Published online Dec 28, 2014. doi: 10.3748/wjg.v20.i48.18338
Revised: July 12, 2014
Accepted: August 13, 2014
Published online: December 28, 2014
Processing time: 211 Days and 21.2 Hours
AIM: To clarify the molecular mechanism involved in pathogenesis of colorectal cancer as well as clinical significance of genetic analysis of histological samples.
METHODS: A total of 480 blood and tissue specimens were collected in our hospital from January 2011 to October 2012. In the observation group, there were 120 blood specimens and 120 intestinal tract tissue specimens collected from patients with neoplastic intestinal polyps. In the control group I there were 80 blood specimens and 80 intestinal tract tissue specimens collected from patients with colorectal cancer. In the control group II there were 40 blood specimens and 40 intestinal tract tissue specimens collected from healthy individuals. The gene segments were amplified using PCR and DNA gel electrophoresis along with DNA sequence analysis were employed for the detection of the following single nucleotide polymorphisms (SNPs): K-RAS codons 12 and 13; hMLH1 (human mutS homolog 1) gene missense mutation at Va1384Asp; hMSH2 (human mutS homolog 2) gene missense mutation at 2783C/A.
RESULTS: The mutation rate of the SNP at Va1384Asp locus of the hMLH1 gene from blood and tissue specimens in the observation group showed no statistical difference from those in the control group I. The mutation rates of SNPs in codons 12 and 13 of K-RAS and at 2783C/A locus of the hMSH2 gene were significantly lower in the observation group than in the control group I (χ2 = 15.476, 29.670, 10.811, 16.618, 33.538, 7.898, P < 0.05). The mutation rate of SNP at Va1384Asp locus of the hMLH1 gene was significantly higher in the observation group when compared to the control group II (χ2 = 10.486, 4.876, P < 0.05). The mutation rates of SNPs in codons 12 and 13 of K-RAS and at 2783C/A locus of the hMSH2 gene did not show any statistical difference from those in the control group II.
CONCLUSION: There may be important clinical significance and relevance between neoplastic intestinal polyps and colorectal cancer in terms of the mechanisms involved in the pathogenesis.
Core tip: This study selects the blood and tissue samples of neoplastic polyps, colorectal cancer and normal tissues for related gene sequencing. By comparing the differences in SNPs, the genetics and pathogenesis of colorectal cancer are analyzed.
-
Citation: Yan ZH, Cui LH, Wang XH, Li C, He X. Comparative study of mutations in SNP loci of
K-RAS ,hMLH1 andhMSH2 genes in neoplastic intestinal polyps and colorectal cancer. World J Gastroenterol 2014; 20(48): 18338-18345 - URL: https://www.wjgnet.com/1007-9327/full/v20/i48/18338.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i48.18338
As a primary malignant neoplastic disease of the digestive system, colorectal cancer has a high incidence rate of 13.29 per 100000 in China[1], second only to primary liver cancer. Colorectal cancer is also one of the leading causes of cancer death[2,3]. Hereditary nonpolyposis colorectal cancer (HNPCC) accounts only for 5%-15% of all colorectal cancers[4], while the rest (more than 80%) of colorectal cancer cases evolve from neoplastic polyps. Since neoplastic polyps are precancerous lesions that can develop into colorectal cancer, previous studies have confirmed that epigenetic phenomena caused by DNA hypermethylation may be early indicators of tumorigenesis[5-8]. Increasing attention has been paid recently to the fundamental and clinical studies on molecular genetics and tissue metabolism which are related to single nucleotide polymorphism (SNP) loci[9]. Here we report the comparison of mutations in the K-RAS, hMLH1 (human mutL homolog1) and hMSH2 (human mutS homolog 2) genes in blood and intestinal tissue specimens collected from patients with neoplastic intestinal polyps and those with colorectal cancer.
All specimens were collected at our hospital from January 2011 to October 2012. A total of 120 blood specimens and 120 colon biopsy specimens were collected from patients with neoplastic intestinal polyps (the observation group). Eighty blood specimens and 80 colon biopsy specimens were collected from patients with colorectal cancer (control group I), and 40 blood specimens and 40 colon biopsy specimens were collected from healthy individuals (control group II). In the observation group (120 patients), there were 71 males and 49 females with an average age of 38.3 ± 10.4 years (range: 24-68 years). The histological classification of the group was defined as follows: 85 cases of tubular adenoma, 17 cases of villous adenoma and 18 cases of mixed adenoma. Among the 80 patients with colorectal cancer (control group I), there were 49 males and 31 females (average age, 42.6 ± 9.1 years; range: 33-72 years) with the following histological classification: 42 cases of tubular adenoma, 27 cases of mucinous adenocarcinoma, 8 cases of papillary adenocarcinoma and 3 cases of other types. Among the 40 cases of healthy individuals (control group II), there were 26 males and 14 females with an average age of 34.6 ± 6.3 years (range: 22-55 years). The detailed data are listed in Table 1. Peripheral blood was collected from each patient and focal tissue biopsy was conducted. All specimens were stored at -20 °C for further analysis. Patients with a family history of familial adenomas, polyps or HNPCC were excluded from this study. Our study was performed in accordance with the guidelines of the Ethics Committee of Navy General Hospital of PLA. Informed consent was obtained from all patients. The research protocol was approved by the institutional review board of the Ethics Committee of Navy General Hospital of PLA.
