Published online Dec 21, 2014. doi: 10.3748/wjg.v20.i47.17883
Revised: June 5, 2014
Accepted: June 14, 2014
Published online: December 21, 2014
Processing time: 229 Days and 22.7 Hours
AIM: To investigate the differentiated whole genome expression profiling of gastric high- and low-grade intraepithelial neoplasia and early-stage adenocarcinoma.
METHODS: Gastric specimens from an upper magnifying chromoendoscopic targeted biopsy were collected from March 2010 to May 2013. Whole genome expression profiling was performed on 19 low-grade intraepithelial neoplasia (LGIN), 20 high-grade intraepithelial neoplasia (HGIN), 19 early-stage adenocarcinoma (EGC), and 19 chronic gastritis tissue samples using Agilent 4 × 44K Whole Human Genome microarrays. Differentially expressed genes between different types of lesions were identified using an unpaired t-test and corrected with the Benjamini and Hochberg false discovery rate algorithm. A gene ontology (GO) enrichment analysis was performed using the GeneSpring software GX 12.6. The differentially expressed gene was verified using a real-time TaqMan® PCR assay with independent tissue samples, including 26 LGIN, 15 HGIN, 14 EGC, and 20 chronic gastritis. The expression of G0S2 were further validated by immunohistochemical staining (IHC) in 24 LGIN, 40 HGIN, 30 EGC and 61 chronic gastritis specimens.
RESULTS: The gene expression patterns of LGIN and HGIN tissues were distinct. There were 2521 significantly differentially expressed transcripts in HGIN, with 951 upregulated and 1570 downregulated. A GO enrichment analysis demonstrated that the most striking overexpressed transcripts in HGIN compared with LGIN were in the category of metabolism, defense response, and nuclear factor κB (NF-κB) cascade. While the vast majority of transcripts had barely altered expression in HGIN and EGC tissues, only 38 transcripts were upregulated in EGC. A GO enrichment analysis revealed that the alterations of the immune response were most prominent in the progression from HGIN to EGC. It is worth noting that, compared with LGIN, 289 transcripts were expressed at higher levels both in HGIN and EGC. A characteristic gene, G0/G1 switch 2 (G0S2) was one of the 289 transcripts and related to metabolism, the immune response, and the NF-κB cascade, and its expression was validated in independent samples through real-time TaqMan® PCR and immunohistochemical staining. In real-time PCR analysis, the expression of G0S2 was elevated both in HGIN and EGC compared with that in LGIN (P < 0.01 and P < 0.001, respectively). In IHC analysis, G0S2 immunoreactivity was detected in the cytoplasmic of neoplastic cells, but was undetectable in chronic gastritis cells. The G0S2 expression in HGIN was higher than that of LGIN (P = 0.012, χ2 = 6.28) and EGC (P = 0.008, χ2 = 6.94).
CONCLUSION: A clear biological distinction between gastric high- and low-grade intraepithelial neoplasia was identified, and provides molecular evidence for clinical application.
Core tip: This is the first study to perform a comprehensive detection of the gene expression profiling of gastric low-grade and high-grade intraepithelial neoplasia and early-stage adenocarcinoma. This study collected precise samples and reports a clear distinction of gene expression profiles between gastric low-grade and high-grade intraepithelial neoplasia, thus providing molecular evidence for their different clinical application. The characteristic upregulated genes during gastric early carcinogenesis were involved in metabolism and the immune response and nuclear factor-κB pathway, whose expression was validated in independent samples through real-time TaqMan® polymerase chain reaction and immunohistochemical staining.
- Citation: Xu X, Feng L, Liu Y, Zhou WX, Ma YC, Fei GJ, An N, Li Y, Wu X, Yao F, Cheng SJ, Lu XH. Differential gene expression profiling of gastric intraepithelial neoplasia and early-stage adenocarcinoma. World J Gastroenterol 2014; 20(47): 17883-17893
- URL: https://www.wjgnet.com/1007-9327/full/v20/i47/17883.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i47.17883
Gastric cancer is the fourth most prevalent malignant disease and the second most common cause of cancer-related deaths worldwide. Early gastric cancer (EGC) is confined to the mucosa and submucosa of the stomach with or without lymph node metastasis. The 5-year survival rates between advanced-stage and early-stage gastric cancer are extremely different (10% and 90%, respectively)[1]. Unlike advanced-stage patients, patients with early-stage gastric cancer and precancerous lesions usually have no symptoms. Precancerous lesions are histological abnormalities and are more likely to occur with cancer. According to the WHO classification of tumors of the digestive system[2], the terms low-grade intraepithelial neoplasia/dysplasia (LGIN) and high-grade intraepithelial neoplasia/dysplasia (HGIN) are recommended for gastric precancerous lesions. Enhanced endoscopy increases the detection of asymptomatic gastric cancer or precancerous lesions for early intervention. Though they have a high incidence, Japan and South Korea have a low mortality-to-incidence ratio (0.43/0.35) that benefits from a population-based screening program[3]. Therefore, early detection and an appropriate diagnosis of gastric intraepithelial neoplasia and early-stage cancer are associated with improved outcomes for patients. Furthermore, it is important to understand more about these stages both clinically and biologically.
