Published online Jun 21, 2015. doi: 10.3748/wjg.v21.i23.7208
Peer-review started: November 19, 2014
First decision: December 11, 2014
Revised: December 26, 2014
Accepted: March 27, 2015
Article in press: March 27, 2015
Published online: June 21, 2015
Processing time: 212 Days and 18.5 Hours
AIM: To study the expression of long noncoding RNAs (lncRNAs) in hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC).
METHODS: The lncRNA profiles between HBV-related HCC tissues and corresponding normal liver tissues were generated using microarray analysis. Datasets were analyzed using multiple algorithms to depict alterations in gene expression on the basis of gene ontology (GO), pathway analysis, and lncRNA levels.
RESULTS: The microarray revealed that 1772 lncRNAs and 2508 mRNAs were differently expressed. The pathway analysis demonstrated that the cell cycle, cytokine-cytokine receptor interaction, chemokine signaling pathway, and phosphoinositide 3-kinase-protein kinase B signaling pathway may play important roles in HCC. Several GO terms, such as cell cycle, DNA replication, immune response, and signal transduction, were enriched in gene lists, suggesting a potential correlation with HBV-related HCC. The upregulated large intergenic noncoding RNA ULK4P2 was physically combined with enhancer of zeste homolog 2. Therefore, the lncRNAs may participate in regulating HBV-related HCC.
CONCLUSION: lncRNAs play important roles in HCC, future studies should verify whether large intergenic noncoding ULK4P2 functions by combining with enhancer of zeste homolog 2 in HCC.
Core tip: This manuscript examines the differential expression of long noncoding RNAs in hepatitis B virus-related hepatocellular carcinoma using gene ontology and pathway analyses, and constructing a long noncoding RNA-mRNA network to research the data. Furthermore, RNA immunoprecipitation revealed that the large intergenic noncoding RNA ULK4P2 physically combined with enhancer of zeste homolog 2 (EZH2). EZH2 plays an important role in many cancer types and is critical for cancer cell proliferation, survival, and metastasis, and drug resistance. The combination of ULK4P2 and EZH2 provides a novel avenue for further study of ULK4P2 in hepatitis B virus-related hepatocellular carcinoma.
- Citation: Yu TT, Xu XM, Hu Y, Deng JJ, Ge W, Han NN, Zhang MX. Long noncoding RNAs in hepatitis B virus-related hepatocellular carcinoma. World J Gastroenterol 2015; 21(23): 7208-7217
- URL: https://www.wjgnet.com/1007-9327/full/v21/i23/7208.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i23.7208
Hepatocellular carcinoma (HCC) is a common human cancer in many countries, especially China[1]. The mortality rate of HCC is third among cancer-related deaths[2], and hepatitis B virus (HBV) infection is associated with almost half of all HCC cases. A striking similarity exists between the geographic distribution of the rates of chronic HBV infection and HCC[3]. However, the mechanism of HBV in HCC has not yet been clarified, and the five-year survival rates of HCC patients remain poor. Therefore, understanding the underlying mechanism of HBV in HCC is crucial to provide accurate information for the early screening, clinical diagnosis, targeted molecular therapy, and prognosis of patients.
Although a significant portion of the human genome is transcribed, protein-coding genes account for only 2% of the genomic sequence[4]. In recent years, the nonprotein-coding portion of the genome has become important in the basic biology and major pathologies of cancer[5]. Noncoding RNAs include microRNAs and long noncoding (lnc)RNAs; the microRNAs has been well studied and are related to cell differentiation and cancers in recent publications[6]. With the development of lncRNA microarrays, high-throughput sequencing, and bioinformatics, an increasing number of lncRNAs have been discovered and have attracted considerable attention in medical molecular biology. lncRNAs are a class of noncoding RNA transcripts longer than 200 nucleotides, with no or little protein-coding capacity. Recent studies have shown that more genomic sequences have been transcribed into lncRNAs than protein-coding RNAs[7]. Although lncRNAs are among the least well understood of the noncoding RNAs, they cannot be completely dismissed as mere transcriptional “noise’’[8].
lncRNAs regulate gene expression at the epigenetic, transcriptional, and post-transcriptional levels, and are involved in many biologic functions, including genomic imprinting, chromosome dosage-compensation, X-chromosome silencing, chromosome modification, intranuclear transport, transcriptional activation, and interference[9]. lncRNAs are abnormally expressed in cancer cells[10,11], and several lncRNAs play important roles in carcinogenesis. However, only a small fraction of lncRNAs is currently well understood.