| Group | Number | Sex (n) | Age (yr) | |||
| (n) | Male | Female | Range | mean ± SD | ||
| Observation group (neoplastic polyps) | Tubular adenoma | 85 | 54 | 31 | 27-68 | 39.2 ± 7.6 |
| Villous adenoma | 17 | 9 | 8 | 24-65 | 36.5 ± 11.2 | |
| Mixed adenoma | 18 | 8 | 10 | 29-66 | 37.4 ± 10.9 | |
| Control group I (colorectal cancer) | Tubular adenocarcinoma | 42 | 24 | 18 | 37-72 | 44.6 ± 11.3 |
| Mucinous adenocarcinoma | 27 | 17 | 10 | 34-70 | 40.2 ± 9.6 | |
| Papillary adenocarcinoma | 8 | 5 | 3 | 33-58 | 41.1 ± 10.3 | |
| Other types1 | 3 | 3 | 0 | 45-52 | 48.3 ± 7.1 | |
| Control group II (normal) | 40 | 26 | 14 | 22-55 | 34.6 ± 6.3 | |
Genomic DNA extraction kit (SBS Genetech, China), PCR amplifier PTC220 (US BIO-RAD Company, United States), DYY-10C electrophoresis apparatus (Beijing Liuyi Instrument Factory, China), Bio-Rad Gel Doc XR System Gel Imaging System, ABI 3730XL Sequencer, Eppendorf 5417R/5810R centrifuge, and CodonCode Aligner sequence alignment analysis software were used in this study.
Collection of blood specimens: All the subjects were instructed to avoid spicy or excitant food one week prior to blood collection. They were also prohibited from drinking and asked to keep an empty stomach (fasting for 6 to 8 h) on the morning of blood collection. Venous blood (5 mL) was drawn from the median cubital vein and stored in an anticoagulant tube (109 mmol/L sodium citrate, 0.4 mL). The collected blood specimens were either immediately sent to the laboratory for analysis or saved at -20 °C. Before analysis, each blood specimen was thawed to room temperature and mixed well. DNA from the blood specimens was extracted using complete blood cells.
Collection of intestinal tissue specimens: All the patients underwent regular examinations and complete inspections including electrocardiogram, blood coagulation and immunity function after admission. Patients were asked to keep a soft liquid diet one day prior to the medical examination. They were orally fed the cathartic agent of the liquor mixed with 20% mannitol (500 mL) and 5% glucose in normal saline (1000 mL) after supper in order to clean the intestine. They were also asked to stop eating on the morning of the examination and keep an empty stomach (for 6-8 h). The method of electrocoagulation assisted excision with nylon loop ligature was applied to conduct tissue biopsy on the tissue lesions of patients with neoplastic polyps and colorectal cancer. For healthy individuals, tissue samples at the 3, 6, 9 and 12 o’clock positions of the intestinal mucosa were taken. All the specimens were stored in a 10% neutral formaldehyde solution until further analysis. The tissue specimens were grinded and DNA was extracted using the phenol-chloroform method.
DNA extraction: DNA was extracted from the centrifuged blood and grinded tissue specimens using the genomic DNA extraction kit. The purity of the extracted DNA was tested using an ultraviolet spectrophotometer.
Primer design for target genes: Primer3 online software was used to design primers for K-RAS codons 12 and 13, hMLH1 gene at Va1384Asp and hMSH2 gene at 2783C/A loci. The primer sequences used in this study are as follows: K-RAS gene: 5′-CGTCTGCAGTCAACTGGAATT-3′ (forward) and 5′-CCTGACATACTCCCAAGGA-3′ (reverse); hMLH1 gene at Va1384Asp locus: 5′-TGTGTGATATGTTTAGATGGAAATGA-3′ (forward) and 5′--TTGAAGTCACACTGCGAAGAA-3′ (reverse); hMSH2 gene at 2783C/A locus: 5′-TCGGGCAGAATTGCTTCTAT-3′ (forward) and 5′-ATTCCAGCACCATTCCAGAG-3′ (reverse).
