronic hepatitis B and D. J Hepatol. 2010;52:658-664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 151] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 12. | Chu CJ, Hussain M, Lok AS. Hepatitis B virus genotype B is associated with earlier HBeAg seroconversion compared with hepatitis B virus genotype C. Gastroenterology. 2002;122:1756-1762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 354] [Cited by in RCA: 359] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 13. | Kao JH, Chen PJ, Lai MY, Chen DS. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology. 2000;118:554-559. [PubMed] |
| 14. | Wai CT, Chu CJ, Hussain M, Lok AS. HBV genotype B is associated with better response to interferon therapy in HBeAg(+) chronic hepatitis than genotype C. Hepatology. 2002;36:1425-1430. [PubMed] |
| 15. | Mahtab MA, Rahman S, Khan M, Karim F. Hepatitis B virus genotypes: an overview. Hepatobiliary Pancreat Dis Int. 2008;7:457-464. [PubMed] |
| 16. | Sánchez-Tapias JM, Costa J, Mas A, Bruguera M, Rodés J. Influence of hepatitis B virus genotype on the long-term outcome of chronic hepatitis B in western patients. Gastroenterology. 2002;123:1848-1856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 312] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 17. | Flink HJ, van Zonneveld M, Hansen BE, de Man RA, Schalm SW, Janssen HL. Treatment with Peg-interferon alpha-2b for HBeAg-positive chronic hepatitis B: HBsAg loss is associated with HBV genotype. Am J Gastroenterol. 2006;101:297-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 202] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 18. | Sheldon J, Ramos B, Toro C, Ríos P, Martínez-Alarcón J, Bottecchia M, Romero M, Garcia-Samaniego J, Soriano V. Does treatment of hepatitis B virus (HBV) infection reduce hepatitis delta virus (HDV) replication in HIV-HBV-HDV-coinfected patients? Antivir Ther. 2008;13:97-102. [PubMed] |
| 19. | Chu CJ, Keeffe EB, Han SH, Perrillo RP, Min AD, Soldevila-Pico C, Carey W, Brown RS, Luketic VA, Terrault N. Hepatitis B virus genotypes in the United States: results of a nationwide study. Gastroenterology. 2003;125:444-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 229] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 20. | Pérez-Olmeda M, Núñez M, García-Samaniego J, Ríos P, González-Lahoz J, Soriano V. Distribution of hepatitis B virus genotypes in HIV-infected patients with chronic hepatitis B: therapeutic implications. AIDS Res Hum Retroviruses. 2003;19:657-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 21. | Chan HL, Wong VW, Tse AM, Tse CH, Chim AM, Chan HY, Wong GL, Sung JJ. Serum hepatitis B surface antigen quantitation can reflect hepatitis B virus in the liver and predict treatment response. Clin Gastroenterol Hepatol. 2007;5:1462-1468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 286] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 22. | Thompson AJ, Nguyen T, Iser D, Ayres A, Jackson K, Littlejohn M, Slavin J, Bowden S, Gane EJ, Abbott W. Serum hepatitis B surface antigen and hepatitis B e antigen titers: disease phase influences correlation with viral load and intrahepatic hepatitis B virus markers. Hepatology. 2010;51:1933-1944. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 339] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 23. | Taylor JM. Hepatitis delta virus. Virology. 2006;344:71-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 141] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 24. | Pollicino T, Raffa G, Santantonio T, Gaeta GB, Iannello G, Alibrandi A, Squadrito G, Cacciola I, Calvi C, Colucci G. Replicative and transcriptional activities of hepatitis B virus in patients coinfected with hepatitis B and hepatitis delta viruses. J Virol. 2011;85:432-439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 87] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 25. | Grabowski J, Wedemeyer H. Hepatitis delta: immunopathogenesis and clinical challenges. Dig Dis. 2010;28:133-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 45] [Article Influence: 3.0] [Reference Citation Analysis (0)] |