In the Correa cascade of multi-step gastric carcinogenesis[4], an inflammation-metaplasia-dysplasia-carcinoma sequence indicates that dysplasia may be a critical point for malignant transformation. A cohort study demonstrated that 24.9% of patients with severe dysplasia and 2.1% of patients with mild-to-moderate dysplasia received a diagnosis of gastric cancer within 1 year of follow-up after the initial diagnosis[5]. Most patients harboring lesions that are classified as high-grade dysplasia are at high risk of either synchronous invasive carcinoma or its rapid development[6]. Based on the potential transition between and morphological similarity of dysplasia and carcinoma, the hypothesis that they are biologically related is reasonable. Previous studies of the genomic copy number aberration of gastric precancerous lesions and carcinoma in situ (CIS) have provided the most prominent 8q gain, which was detected most frequently in both HGIN and CIS but was undetected in LGIN using array comparative genomic hybridization[7]. Therefore, evidence has shown that molecular variations in gastric carcinogenesis have already appeared in precancerous lesions or EGC.
According to the revised Vienna classification of gastrointestinal epithelial neoplasia, the clinical management of endoscopic follow-up is recommended for category 3 (LGIN), while endoscopic or surgical local resection is recommended for category 4 (HGIN). LGIN and HGIN apparently have different clinicopathological characteristics; however, little is known about their biological characteristics. Previous gene expression profiling studies on gastric precancerous lesions did not detail the differences between LGIN and HGIN.
In this study, the gene expression profiling of gastric high- and low-grade intraepithelial neoplasia and early-stage adenocarcinoma were investigated to explore the molecular alterations in the malignant progression of gastric neoplasia. A clear distinction of the gene expression profiles between HGIN and LGIN were identified, thus providing molecular evidence for their different clinical relevance. The microarray data were validated by quantitative real-time polymerase chain reaction (PCR) in an independent group of patients, and followed by immunohistochemical (IHC) staining. Interestingly, characteristic upregulated genes during gastric early carcinogenesis were involved in metabolism and the immune response and the nuclear factor κB (NF-κB) pathway.
Subjects were recruited from Peking Union Medical College Hospital (PUMCH) and Qinghai Provincial People’s Hospital, and provided 137 samples and 15 samples, respectively, between March 2010 and May 2013. Gastric specimens from an upper magnifying chromoendoscopic targeted biopsy were collected. The samples used for pathological diagnosis and for this experiment in each patient were very similar. According to the WHO Classification of Tumors of the Digestive System, the samples can be grouped into 4 categories: LGIN (8148/0), HGIN (8148/2), EGC (8140/3), and the chronic gastritis group.
The pathological diagnosis of chronic gastritis was based on the Sydney classification and considered as controls. EGC was confined to the mucosa or submucosa as determined by surgery or endoscopic submucosal dissection (ESD) after biopsy. This study consisted of a discovery phase and a validation phase with 77 and 75 tissue samples, respectively. In the discovery phase, gene expression profiling was performed on 19 LGIN, 20 HGIN, 19 EGC, and 19 chronic gastritis tissue samples using microarrays. In the validation phase, independent tissue samples from 26 LGIN, 15 HGIN, 14 EGC, and 20 chronic gastritis patients were used in a real-time TaqMan® PCR assay (Applied Biosystems, CA, Unites States). The clinicopathological characteristics of the patients in the different groups were evaluated in terms of gender and age. The inclusion criteria were: voluntary participation in the study with informed consent and a definite pathological diagnosis by 2 pathologists. The pathologists reviewed all cases from the 2 different hospitals according to the same criteria and agreed with all the diagnosis. This study was approved by the Ethics Committee of PUMCH and also received institutional approval; the experiments were carried out in accordance with the World Medical Association Declaration of Helsinki Ethical Principles for Medical Research[8].