PCR amplification reaction: PCR reaction mix consisted of 2.5 μL 10 × PCR buffer, 16.775 μL double distilled water (dd H2O), 1.0 μL forward primer (10 μmol/L), 1.0 μL reverse primer (10 μmol/L), 2 μL template DNA, 1.6 μL 4 × dNT (2.5 mmol/L), 0.125 μL Taq enzyme (5 U/μL) and MgCl2 solution (1.5 mmol/L). PCR reaction conditions were as follows: initial denaturation at 95 °C for 4 min, 30 cycles of 95 °C for 30 s, annealing (Ta) at 58 °C for 30 s, and amplification at 72 °C for 30 s, and a final extension at 72 °C for 10 min. The PCR amplification products were separated by 1% agarose gel electrophoresis (AGE) and imaged using the Gel Doc XR gel imaging system (Bio-Rad Company, United States).
DNA sequence analysis: 20 μL PCR product was sent to Beijing Yiming Fuxing Biotechnology (Beijing, China) for sequencing. The sequencing was conducted in both directions (forward and reverse) with respect to K-RAS codons 12 and 13, hMLH1 gene at Va1384Asp and hMSH2 gene at 2783C/A loci.
All data were analyzed using SPSS17.0 statistical software. The χ2 test for 2 × 2 fourfold table was used to compare the rates of mutations at various gene loci, with P < 0.05 considered statistically significant. When the total specimen size was ≥ 40, but one of the theoretical frequencies was 1 ≤ T ≤ 5 or the χ2 value was slightly greater than 3.84 before verification, the correction formula was used to enumerate the χ2 value. If still P < 0.05, it suggested that the verification results were consistent and that the difference was statistically significant.
The rates of mutations in the codons 12 and 13 of the K-RAS gene and the missense mutation of the hMSH2 gene at 2783C/A in the blood specimens of the observation group (neoplastic polyps) were significantly different (χ2 = 15.476, 29.670 and 10.811, respectively, P < 0.05) compared to the rates of mutations of relevant target gene loci in the control group I (colorectal cancer). No statistically significant difference was observed in the rates of mutations in the codons 12 and 13 of the K-RAS and the missense mutation 2783C/A in the hMSH2 gene in the blood specimens between the observation group (neoplastic polyp) and control group II (healthy population). The rate of missense mutation Va1384Asp in the hMLH1 gene showed a significant difference between the experimental group and the control group II (χ2 = 10.486, P < 0.05), although no statistical difference was observed between the experiment group and the control group I. The detailed results are listed in Table 2.
| K-RAS-12 | K-RAS-13 | hMLH1-Va1384Asp | hMSH2-2783C/A | |||||
| + | - | + | - | + | - | + | - | |
| Observation group | 3 | 117 | 0 | 120 | 11 | 109 | 0 | 120 |
| Control group I | 15 | 65 | 18 | 62 | 10 | 70 | 7 | 73 |
| χ2 | 15.476 | 29.670 | 0.568 | 10.811 | ||||
| P value | 0.000 | 0.000 | 0.451 | 0.001 | ||||
| Control group II | 1 | 39 | 0 | 40 | 1 | 39 | 0 | 40 |
| χ2 | 0.000 | - | 10.486 | - | ||||
| P value | 1.000 | - | 0.001 | - | ||||
The results are summarized in Table 3. The rates of mutations in the codons 12 and 13 of the K-RAS gene and the missense mutation 2783C/A in the hMSH2 gene in the tissue specimens of the observation group (neoplastic polyps) showed statistical differences (χ2 = 16.618, 33.538, 7.898, respectively, P < 0.05) when compared to the rates of mutations of relevant target gene loci in the control group I (colorectal cancer), but they showed no significant difference when compared with the rates of mutations of relevant target gene loci in the control group II (healthy population). The rate of missense mutation of the hMLH1 gene at Va1384Asp showed a significant difference between the experimental group and the control group II (χ2 = 3.883, P < 0.05), while no statistical difference was observed between the experiment group and the control group I.