The molecular mechanisms underlying HBV-related HCC are not clear, and the contributions of lncRNAs in HCC have only been gradually elucidated, including of HEIH, which is an lncRNA that is highly and specifically expressed in HCC tissues[1]. The downregulated lncRNA GAS5 is associated with HCC prognosis[12]. The lncRNA ATB, a highly expressed gene in HCC, participates in the epithelial-mesenchymal transition induced in HCC metastases and is associated with poor prognosis. However, the important roles of lncRNAs in HCC have yet to be elucidated. In the present study, lncRNA microarrays were used to detect differentially expressed lncRNAs between HBV-related HCC tissues and nontumor tissues. In addition, gene ontology (GO) analysis, pathway analysis, and an lncRNA-mRNA network were used to predict the functions of these abnormally expressed lncRNAs in HCC.
Tissue samples from five HBV-related HCC patients were used for the microarray analysis, and tissue samples from 14 HCC patients who had hepatectomy in the Renmin Hospital of Wuhan University between May 2013 and May 2014 were used for data validation. After removal from the body, the tissues were snap frozen in liquid nitrogen within 30 min and then stored at -80 °C until use. All patients provided written informed consent. The study was approved by the Human Research Ethics Committee of Renmin Hospital of Wuhan University. Table S1 lists the characteristics of the patients.
To extract RNA, frozen tissues were ground into powder with TRIzol reagent (Invitrogen of Thermo Fisher Scientific, Waltham, MA, United States). RNA purification was performed with an RNA-containing aqueous phase using the RNeasy minikit (Qiagen, Venlo, Limburg, Netherlands). Quantification and quality evaluation were performed using a Nanodrop and Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States), respectively.
Arraystar Human LncRNA Microarray V3.0 (Arraystar, Rockville, MD), an updated version of Microarray V2.0, was designed for the global profiling of human lncRNAs and protein-coding transcripts. Approximately 30586 lncRNAs and 26109 coding transcripts can be detected by the third-generation lncRNA microarray. lncRNAs were carefully constructed using well-respected public transcriptome databases (Refseq, UCSC Known Genes, and Genecode) and landmark publications. Each transcript was represented by a specific exon or splice junction probe that can accurately identify individual transcripts. Positive probes for housekeeping genes and negative probes were also printed onto the array for hybridization quality control. Data were extracted and normalized using GeneSpring GX v11.5.1 software (Agilent Technologies). Differentially expressed lncRNAs with statistical significance were identified through Volcano Plot filtering and hierarchical clustering. Differentially expressed genes were identified through the random variance model, and P values were calculated using the paired t-test. The significance thresholds set for the up- and downregulated genes were fold change ≥ 2.0 and P≤ 0.05.
Total RNA was reverse transcribed using a Fermentas RT reagent kit (Perfect Real Time; Thermo Fisher Scientific) according to the manufacturer’s instructions. Four distinctively upregulated lncRNAs were randomly selected to validate their expression levels through quantitative real-time reverse-transcription (qRT)-PCR using SYBR Green assays (TaKaRa, Otsu, Shiga, Japan). GAPDH was used as an internal control. Table S2 shows the primer sequences.
An lncRNA-mRNA network was developed to identify the interactions between mRNA and lncRNA[13]. The network was built according to the normalized signal intensities of specific expression levels of genes and lncRNAs (for each gene-lncRNA, gene-gene, or lncRNA-lncRNA pair). Pearson’s correlation was calculated and significantly correlated pairs were used to construct the network.
The main functions of the differentially expressed genes were analyzed using GO analysis, a key functional classification of NCBI, GO can organize genes into hierarchical categories and uncover the gene regulatory network on the basis of biologic processes and molecular functions[14].