Formalin-fixed paraffin-embedded blocks of 155 specimens were obtained from patients who underwent ESD in the Departments of Gastroenterology or underwent gastrectomy in the Department of General Surgery at PUMCH between September 2010 and September 2013. Patient age ranged from 39 to 78 years with a mean of 56 years, and the male-to-female ratio was 1.47. The pathological diagnosis of 61 chronic gastritis was based on the Sydney classification. A total of 94 neoplasia were diagnosed by hematoxylin and eosin staining according to the WHO Classification of Tumors of the Digestive System, with 24 specimens classified as LGIN, 40 as HGIN, and 30 as EGC.
The samples were stored in RNAlater® Solution immediately after biopsy during upper endoscopy. The samples were incubated in RNAlater® Solution overnight at 4 °CC and then transferred to -80 °C. Total RNA was extracted using the RNeasy Mini Kit (Qiagen, MD, Unites States). The concentration was measured by ND-1000 UV-VIS spectrophotometry (NanoDrop Technologies, DE, Unites States). The quality of the purified RNA (RNA integrity number, RIN) was determined using the RNA 6000 LabChip Kit and Agilent 2100 Bioanalyzer (Agilent, CA, Unites States). RNA samples with an A260/280 ratio greater than 1.8 and an RIN greater than 6.5 were included.
Sample labeling, hybridization, washing, and scanning steps were performed at the Cancer Institute and Chinese Academy of Medical Science according to the manufacturer’s instructions. Initially, 100 ng total RNA was added to the reaction to generate 1.65 μg Cy3-labeled cRNA using the Low Input Quick Amp Labeling Kit (Agilent). Then, the cRNA was hybridized to an Agilent 4 × 44 K Whole Human Genome microarray. After hybridization, the slides were washed and then scanned with the Agilent G2505B Microarray Scanner System. The fluorescent intensities of the scanned images were extracted and preprocessed with the Agilent Feature Extraction Software (version 9.1). The microarray data were submitted to the National Center for Biotechnology Information Gene Expression Omnibus database (GSE55696). Data normalization, quality control, principal component analysis, and filter by flags were performed using GeneSpring software GX 12.6 (Agilent Technologies), and only the detected flags were acceptable.
First, 500 ng total RNA was reverse transcribed using SuperScript™ IIReverse Transcriptase, and cDNA was produced. The gene expression was analyzed on the MX3005P™ QPCR System (Stratagene) using TaqMan® Gene Expression Assay probes for G0S2 (Hs00274783_s1) and POLR2A (Hs00172187_m1) according to the manufacturer’s protocol (Applied Biosystems, CA, Unites States). The PCR program was initiated at 50 °C for 2 min and at 95 °C for 10 min before 45 thermal cycles, each at 95 °C for 15 s and at 60 °C for 1 min. The gene expression levels of G0S2 were assessed relative to those of POLR2A. The equation -ΔCTG0S2 = -(CTG0S2 - CTPOLR2A) represents the expression level of G0S2, i.e., a greater -ΔCTG0S2 value indicates a higher level of G0S2 expression.
IHC analysis included a polyclonal antibody for G0S2 (Novus, CO, Unites States; dilution 1:100). Briefly, paraffin blocks were cut into 3-μm thickness sections. These sections were deparaffinized in xylene and rehydrated in a descending ethanol-to-water gradient series. Endogenous peroxidase was blocked by exposure to 3% H2O2 for 10 min. For antigen retrieval, sections were subjected to microwave heat in citrate buffer (pH 9.0) with ebullition for 5 min. After cooling to room temperature, sections were incubated with a polyclonal antibody for G0S2 at room temperature for 2 h. Anti-rabbit immunoglobulin G labeled with biotin (OriGene Technologies, Beijing, China) was used as a secondary antibody for the detection of primary antibodies and was incubated at room temperature for 30 min. The tissue sections were ready for chromogen reaction with 0.02% diaminobenzidine, and then were counterstained with hematoxylin. All samples were processed under the same conditions. Human seminiferous duct cells in testis tissue served as positive control following the manufacturer’s protocol.
All immunostained slides were evaluated independently by 2 pathologists (Zhou WX and Li Y). The percentage of G0S2 positive cells was calculated and scored as follows: score 0 = undetectable staining and from 1% to 29% of positive cells; 1 = from 30% to 59% of positive cells; 2 = from 60% to 89% of positive cells; 3 = ≥ 89% of positive cells. The intensity of cytoplasmic staining was also evaluated for G0S2 and scored from 1 to 3 (from low, through medium to high). For statistical analysis, multiplying the percentage and intensity scores provided a composite expression score (0-9). A composite score of 1-4 was classified as negative and 5-9 was ranked as positive.