| K-RAS-12 | K-RAS-13 | hMLH1-Va1384Asp | hMSH2-2783C/A | |||||
| + | - | + | - | + | - | + | - | |
| Observation group | 7 | 113 | 1 | 119 | 19 | 101 | 3 | 117 |
| Control group I | 21 | 59 | 22 | 58 | 15 | 65 | 10 | 70 |
| χ2 | 16.618 | 33.538 | 0.289 | 7.898 | ||||
| P value | 0.000 | 0.000 | 0.591 | 0.005 | ||||
| Control group II | 1 | 39 | 0 | 40 | 1 | 39 | 0 | 40 |
| χ2 | 0.702 | 0.335 | 3.883 | 1.019 | ||||
| P value | 0.361 | 0.750 | 0.048 | 0.419 | ||||
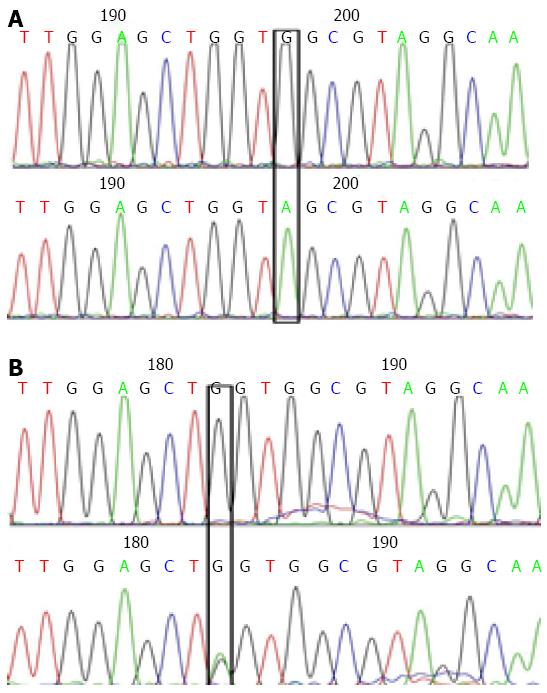
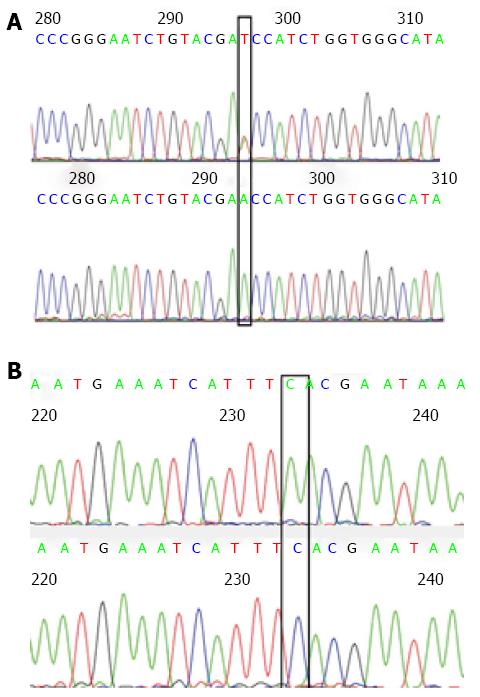
As indicated in Figures 1 and 2, in a typical sequencing peak map of codons 12 and 13 of the K-RAS gene, the bases at loci 182 and 197 were both G (black), suggesting a normal wild type gene. After mutation, the original G peaks at loci 182 and 197 were replaced by A peaks (green). No displacement or deletion occurred in adjacent base loci, and the detection and analysis showed that G content was greater than A content, indicating that the gene is a mutant gene type. In a typical sequencing peak map of the hMLH1 gene at Val384Asp297 locus, T peak (brown) indicates a normal wild type gene. After mutation, the original sequencing peak at Val384Asp297 locus disappeared, and the A peak showing the missense mutation (green) can be seen. No displacement or deletion was observed in adjacent base loci, and the detection and analysis showed that T content greater than A content, indicating a mutant gene type. In a typical sequencing peak map of hMSH2 gene at 2783C/A233 locus, the base is C (blue) indicating a normal wild type gene. After mutation, the original sequencing peak at 2783C/A233 locus disappeared and the A peak (green) indicating a missense mutation was observed. No displacement or deletion in adjacent base loci occurred and the detection and analysis showed that C content greater than A content, indicating a mutant gene type.
In recent years, the biological relationship between the mutations in the SNP loci and pathogenesis of diseases is being gradually recognized and clarified. As a member of the RAS protein superfamily, K-RAS jointly with other molecules constitutes the molecular switch regulating the GTPases and is responsible for the transition of extracellular signals, so as to trigger a series of cellular processes like proliferation, survival and differentiation[10,11]. Once the K-RAS gene has been mutated to a constitutively active form, it mediates signal transduction through the K-RAS-BRAF-MEK pathway and plays a role in inhibiting the epidermal growth factor receptor. Therefore, k-ras has been extensively studied, as it is one of the first cell markers found in tumors[12,13]. Numerous fundamental and clinical studies have shown increased[14-17] mutation rates of the K-RAS gene in human tumor cells (as high as 30% in some tumor cells). The most common type of K-RAS gene mutation is a locus-specific mutation, and the mutation loci primarily exist on the 12th and 13th codons of exon 2 (accounting for 90%) and the 61st codon of exon 3. Specific mutation types observed are: GGT>GAT (G12D), GGT>GTT (G12V), and GGC>GAC (G13D)[18]. The RAS protein-mediated signal transduction of GTPase has a clear feedback control as GTP hydrolysis results in the inactivation of RAS protein. In an event of mutations in the SNP loci, activation of oncogenes results in the stabilization of RAS protein in a GTP binding activated form. This in turn affects the downstream signal transduction pathways resulting in malignant cell proliferation or differentiation, forming the basis for the occurrence of malignant tumors[19-21].