Pathway analysis was used to determine the main pathway of the differentially expressed genes according to KEGG, Biocarta, and Reatome. Fisher’s exact test and χ2 tests were used to select the main pathway, and the significance threshold was defined with P value and FDR[15].
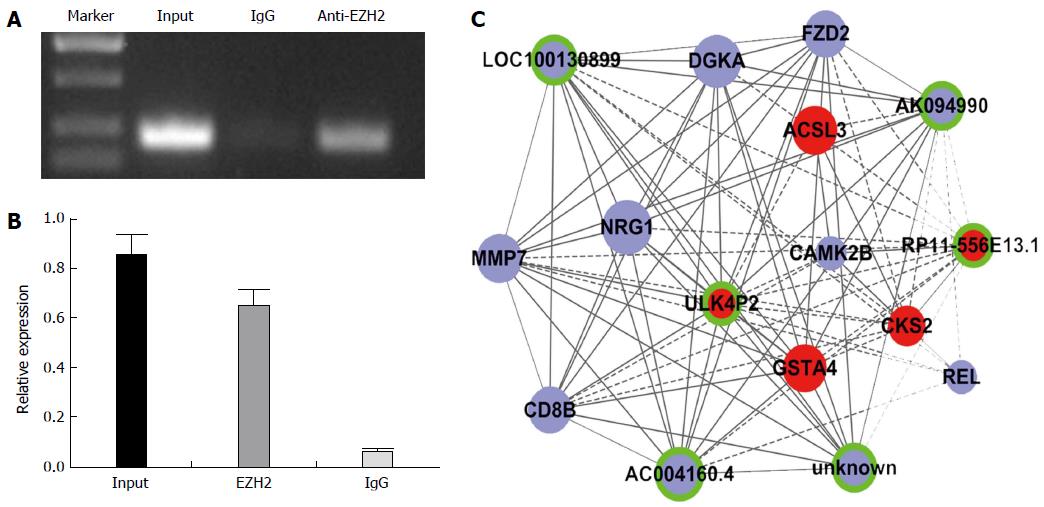
RNA immunoprecipitation experiments were performed using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore Corp, Billerica, MA, United States) according to the manufacturer’s instructions using enhancer of zeste homolog 2 (EZH2) antibodies (Cell Signaling Technology Inc., Danvers, MA, United States). The coprecipitated RNAs were detected via qRT-PCR. RIP assays were performed in biologic triplicates.
All statistical data were analyzed using SPSS 17.0 software (SPSS Inc., Chicago, IL, United States). Differences in lncRNA expression between the tumor and corresponding non-tumor tissues were analyzed using Student’s t-tests. Statistical significance was considered at P < 0.05.
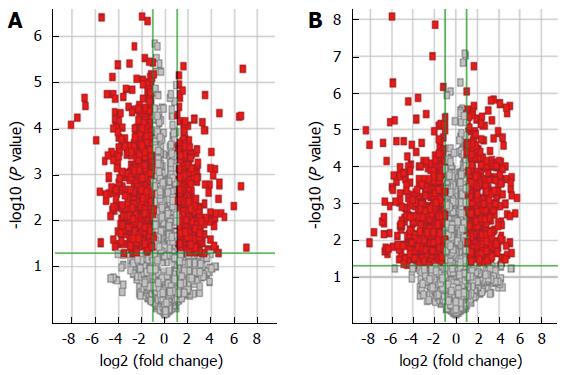
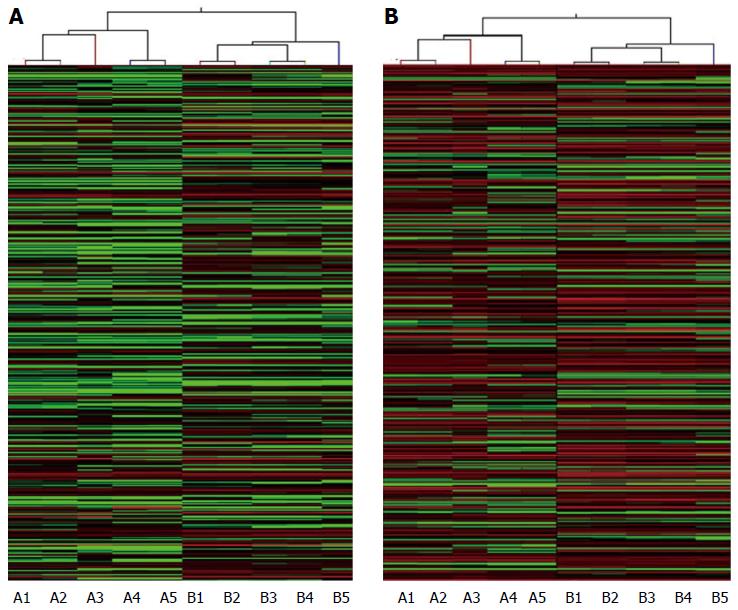
Five HBV-related patients (HBV surface antigen-positive) were selected to visualize the differentially expressed lncRNAs and mRNAs. Volcano plot analysis was used to directly detect lncRNAs and mRNAs abnormally expressed between the HCC tissues and corresponding non-tumor tissues (Figure 1). Hierarchical clustering is a simple and commonly used clustering technique to analyze gene expression data. Figure 2 shows the differentially expressed lncRNAs and mRNAs. Data analysis revealed 1772 differently expressed lncRNAs (Table S3) and 2508 differentially expressed mRNAs (Table S4) in HCC tissues compared with corresponding normal tissues. Of the 1772 differentially expressed lncRNAs, 637 were