The statistical analyses of the gene expression microarray data were performed using an unpaired t-test and corrected with the Benjamini and Hochberg false discovery rate (FDR) algorithm. The statistical significance was assessed at P < 0.05 to identify genes that were differentially expressed between different types of lesions and to perform a gene ontology (GO) enrichment analysis using the GeneSpring software GX 12.6. Hierarchical clustering was performed using R (http://www.r-project.org/). SPSS (version. 18.0) software (SPSS Inc., Chicago, IL, Unites States) was used for statistical analysis of the clinical features, the qPCR results, and the IHC results. The χ2 test was used to compare the categorical variables. Regarding the numerical variables, comparisons of parameters between 2 groups were made using an unpaired t-test, and those among 4 groups were performed using one-way analysis of variance followed by least significant difference. P < 0.05 was considered statistically significant.
In total, 152 tissue samples were randomly divided into 2 sets, with no significant differences with respect to age (P = 0.14), gender (P = 0.48), or other clinicopathological features; the sets were used in the discovery and validation phases. The clinical characteristics of all patients are summarized in Table 1. The mean age of the patients was 57.76 ± 9.30 years. Regarding the EGC group, the 2 sets showed no significant differences in terms of depth of invasion (P = 0.31) or differentiation (P = 0.56) (Table 2).
| Discovery phase(n = 77) | Validation phase(n = 75) | P value | |
| Gender | 0.48 | ||
| Male | 46 | 49 | |
| Female | 31 | 26 | |
| Age (yr) | 0.14 | ||
| Mean ± SE (range) | 58.87 ± 9.13 (36-78) | 56.61 ± 9.39 (36-79) |
| Discovery phase(n = 19) | Validation phase(n = 14) | P value | |
| Depth of invasion | 0.31 | ||
| Tunica mucosa | 6 | 2 | |
| Muscularis mucosae | 8 | 5 | |
| Submucosa | 5 | 7 | |
| Differentiation | 0.56 | ||
| Well | 5 | 4 | |
| Moderate | 4 | 5 | |
| Poor | 10 | 5 |
The whole genomic expression profiles of 19 LGIN and 20 HGIN were analyzed. The transcripts whose expression level differed significantly between LGIN and HGIN were explored. In total, 2521 significantly differentially expressed transcripts (951 upregulated and 1570 downregulated in HGIN) were identified and may be involved in the development of HGIN.
To identify the functionally relevant gene clusters, a GO enrichment analysis of the 951 and 1570 above-mentioned transcripts was performed. There were no enriched GO terms among the 1570 downregulated transcripts. The enriched GO terms in the 951 upregulated transcripts were associated with metabolism, the defense response, and the NF-κB cascade, which are shown in Table 3. It was hypothesized that the upregulated transcripts contributed the most relevant functions and processes. The biological distinctions between HGIN and LGIN were primarily involved in metabolism, the defense response, and the NF-κB cascade (Table 3).
| GO accession | GO term | P value | Genes involved |
| GO:0043170|GO:0043283 | Macromolecule metabolic process | 4.01 × 10-9 | 272 |
| GO:0044260|GO:0034960 | Cellular macromolecule metabolic process | 3.84 × 10-9 | 247 |
| GO:0006952|GO:0002217|GO:0042829 | Defense response | 9.44 × 10-8 | 63 |
| GO:0005515|GO:0045308 | Protein binding | 1.09 × 10-7 | 358 |
| GO:0044422 | Organelle part | 3.35 × 10-7 | 292 |
| GO:0044428 | Nuclear part | 4.51 × 10-7 | 145 |
| GO:0044446 | Intracellular organelle part | 4.17 × 10-7 | 288 |
| GO:0070013 | Intracellular organelle lumen | 5.50 × 10-6 | 144 |
| GO:0043122 | Regulation of I-κB kinase/NF-κB cascade | 6.79 × 10-6 | 21 |
| GO:0043123 | Positive regulation of I-κB kinase/NF-κB cascade | 9.53 × 10-6 | 18 |
Gene expression analyses were performed in 20 HGIN and 19 EGC to reveal the molecular differences between these tissues. Unlike the marked differences between HGIN and LGIN, there were only 58 differentially expressed transcripts between HGIN and EGC, including 38 upregulated and 20 downregulated transcripts in EGC. No GO term was enriched based on the 20 downregulated transcripts. Although the expression differences between HGIN and EGC were not great, it is worth noting that the GO enrichment analysis result of 38 upregulated transcripts was relatively prominent in the progression from HGIN to EGC. The molecular differences between HGIN and EGC mainly focused on the immune response, as shown in Table 4.