The hMLH1 and hMSH2 genes, as two most important genes in the mismatch repair gene (MMR) genes, have significant hereditary susceptibility and are involved in the base mismatch repair, fidelity of DNA replication, maintenance of gene stability and the reduction of spontaneous mutations at independent loci in the DNA replication process, so as to achieve the purpose of ensuring the biological stability of DNA. The lack of DNA-MMR protein increases both instability of repetition during DNA replication, as well as errors by DNA polymerase[22]. Their inactivation increases the incidence of base mismatch during DNA replication and leads to events like the occurrence of incipient tumors[23,24]. It is important to point out that, as an MMR-defective tumor marker, microsatellite instability (MSI) shows the distinct characteristic of an increased number of 1-4 bp repeat sequences[25]. MSI can rapidly cause frameshift mutations in the repeated sequences in target cancer suppressor genes, accounting for about 15% of sporadic CRCs[26]. The studies performed by Kim et al[27] and Li et al[28] in the case of sporadic colorectal cancer showed that although the allele frequency of SNPs was relatively low, these polymorphisms were closely related to the occurrence of colorectal cancer. Further studies are still required to explore the correlation between colorectal cancer and the location of SNPs. At present, studies on the role of hMSH2 in CRC are very limited. As reported in relevant articles[29-32], hMSH2 mutation is closely related to p53 mutation. The hMSH2 mutation may directly cause p53 mutation. Studies conducted by Zhang et al[33] have shown that the incidence of MSH2 promoter methylation in sporadic CRC can be as high as 18.3% (212/1160 cases). The reduction of MSH2 expression in CRC patients is likely mainly caused by gene mutations of hMSH2, including missense mutations and also possibly mutations at the SNP site[34,35]. The hypermethylation of hMLH1 promoter may result in the loss of expression of the hMLH1 gene[25].
In this study, the blood and tissue specimens of 120 patients with tumorous polyp of the intestinal tract were collected to establish an observation group, while the blood and tissue specimens of 80 patients with colorectal cancer and 40 healthy individuals were collected to establish the control groups I and II, respectively. The PCR amplification and sequencing of the target genes (K-RAS, hMLH1 and hMSH2) were conducted. As indicated by the results of blood specimens, except the hMLH1 gene at Va1384Asp which showed no statistical difference (χ2 = 0.568, P > 0.05) from that of patients with colorectal cancer in terms of the missense mutation rate, the mutation rates at SNP loci of the K-RAS and hMSH2 genes of the specimens from patients with neoplastic intestinal polyp were all significantly lower than those of the specimens from patients with colorectal cancer (χ2 = 15.476, 29.670, 10.811, P < 0.05). The results from the tissue specimens were consistent with those from the blood specimens, i.e., as far as the missense mutation rate of the hMLH1 gene at Va1384Asp was concerned, no statistical difference (χ2 = 0.289, P > 0.05) was seen between the observation group (neoplastic intestinal polyp) and the control group I (colorectal cancer). Consistent with the incidence of hMLH1 mutations in CRC shown by Esteller et al[36] and Psofaki et al[37] (1160 cases, 18.3%), the study had also pointed out that, in adenomas that do not necessarily progress to cancer, their hMLH1 mutation rate was still higher than the hMSH2 mutation rate in samples from the same period. However, Lee et al[38] have put forward a contradictory conclusion: besides mutations in the hMLH1 gene, CpG island methylation in hMSH2 and MGMT is more closely correlated to the traditional adenoma-tumorigenesis pathway. Therefore, whether the appearance of SNPs ultimately leads to the epigenetic phenomenon of hMLH1 needs to be verified in further studies. On one hand, the mutation rates of the SNP loci of the K-RAS and hMSH2 in the blood and tissue specimens of patients with neoplastic intestinal polyp showed no statistical difference when compared to the healthy individuals. On the other hand, the mutation rate of the hMLH1 gene at Va1384Asp locus in the blood and tissue specimens of patients with neoplastic intestinal polyps was significantly higher than that of the healthy population (where χ2 = 10.486, 4.876, P < 0.05), indicating that the mutation rate of the SNP locus Va1384Asp of the hMLH1 gene shows similarities between the observation group and the control group I as well as differences between the observation group and the control group II. What also needs to be pointed out is that, considering that there is still controversy with respect to the efficacy of immunocytochemical analysis of the MMR gene as an alternative molecular marker and also considering the special hereditary definition of SNPs in target genes[39,40], the present study has adopted direct sequencing to achieve more direct results and avoid the influence of other genetic mechanisms. Considering the role of neoplastic polyps as an important precancerous lesion of colorectal cancer, it has been indicated that the genetic and histological changes must have been present during the course of development of the disease. At present, the molecular mechanisms of the pathogenesis of neoplastic polyps are not clear. However, recent studies have shown that the surface migration and atypical hyperplasia of deep crypt cells play a primary role in the pathogenesis of neoplastic polyps[41]. Taking into account the biological and genetic functions of target genes, we suggest that the differences