upregulated and 1135 were downregulated. Of the 2508 differentially expressed mRNAs, 1194 were upregulated and 1314 mRNAs were downregulated. The distinctively expressed lncRNAs in HCC are listed in Table 1, several of which, particularly MEG3 and GAS5, reportedly play roles in cancer development.
| lncRNA | Fold change | Regulation | RNA length | Chromosome |
| ENST00000577848 | 12.43885 | Up | 1455 | 18 |
| ENST00000426413 | 101.25230 | Up | 1577 | 7 |
| NR_024478 | 14.71172 | Up | 2282 | 6 |
| uc010kwq.2 | 12.91793 | Down | 4744 | 7 |
| TCONS_00005259 | 10.69086 | Down | 3742 | 2 |
| uc002nbr.3 | 29.97769 | Down | 1413 | 19 |
| TCONS_00019684 | 19.73914 | Down | 1147 | 11 |
| NR_027133 | 12.69278 | Down | 2597 | 15 |
| NR_033957 | 17.82503 | Down | 2877 | 10 |
| MEG3 | 10.96779 | Down | 1351 | 14 |
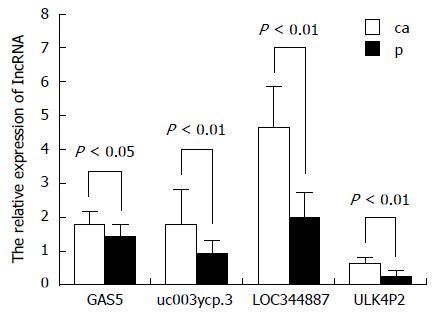
To validate the microarray data, four upregulated lncRNAs were randomly selected and analyzed for their expression levels in tissue samples from 14 HBV-related HCC patients. The corresponding non-tumor tissues were analyzed via qRT-PCR (Figure 3), in which the obtained results are consistent with the microarray data.
According to the relationship between lncRNAs and their associated protein-coding genes, lncRNAs can be classified into natural antisense, intronic antisense, bidirectional, exon-sense overlapping, intron-sense overlapping, and intergenic. In order to systematically predict the function of lncRNAs, lncRNA subgroup analyses were performed, including antisense lncRNA, large intergenic noncoding (linc)RNA, Hox Loci lncRNA, T-UCR, and enhancer-like lncRNAs analyses based on those classifications. The profiles concerning enhancer-like lncRNAs (Table S5), HOX Loci lncRNAs (Table S6), and lincRNAs were examined (Table S7).