| GO accession | GO term | P value | Genes involved |
| GO:0050776 | Regulation of the immune response | 2.27 × 10-8 | 9 |
| GO:0050851 | Antigen receptor-mediated signaling pathway | 4.24 × 10-7 | 5 |
| GO:0002429 | Immune response-activating cell surface receptor signaling pathway | 5.82 × 10-7 | 5 |
| GO:0050863 | Regulation of T cell activation | 6.90 × 10-7 | 6 |
| GO:0002768 | Immune response-regulating cell surface receptor signaling pathway | 7.10 × 10-7 | 5 |
| GO:0050778 | Positive regulation of the immune response | 3.23 × 10-7 | 7 |
| GO:0002684 | Positive regulation of the immune system process | 3.92 × 10-7 | 8 |
| GO:0001772 | Immunological synapse | 1.95 × 10-6 | 3 |
| GO:0051249 | Regulation of lymphocyte activation | 2.53 × 10-6 | 6 |
| GO:0042102 | Positive regulation of T cell proliferation | 2.81 × 10-6 | 4 |
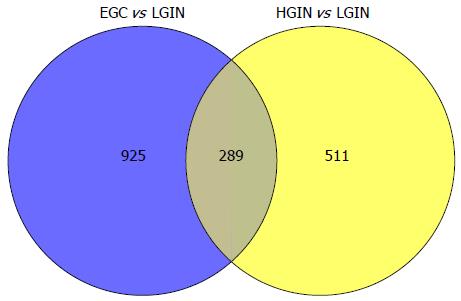
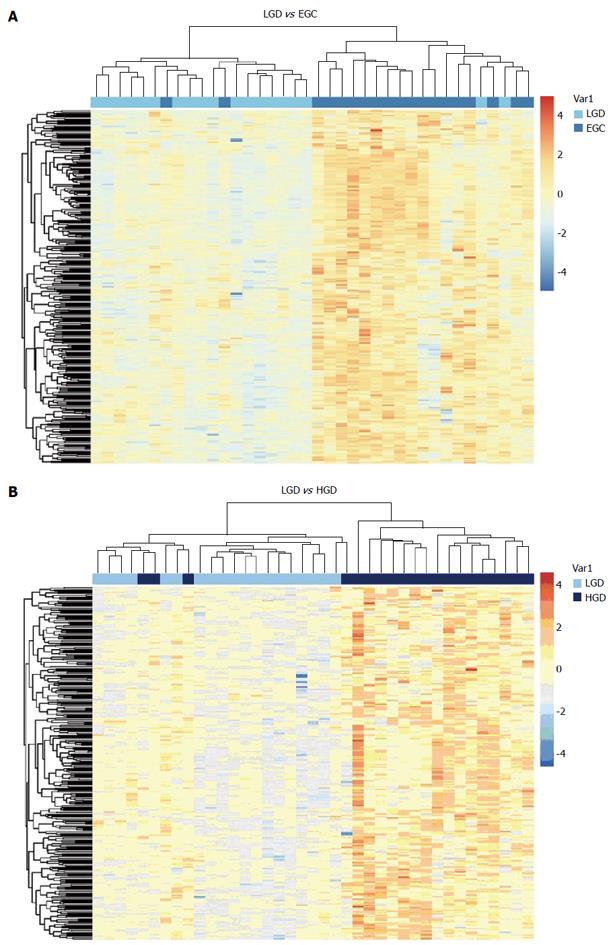
When compared with LGIN, among the 951 upregulated genes in HGIN, 289 transcripts (Figure 1) were expressed at a similarly high level in EGC and were involved in the GO enrichment analysis. Indeed, the 289 transcripts could separate both HGIN and EGC from the LGIN class. The heat map in Figure 2 shows their clear separation. These transcripts that were overexpressed in HGIN compared with LGIN were also overexpressed at similar or even higher levels in EGC. In terms of GO enrichment, some of the alterations associated with HGIN compared with LGIN, such as metabolism, the defense response, and the NF-κB cascade, were still retained in EGC.
G0S2 was related to metabolism, the defense response, and the NF-κB cascade and was overexpressed in both HGIN and EGC compared with LGIN.