in SNP locus mutations of the KAS, hMLH1 and hMSH2 genes between patients with neoplastic polyp and patients with colorectal cancer can be explained by the differences in the functional role of these genes. The KAS gene is primarily involved in cellular differentiation, proliferation and energy metabolism, while the hMLH1 gene is an important gene in the mismatch repair system. The results of the study indicate that the hMLH1 gene might have been subjected to alteration during molecular events in the early stages of colorectal cancer or even in the precancerous lesions of neoplastic polyps. Although the SNP locus mutation of the K-RAS gene has been shown to be an early molecular event in the onset of colorectal cancer, it was demonstrated in this study that the SNP locus mutation of the K-RAS gene might not play an important role in patients with intestinal neoplastic polyps. Similarly, the hMSH2 gene, which was also considered to be an important gene in the mismatch repair system, was not significantly changed in the neoplastic polyps. This might be explained by the differences between the “second hit” mechanisms of the hMLH1 and hMSH2 genes; however, the specific mechanism needs to be further investigated. We also suggest that the hypermethylation of the hMLH1 gene is either easily facilitated or the modification occurs early during the course of disease progression. A study conducted by Xiang et al[42] on the role of mismatch repair proteins (hMLH1 and hMSH2) in intestinal polyps and sporadic colorectal cancer showed that the negative expression rates of hMLH1 and hMSH2 proteins were 4.76% and 0 in inflammatory intestinal polyps, 21.66% and 8.92% in adenomatous polyp tissue of the intestinal tract, and 27.27% and 13.64% in sporadic colorectal cancer tissue, respectively. These results demonstrated the clinical significance of the negative expression of mismatch repair protein hMLH1 caused by the mutation of the hMLH1 gene in both neoplastic intestinal polyps and colorectal cancer. Additionally, in this study, the negative expression rate of hMLH1 protein was greater than the mutation rate of SNPs for the hMLH1 gene at Va1384Asp locus. As mentioned in the study by Xiang et al[42] the negative expression of mismatch repair proteins hMLH1 and hMSH2 cannot be attributed to a single factor of mutations in the SNP locus, but other factors such as protein translation also play an important role. It is also suggested that, the mutations in the SNP loci of the target genes were not a determining factor for the increased susceptibility to developing colorectal cancer and the exact role of mutations in the SNP loci and their involvement in the pathogenesis of colorectal cancer needs to be further evaluated.
In conclusion, there are similarities and differences between the mutation rates of the K-RAS, hMLH1 and hMSH2 genes in the blood and tissue biopsy specimens of patients with neoplastic intestinal polyps and those with colorectal cancer. Our study indicates that there may be a close relevance between neoplastic intestinal polyps and colorectal cancer in terms of the genetic alterations and pathogenic mechanisms, which may have important clinical significance.
The development process of colorectal cancer relates to a complex process with multi-gene, multi-stage, multi-step changes, and mostly follows the mode of “normal intestinal epithelium → hyperplasia and micro adenoma → early adenoma → mid adenoma → advanced adenoma → colorectal cancer → metastasis". At the same time, with the development of molecular genetics, colorectal cancer prevention research is no longer limited to tissue pathology, and the significance of biological genetics between neoplastic polyps and colorectal cancer, as well as the disease prevention, epidemiology and etiology are paid more attention gradually. Related research has made a breakthrough in the pathogenesis and genetic susceptibility of colorectal cancer, which provides a new idea for the clinical prevention and treatment interventions.
Single nucleotide polymorphism (SNP) is of high value in many research fields, such as genetic analysis, diagnosis and treatment of diseases, biomedical research, drug development and population ecology evaluation. At present, with the development of genetics, molecular biology and detection technology, SNP has played a more important role in the correlation research of tumor pathogenesis, prognosis evaluation and judgment. Especially, the DNA mismatch repair system and the K-RAS gene SNP polymorphism have close relation with colorectal cancer susceptibility, so they are significant in early genetic events of colorectal cancer.
The target gene SNP sequencing is conducted using blood and tissue samples of neoplastic polyps, colorectal cancer and normal tissues. By directly comparing and analyzing the statistically differences of results, the study further clarifies the significance of different genes and genetic events in the pathogenesis of colorectal cancer.
By comparing the differences of target gene SNPs in different stages, this study further proves the potential clinical value of molecular markers and molecular diagnosis, thus settling a theoretical basis and exploring new clinical ideas for screening groups with high tumor risk as well as establishing an effective early warning mechanism.
Through conducting SNP sequencing using blood and tissue samples of neoplastic polyps, colorectal cancer and normal tissues, this study compares and analyzes the results theoretically, explores the role of SNPs in different genes in the pathogenesis of colorectal cancer, and provides new research directions for molecular genetics and pathogenesis of colorectal cancer.