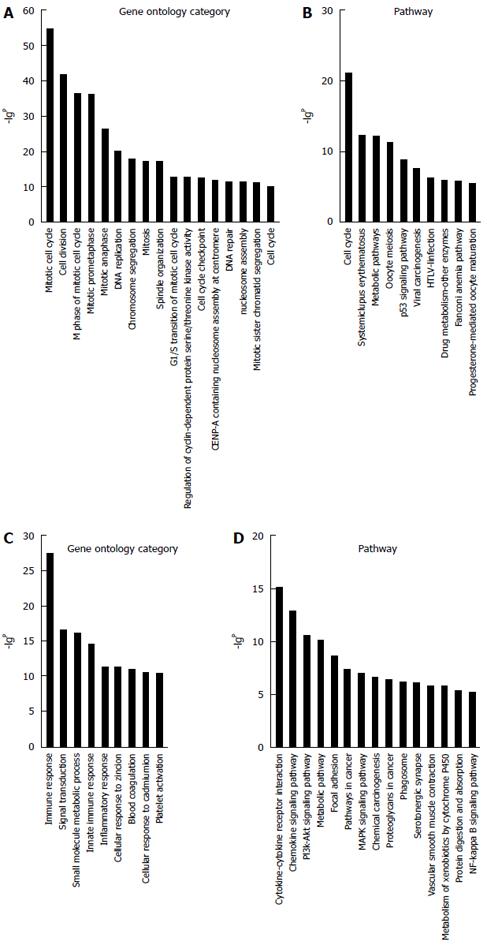
Due to the lack of a comprehensive annotation database for lncRNAs, a profile of mRNAs in HCC was constructed to indirectly predict the function of the lncRNAs. A total of 1270 filtered mRNAs (threefold change) and 392 lncRNAs (fivefold change) were included in GO and Signaling pathway analyses (Figure 4). The GO analysis showed the distinctive functions of up- and downregulated involve the cell cycle, cell division, immune response, and DNA replication. The Pathway analysis showed that aberrantly genes are involved in the cell cycle, mitogen-activated protein kinase pathway, phosphoinositide 3-kinase-protein kinase B signaling pathway, and cytokine-cytokine receptor interaction.
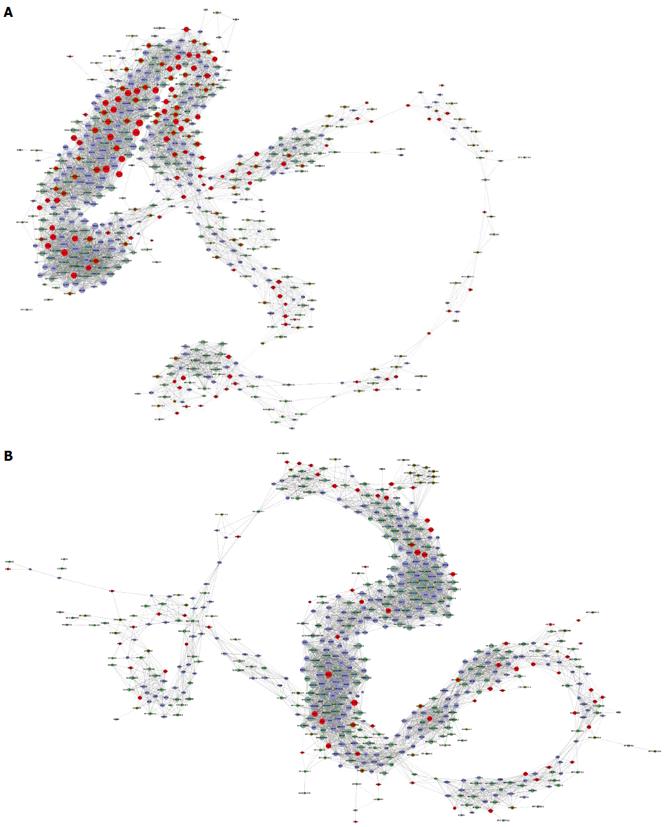
Coexpression network analysis was performed between the filtered 392 lncRNAs and the filtered 348 mRNAs with significant enrichment in GO and Signaling pathway analyses. The network structure of the HCC tissues and corresponding non-tumor tissues samples was markedly different (Figure 5).