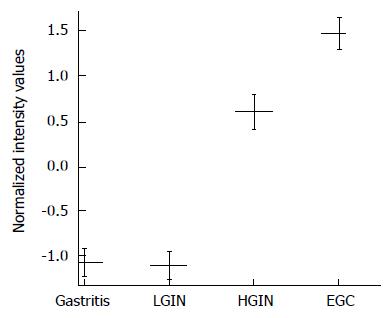
As shown in Table 5, the expression level of G0S2 in gastric EGC tissue was 6.00-fold higher than in gastric LGIN tissue (unpaired t-test with FDR correction Pcorr < 0.001) (Table 5). The expression level of G0S2 in gastric HGIN tissue was 3.28-fold higher than in gastric LGIN tissue (Pcorr < 0.05). Between HGIN and early-stage adenocarcinoma, the expression differences of G0S2 were not statistically significant (Pcorr > 0.05). However, the expression of EGC was 1.83 fold higher than that of HGIN (Figure 3).
| Unpaired t-test of G0S2 | P value (corr) | P value | FC (abs) | Direction | Probe name | Gene symbol description |
| HGIN vs LGIN | 0.021 | 0.001 | 3.28 | Up in HGIN | A_23_P74609 | Homo sapiens G0/G1 switch 2 (G0S2), mRNA NM_015714 |
| EGC vs LGIN | < 0.001 | < 0.001 | 6.00 | Up in EGC | ||
| HGIN vs Gastritis | 0.005 | 0.001 | 3.19 | Up in HGIN | ||
| EGC vs Gastritis | < 0.001 | < 0.001 | 5.83 | Up in EGC | ||
| EGC vs HGIN | 0.463 | 0.103 | 1.83 | Up in EGC | ||
| LGIN vs Gastritis | 0.946 | 0.927 | 1.03 | Down in LGIN |
Compared with gastritis, the overexpression of G0S2 in HGIN and EGC was statistically significant (Pcorr < 0.01). The difference between LGIN and gastritis was not statistically significant. The expression of G0S2 was relatively higher in the malignant lesions than in the benign lesions (Figure 3).
To validate the above results, qPCR of G0S2 was performed to quantify the RNA levels in independent tissue samples. The expression of G0S2 was elevated in HGIN and early-stage adenocarcinoma compared with LGIN, which was identical to the microarray results (Figure 4). Compared with LGIN, the difference in G0S2 expression was significant both in EGC and HGIN (P < 0.001 and P < 0.01, respectively) (Figure 4). Similarly, compared with gastritis, the difference in G0S2 expression was significant in EGC and HGIN (P < 0.001 and P < 0.05, respectively).
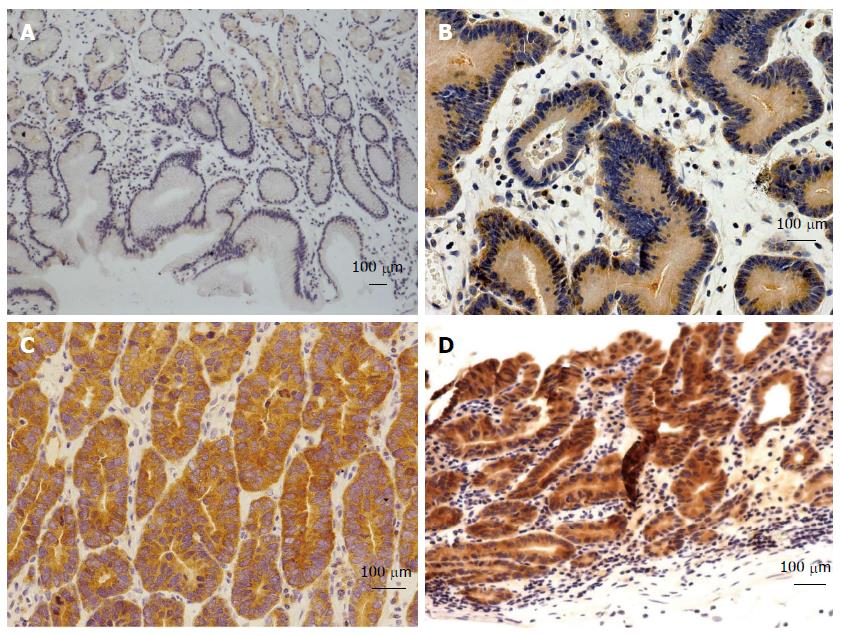
G0S2 immunoreactivity was detected in the cytoplasmic of neoplastic cells, but was undetectable (Figure 5A) in chronic gastritis cells (gastritis vs LGIN, P = 0.005; gastritis vs HGIN, P < 0.001; gastritis vs EGC, P = 0.004). The expression rates of G0S2 were 12.5% in LGIN (Figure 5B), 42.5% in HGIN (Figure 5C), and 13.3% in EGC (Figure 5D). The G0S2 expression in HGIN was higher than that of LGIN (P = 0.012, χ2 = 6.28) and EGC (P = 0.008, χ2 = 6.94) (Table 6).