P- Reviewer: Ponzetti A, Sinagra E S- Editor: Qi Y L- Editor: Wang TQ E- Editor: Wang CH
| 1. | Li JG, Ua JJ, Uang DM. SH2-B expression in colon cancer and its clinical significance. Zhongguo Xiandai Yixue Zazhi. 2010;20:2004-2007. |
| 2. | Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277-300. [PubMed] |
| 3. | László L. Predictive and prognostic factors in the complex treatment of patients with colorectal cancer. Magy Onkol. 2010;54:383-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 4. | Rodriguez-Bigas MA, Boland CR, Hamilton SR, Henson DE, Jass JR, Khan PM, Lynch H, Perucho M, Smyrk T, Sobin L. A National Cancer Institute Workshop on Hereditary Nonpolyposis Colorectal Cancer Syndrome: meeting highlights and Bethesda guidelines. J Natl Cancer Inst. 1997;89:1758-1762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 772] [Cited by in RCA: 713] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 5. | Bai AH, Tong JH, To KF, Chan MW, Man EP, Lo KW, Lee JF, Sung JJ, Leung WK. Promoter hypermethylation of tumor-related genes in the progression of colorectal neoplasia. Int J Cancer. 2004;112:846-853. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 61] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 6. | Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141-196. [PubMed] |
| 7. | Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1657] [Cited by in RCA: 1607] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 8. | Hu X, Zhang Z, Ma D, Huettner PC, Massad LS, Nguyen L, Borecki I, Rader JS. TP53, MDM2, NQO1, and susceptibility to cervical cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:755-761. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 46] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 9. | Fang KH, Chang XT. Research progress on relationship between gene single nucleotide polymorphisms and tumor. Zhonghua Zhongliu Fangzhi Zazhi. 2011;18:151-155. |
| 10. | Colicelli J. Human RAS superfamily proteins and related GTPases. Sci STKE. 2004;2004:RE13. [PubMed] |
| 11. | Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2345] [Cited by in RCA: 2382] [Article Influence: 108.3] [Reference Citation Analysis (0)] |
| 12. | Lièvre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Côté JF, Tomasic G, Penna C, Ducreux M. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992-3995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1669] [Cited by in RCA: 1699] [Article Influence: 89.4] [Reference Citation Analysis (0)] |
| 13. | Yuan Y, Hu HG, Ye XX, Shen H, Zheng S. [K-ras gene mutation in colorectal cancer and its clinicopathologic significance]. Zhonghua Wai Ke Zazhi. 2010;48:1247-1251. [PubMed] |
| 14. | Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459-465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1287] [Cited by in RCA: 1378] [Article Influence: 62.6] [Reference Citation Analysis (0)] |
| 15. | Quinlan MP, Quatela SE, Philips MR, Settleman J. Activated Kras, but not Hras or Nras, may initiate tumors of endodermal origin via stem cell expansion. Mol Cell Biol. 2008;28:2659-2674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 72] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 16. | Minoo P, Zlobec I, Peterson M, Terracciano L, Lugli A. Characterization of rectal, proximal and distal colon cancers based on clinicopathological, molecular and protein profiles. Int J Oncol. 2010;37:707-718. [PubMed] |
| 17. | Onozato W, Yamashita K, Yamashita K, Kuba T, Katoh H, Nakamura T, Sato T, Ihara A, Okayasu I, Watanabe M. Genetic alterations of K-ras may reflect prognosis in stage III colon cancer patients below 60 years of age. J Surg Oncol. 2011;103:25-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 18. | Ma ES, Wong CL, Law FB, Chan WK, Siu D. Detection of KRAS mutations in colorectal cancer by high-resolution melting analysis. J Clin Pathol. 2009;62:886-891. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 38] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 19. | Wu WH, Tang YZ, Xiao LB. Detection of K-ras gene mutations in cancer tissues,preoperative carcinoembryonic antigen in serums of patients with colorectal cancer and their correlation with clinical pathological characteristics. Zhonghua Putongwai Kexue Zazhi (Dianziban). 2010;4:350-354. |
| 20. | Catela Ivkovic T, Loncar B, Spaventi R, Kapitanovic S. Association of H-ras polymorphisms and susceptibility to sporadic colon cancer. Int J Oncol. 2009;35:1169-1173. [PubMed] |
| 21. | Zhang Y, Jin M, Liu B, Ma X, Yao K, Li Q, Chen K. Association between H-RAS T81C genetic polymorphism and gastrointestinal cancer risk: a population based case-control study in China. BMC Cancer. 2008;8:256. [PubMed] |