EZH2 is a component of the polycomb repressive complex 2 (PRC2). EZH2 has been reported in numerous cancers, including HCC[16]. It is reported that 20% lincRNAs can combine with EZH2, including HOTAIR, HEIH, and H19[17,18] which may regulate gene expression in cancers. Thus, we hypothesized that EZH2 may also have some relationship with lincRNAs that are differentially expressed in HBV-related HCC. Significantly expressed lincRNAs were selected to validate their combination with EZH2 using a RIP assay. The data showed that ULK4P2 is physically combined with EZH2 (Figure 5). Next, a ULK4P3-mRNA subnetwork was constructed to characterize the role of ULK4P2 (Figure 6). In the coexpression network, ULK4P2 is connected to six lncRNAs and nine mRNAs that are enriched for gene products involved in tumor cell proliferation and metastasis.
Recently, studies have shown that more genomic sequences are transcribed into lncRNAs than protein-coding RNAs, and lncRNAs are being characterized at a rapid pace. With an increase in the number of well-characterized cancer-associated lncRNAs, the study of lncRNAs in cancer is now generating new hypotheses about the biology of cancer cells. For HCC, deregulated expression of both protein-coding genes and microRNAs has been suggested to have considerable potential for predicting the prognosis of HCC patients[19]. However, the research concerning lncRNAs in HBV-related HCC is still in a preliminary stage.
In this study, the aberrant lncRNAs in five HBV-related HCC and corresponding non-tumor tissues were screened using lncRNA microarray V3.0, which is designed for the global profiling of human lncRNAs and protein-coding transcripts, and it is updated from the previous Microarray V2.0. The lncRNAs are carefully constructed using the most highly respected public transcriptome databases (Refseq, UCSC knowngenes, Genecode, etc.), as well as landmark publications. Each transcript is represented by a specific exon or splice junction probe that can accurately identify an individual transcript. Positive probes for housekeeping genes and negative probes are also printed onto the array for hybridization quality control. Data was extracted and normalized using the GeneSpring GX v11.5.1 software package (Agilent Technologies) to guarantee the accuracy, which showed that there are 637 upregulated lncRNAs and 1135 downregulated. Four distinctively expressed lncRNAs were randomly selected from the microarray data to validate their reliability in 14 HBV-related HCC samples; the result is consistent with microarray data. Meanwhile, the differentially expressed mRNAs were also examined.
Recently, some classes of lncRNAs, such as lincRNAs and HOX lncRNAs, have been identified with specific functions in human cells[20]. Thus the data are displayed as three clusters of lncRNAs.
lncRNAs with enhancer-like function were identified using GENCODE annotation of the human genes. Profiling data of all probes for lncRNAs with enhancer-like function revealed that many enhancer-like RNAs were bidirectional, lacked a polyA tail, and had very low copy numbers[21]. Depletion of this type of lncRNA leads to decreased expression of their neighboring protein-coding genes, as enhancer-like lncRNAs can combine with enhancer-binding proteins to regulate gene activation. Furthermore, detailed functional analysis of noncoding RNA adjacent to the Snai1 locus, the master regulator of hematopoiesis, using reporter assays demonstrated a role for this noncoding RNA in an RNA-dependent potentiation of gene expression[22]. These studies suggest a role of enhancer-like lncRNAs in positive regulation of protein-coding genes.
lincRNAs are another class of newly discovered lncRNAs with dysregulated expression in many tumors. lincRNAs have been involved in diverse biologic processes, including cell-cycle regulation, immune surveillance, and embryonic stem cells[23], but their mechanism is elusive. The results of this study show aberrant expression of 374 lincRNAs, which are nearby (distance < 300 bp) coding gene pairs. Recently, it was reported that 20% of lincRNAs are bound by PRC2, and that additional lincRNAs are bound by other chromatin-modifying complexes to regulate gene expression[24]. The mechanism concerning lincRNAs is intriguing, with recently reported novel functions that indicate they work in a more conventional way[25].