In conclusion, HGIN and LGIN are biologically distinct, and the expression of G0S2 is significantly elevated in HGIN and EGC compared with LGIN. As a NF-κB cascade-and metabolism-related gene, G0S2 may be of considerable interest in the malignant transformation of gastric intraepithelial neoplasia.
According to the Vienna classification of GIN, patients with HGIN or carcinoma (CIS, suspicious, intramucosal, or submucosal invasion) should undergo resection (endoscopic or surgical). For patients with LGIN, endoscopic resection or follow-up is indicated[9]. There is evidence that genetic factors are related to gastric cancer and relevant precancerous lesions[10]. Gastric precancerous lesions are currently distinguished into LGIN and HGIN. Several large-scale follow-up studies have revealed that patients with HGIN are at higher risk of developing gastric carcinoma than are patients with LGIN. In this study, the gene expression profiles of gastric LGIN, HGIN, and EGC tissue were analyzed. There were 951 significantly upregulated transcripts and 1570 significantly downregulated transcripts in HGIN compared with LGIN. However, there were no enriched GO terms among the 1570 downregulated transcripts when a GO enrichment analysis was performed. Similarly, there were 38 upregulated and 20 downregulated transcripts in EGC compared with HGIN and no GO term was enriched based on the 20 downregulated transcripts. It was supposed that the transcripts contributed most to gastric early carcinogenesis were upregulated along with the progression from LGIN, through HGIN to EGC. The gene expression patterns between HGIN and LGIN were distinct, and the enriched GO terms in the above-mentioned 951 differentially expressed transcripts were associated with metabolism, the defense response, and the NF-κB cascade. Similarly, the differences in gene expression profiling between EGC and LGIN were significant with the parallel enriched GO terms. Presumably, the biological similarities between HGIN and EGC were prominent when compared with LGIN. However, the molecular differences between HGIN and EGC were not significant and the involved function of their differentially expressed transcripts were mainly focused on the immune response.
The genes that are involved in metabolism, the defense response, and the NF-κB cascade were not only upregulated in HGIN compared with LGIN but were also upregulated at similar or even higher expression levels in EGC. A characteristic gene related to metabolism, the immune response, and the NF-κB cascade, G0S2, was subsequently validated in independent samples using a qPCR test and an IHC test. Although the microarray analysis and qPCR analysis showed that G0S2 expression was higher in EGC and HGIN than in LGIN, IHC staining results showed the expression rate of G0S2 was higher in HGIN but similar in LGIN and EGC. The expression level of mRNA and IHC staining may be not the same.
G0S2 is located on chromosome 1 of the genome and encodes a small basic protein of 103 amino acids. G0S2 is related to the re-entry of cells from the G0 phase to the G1 phase of the cell cycle, implying that the G0S2 gene has a cell proliferative function. Previous studies have indicated that G0S2 is highly induced through NF-κB following tumor necrosis factor-α treatment[11]. G0S2 is a novel target gene of PPARs (peroxisome proliferator-activated receptors, α, β/β, and γ), which are mainly involved in lipid metabolism[12,13]. Activation of PPARγ inhibits cell growth and induces apoptosis in gastric cancer cells. G0S2 is an inhibitor of adipose triglyceride lipase and decreases lipolysis[14-17]. G0S2 is a positive regulator of oxidative phosphorylation[18]. G0S2 was upregulated in inflammatory processes, was involved in T cell quiescence, and inhibited proliferation of T cells. Inhibition of MAPK, PI3K, mTOR and Ca2+/calcineurin pathways abolished G0S2 gene suppression[19-21].
NF-κB transcription factor and its signaling pathway play a key role in controlling the immune response[22-24]. NF-κB provides a mechanistic link between inflammation and cancer[25-27]. NF-κB controls the ability of pre-neoplastic and malignant cells to resist apoptosis-based tumor-surveillance mechanisms, which is crucial for the clinical development of NF-κB inhibitors for cancer therapy[28]. The alterations in certain aspects of lipid metabolism were involved in tumorigenesis, including the synthesis and degradation of lipids that contribute to energy homeostasis and the abundance of lipids with signaling functions[29-32].
Previous studies with HGIN and LGIN were mostly based on several known genes[33,34], such as p53, Her-2, and E-cadherin[35]. Until now, few studies have focused on GIN, especially from the aspect of the whole genome expression profile.