| 22. | Strand M, Prolla TA, Liskay RM, Petes TD. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993;365:274-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 746] [Cited by in RCA: 732] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 23. | Tang CB, Cheng HM, Yang WL, Zhao JH. Relationship between pitpatterns of colorectal polypoid lesions classified by magnifying chromoendoscopy and expression of hMLH1 and hMSH2 proteins in colorectal mucosa. Shijie Huaren Xiaohua Zazhi. 2011;19:596-601. |
| 24. | Zhang GQ, Zhang YK, Lu HY, Li JC. Study on genetic instability of nm23-H1gene and expression of hMLH1, hMSH2 in sporadic gallbladder carcinoma. Zhongguo Bingli Shengli Zazhi. 2007;23:2179-2184. |
| 25. | Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95:6870-6875. [PubMed] |
| 26. | Imai K, Yamamoto H. Carcinogenesis and microsatellite instability: the interrelationship between genetics and epigenetics. Carcinogenesis. 2008;29:673-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 323] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 27. | Kim JC, Roh SA, Koo KH, Ka IH, Kim HC, Yu CS, Lee KH, Kim JS, Lee HI, Bodmer WF. Genotyping possible polymorphic variants of human mismatch repair genes in healthy Korean individuals and sporadic colorectal cancer patients. Fam Cancer. 2004;3:129-137. [PubMed] |
| 28. | Li HC, Feng HY, Zhang XP, Liu R, Ma DW, Qin H, Zhou Y, Yu L. [Association of mismatch repair gene polymorphism with susceptibility to sporadic colorectal cancer in Tianjin region]. Yi Chuan. 2010;32:1241-1246. [PubMed] |
| 29. | Ehrlich M. DNA methylation and cancer-associated genetic instability. Adv Exp Med Biol. 2005;570:363-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 30. | Chen HY, Jiang T, Yuan F. The Methylation Status and Expression of MGMT and hMLH1 in Glioma and Their Clinical Significance. Zhoudu Yike Daxue Xuebao. 2009;30:203-207. |
| 31. | Pang YJ, Wang CX, Feng JB. Expression of hMSH2, Hmlh1 and p53 in the sporadic colorectal carcinoma. Shandong Daxue Xuebao (Yixueban). 2005;45:83-85. |
| 32. | Roger L, Jullien L, Gire V, Roux P. Gain of oncogenic function of p53 mutants regulates E-cadherin expression uncoupled from cell invasion in colon cancer cells. J Cell Sci. 2010;123:1295-1305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 92] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 33. | Zhang H, Fu WL, Huang Q. Mapping of the methylation pattern of the hMSH2 promoter in colon cancer, using bisulfite genomic sequencing. J Carcinog. 2006;5:22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 34. | Belvederesi L, Bianchi F, Galizia E, Loretelli C, Bracci R, Catalani R, Amati M, Cellerino R. MSH2 missense mutations and HNPCC syndrome: pathogenicity assessment in a human expression system. Hum Mutat. 2008;29:E296-E309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 35. | Seifert M, Reichrath J. The role of the human DNA mismatch repair gene hMSH2 in DNA repair, cell cycle control and apoptosis: implications for pathogenesis, progression and therapy of cancer. J Mol Histol. 2006;37:301-307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 41] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 36. | Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225-3229. [PubMed] |
| 37. | Psofaki V, Kalogera C, Tzambouras N, Stephanou D, Tsianos E, Seferiadis K, Kolios G. Promoter methylation status of hMLH1, MGMT, and CDKN2A/p16 in colorectal adenomas. World J Gastroenterol. 2010;16:3553-3560. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 35] [Cited by in RCA: 45] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 38. | Lee KH, Lee JS, Nam JH, Choi C, Lee MC, Park CS, Juhng SW, Lee JH. Promoter methylation status of hMLH1, hMSH2, and MGMT genes in colorectal cancer associated with adenoma-carcinoma sequence. Langenbecks Arch Surg. 2011;396:1017-1026. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 39. | Salahshor S, Koelble K, Rubio C, Lindblom A. Microsatellite Instability and hMLH1 and hMSH2 expression analysis in familial and sporadic colorectal cancer. Lab Invest. 2001;81:535-541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 81] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 40. | Shia J, Klimstra DS, Nafa K, Offit K, Guillem JG, Markowitz AJ, Gerald WL, Ellis NA. Value of immunohistochemical detection of DNA mismatch repair proteins in predicting germline mutation in hereditary colorectal neoplasms. Am J Surg Pathol. 2005;29:96-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 113] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 41. | Lakatos PL, Lakatos L. Risk for colorectal cancer in ulcerative colitis: changes, causes and management strategies. World J Gastroenterol. 2008;14:3937-3947. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 339] [Cited by in RCA: 322] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 42. | Xiang FF, Mao CP. The different expressions of mismatch repair protein (hMLH1 and hMSH2) in colorectal polyps and sporadic colorectal cancer. Zhongguo Linchang Yishi Zazhi. 2012;6:4280-4284. |