Hox genes, a highly conserved subgroup of the homeobox superfamily, have crucial roles in development, regulating numerous processes including apoptosis, differentiation, motility, and angiogenesis. The abnormal expression of HOX genes plays an important role in tumorigenesis, oncogenesis, or tumor suppression[26]. In mammals, 39 HOX transcription factors are clustered on four chromosomal loci, termed HOXA through HOXD[27]. Transcription of many lncRNAs has been observed in human HOX loci, described herein as HOX lncRNAs, which mainly participate in epigenetic regulation. The HOX cluster profiling reported in the present study includes 95 probes designed for the four HOX loci to target lncRNAs and coding transcripts. The results show that HOTAIR, the best-studied HOX lncRNA, is upregulated in cancer tissues compared to corresponding non-cancer tissues, which is consistent with a previous report[28].
To confirm the differential gene expression between HCC and corresponding non-tumor tissues, 348 mRNAs involved in function functions and signal pathways and deregulated lncRNAs were used to construct the lncRNA-mRNA coexpression network. The results show that the network structure of the HCC tissues and corresponding non-tumor tissues samples was markedly different, which indicates that the coexpression patterns of mRNA and lncRNAs between HCC and corresponding non-tumor tissues are different. GO analysis of the filtered differentially expressed mRNAs showed that they are enriched in cell cycle, DNA replication, immune response, and signal transduction pathways. The Signaling pathway analysis showed that cell cycle, cytokine-cytokine receptor interaction and chemokine and phosphoinositide 3-kinase-protein kinase B signaling pathways were correlated with HBV-related HCC.
EZH2 is the core enzymatic subunit of PRC2, and evidence shows that EZH2 is overexpressed in many cancer types and is critical for cancer cell proliferation, metastasis, and drug resistance[29]. To detect a possible combination between the candidate lincRNAs and EZH2, RIP was performed using an EZH2 antibody and nuclear extracts of hepG2 cells. The results demonstrate that the lincRNA ULK4P2 is physically combined with the EZH2 antibody, indicating that it may have function by combining with EZH2 in HCC. ULK4P2 is an lincRNA with little-known function located on chromosome 15. Because the coexpression network may correspond to biologic pathways, and many functions of protein-coding RNAs could be found in NCBI RefSeq[30], an ULK4P2-mRNA subnetwork was constructed, which indicated its relationship with some cell proliferation and metastasis genes. These data suggest that ULK4P2 may have an important role in HBV-related HCC initiation, growth, or metastasis.
Recent advances in microarray technologies and the rapid development have transformed the study of tumors into the genome era. As a result, the unstudied part of noncoding RNAs have been implicated as oncogenes and suppressor genes in tumors. The result in this paper gives us more information to further study lncRNAs in HBV-related HCC.
Hepatocellular carcinoma (HCC) is a common and deadly cancer, but the mechanism regarding hepatitis B virus (HBV)-related HCC is still not entirely understood. Long noncoding RNAs (lncRNAs) are a group of noncoding RNAs that are longer than 200 nucleotides. Recent study of lncRNAs demonstrates their involvement in human disease, including cancers.
lncRNA is a hotspot in the field of RNA, and shows involvement in many human diseases.
The result of the lncRNA microarray provide us with many more lncRNAs study in greater depth; the result may be useful for HBV-related patients. For example, ULK4P2 may participate in HCC by combining with EZH2.
Enhancer of zeste homolog 2 (EZH2) is the core enzymatic subunit of polycomb repressive complex 2, and evidence shows that EZH2 is overexpressed in many cancer types and is critical for cancer cell proliferation, metastasis, and drug resistance. ULK4P2 is a large intergenic noncoding RNA.
The manuscript is well presented and of interest. The design of study is appropriate, the study was done well, and its results can contribute to knowledge of this topic. All parts of the manuscript are well organized and valuable conclusions are provided. References are also appropriate, relevant and updated.
P- Reviewer: Aghakhani A, Cao GW S- Editor: Ma YJ L- Editor: AmEditor E- Editor: Wang CH
| 1. | Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679-1689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 482] [Cited by in RCA: 555] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 2. | El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557-2576. [PubMed] |
| 3. | Chang MH. Cancer prevention by vaccination against hepatitis B. Recent Results Cancer Res. 2009;181:85-94. [PubMed] |
| 4. | Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. A new frontier for molecular medicine: noncoding RNAs. Biochim Biophys Acta. 2005;1756:65-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 50] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 5. | Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3924] [Cited by in RCA: 4443] [Article Influence: 277.7] [Reference Citation Analysis (0)] |
| 6. | Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-838. [PubMed] |
| 7. | Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391-407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1266] [Cited by in RCA: 1470] [Article Influence: 113.1] [Reference Citation Analysis (0)] |
| 8. | Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629-641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4004] [Cited by in RCA: 3940] [Article Influence: 246.3] [Reference Citation Analysis (0)] |
| 9. | Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494-1504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1668] [Cited by in RCA: 1870] [Article Influence: 116.9] [Reference Citation Analysis (0)] |
| 10. | Benetatos L, Vartholomatos G, Hatzimichael E. MEG3 imprinted gene contribution in tumorigenesis. Int J Cancer. 2011;129:773-779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 225] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 11. | Piao HL, Ma L. Non-coding RNAs as regulators of mammary development and breast cancer. J Mammary Gland Biol Neoplasia. 2012;17:33-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 65] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 12. | Tu ZQ, Li RJ, Mei JZ, Li XH. Down-regulation of long non-coding RNA GAS5 is associated with the prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol. 2014;7:4303-4309. [PubMed] |
| 13. | Pujana MA, Han JD, Starita LM, Stevens KN, Tewari M, Ahn JS, Rennert G, Moreno V, Kirchhoff T, Gold B. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat Genet. 2007;39:1338-1349. [PubMed] |
| 14. | Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25-29. [PubMed] |
| 15. | Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277-D280. [PubMed] |
| 16. | Tan JZ, Yan Y, Wang XX, Jiang Y, Xu HE. EZH2: biology, disease, and structure-based drug discovery. Acta Pharmacol Sin. 2014;35:161-174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 167] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 17. | Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333:213-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 400] [Cited by in RCA: 461] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 18. | Li X, Wu Z, Mei Q, Li X, Guo M, Fu X, Han W. Long non-coding RNA HOTAIR, a driver of malignancy, predicts negative prognosis and exhibits oncogenic activity in oesophageal squamous cell carcinoma. Br J Cancer. 2013;109:2266-2278. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 148] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 19. | Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437-1447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 624] [Cited by in RCA: 662] [Article Influence: 41.4] [Reference Citation Analysis (0)] |
| 20. | Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667-11672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2182] [Cited by in RCA: 2352] [Article Influence: 147.0] [Reference Citation Analysis (0)] |
| 21. | Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3236] [Cited by in RCA: 3193] [Article Influence: 245.6] [Reference Citation Analysis (0)] |
| 22. | Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46-58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1483] [Cited by in RCA: 1452] [Article Influence: 96.8] [Reference Citation Analysis (0)] |
| 23. | Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223-227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3467] [Cited by in RCA: 3303] [Article Influence: 206.4] [Reference Citation Analysis (0)] |
| 24. | Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343-349. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2760] [Cited by in RCA: 2526] [Article Influence: 180.4] [Reference Citation Analysis (0)] |
| 25. | Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295-300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1477] [Cited by in RCA: 1574] [Article Influence: 112.4] [Reference Citation Analysis (0)] |
| 26. | Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10:361-371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 570] [Cited by in RCA: 634] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 27. | Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311-1323. [PubMed] |
| 28. | Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, Zheng SS. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243-1250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 530] [Cited by in RCA: 609] [Article Influence: 43.5] [Reference Citation Analysis (0)] |
| 29. | Juan AH, Derfoul A, Feng X, Ryall JG, Dell’Orso S, Pasut A, Zare H, Simone JM, Rudnicki MA, Sartorelli V. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev. 2011;25:789-794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 180] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 30. | Prieto C, Risueño A, Fontanillo C, De las Rivas J. Human gene coexpression landscape: confident network derived from tissue transcriptomic profiles. PLoS One. 2008;3:e3911. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 183] [Cited by in RCA: 189] [Article Influence: 11.1] [Reference Citation Analysis (0)] |