The microarray approach has been widely applied to the investigation of advanced gastric cancer. However, advanced gastric cancer has a poor prognosis and greater heterogeneity with its progression. Several studies have revealed that genetic alterations begin in the early stages of cancer and even in precancerous lesions. Early studies using CGH arrays suggested that an 8q gain in HGIN may play a pivotal role in the development of gastric carcinoma[7]. However, the gene expression levels and functions associated with the copy number status of 8q were not detailed. Much less is known in terms of a comparison among the expression profiles of LGIN, HGIN, and EGC due to the difficulties of early detection and biopsy. Based on the clinicopathological findings and a clear research design, this study collected samples of LGIN, HGIN, and EGC to perform a comprehensive determination of their expression profiles at a whole-genome level. Through this study of gene expression profiles, the biological basis of the clinical differences between HGIN and LGIN was determined.
Despite the limitation that a larger number of samples in each group would have been beneficial, this large-scale study opens the door for further research into precancerous lesions. The asymptomatic characteristics, atypical histology, and lack of specific biomarkers in precancerous lesions[10,36] make them difficult to diagnose early and accurately. Moreover, the performance of the gene in IHC experiments may be useful for early detection of GIN, and clinical application for auxiliary diagnosis of atypical lesions is promising. Furthermore, the gene function-related research requires future study.
In summary, the microarray analysis in this study identified differences in the gene expression between LGIN and HGIN. The obvious molecular distinction between LGIN and HGIN was almost identical in different clinical practices. Metabolism, the immune response, and the NF-κB cascade appear to be important processes underlying malignant transformation.
We are grateful to all of the patients for their participation and to the physicians at the Peking Union Medical College Hospital and the Qinghai Provincial People’s Hospital for their cooperation.
Early detection and an appropriate diagnosis of gastric intraepithelial neoplasia (GIN) and early-stage cancer are associated with improved outcomes for patients. It is important to understand more about these stages both clinically and biologically. However, little is known about their biological characteristics due to the difficulties of early detection and biopsy.
Low-grade GIN (LGIN) and high-grade GIN (HGIN) apparently have different clinicopathological characteristics. Previous gene expression profiling studies on gastric precancerous lesions did not detail the differences between LGIN and HGIN. Based on the potential transition between and morphological similarity of dysplasia and carcinoma, the hypothesis that they are biologically related is reasonable.
This is the first study to perform a comprehensive detection of the gene expression profiling of LGIN and HGIN and early-stage adenocarcinoma. The characteristic upregulated genes during gastric early carcinogenesis were involved in metabolism and the immune response and nuclear factor-κB (NF-κB) pathway, and their expression was validated in independent samples through real-time TaqMan® PCR and immunohistochemical staining.
This study collected specific samples to report the clear distinction of the gene expression profiles between LGIN and HGIN, thus providing molecular evidence for their different clinical relevance. G0S2 was identified as a potential marker to differentiate LGIN and HGIN.
NF-κB transcription factor and its signaling pathway play a key role in controlling the immune response. NF-κB provides a mechanistic link between inflammation and cancer. NF-κB controls the ability of pre-neoplastic and malignant cells to resist apoptosis-based tumor-surveillance mechanisms, which is crucial for the clinical development of NF-κB inhibitors in cancer therapy. G0S2 is related to the re-entry of cells from the G0 phase to the G1 phase of the cell cycle. G0S2 is an inhibitor of adipose triglyceride lipase and decreases lipolysis, and is a positive regulator of oxidative phosphorylation.
This manuscript presents an interesting differential gene expression profiling of gastric LGIN, HGIN and early gastric cancer, and try to explore the molecular alteration in the malignant progression of gastric neoplasia. From their microarray data, authors identified G0S2, a putative lymphocyte G0/G1 switch gene product as a potential marker to differentiate low grade neoplastic lesion from high grade lesion in the stomach.
P- Reviewer: Raggi C, Shi XY, Xu R S- Editor: Gou SX L- Editor: Cant MR E- Editor: Wang CH
| 1. | Butte JM, Torres J, Viviani P, Duarte I, Crovari F, Guzmán S, Cabrera R, Pedemonte J, Llanos O. [Long term survival of patients operated for early gastric cancer]. Rev Med Chil. 2008;136:1424-1430. [PubMed] |
| 2. | Bosman FT, Carneiro F, Hruban RH, Theise ND. WHO classification of tumours of the digestive system. Lyon: IARC 2010; . |
| 3. | Shen L, Shan YS, Hu HM, Price TJ, Sirohi B, Yeh KH, Yang YH, Sano T, Yang HK, Zhang X. Management of gastric cancer in Asia: resource-stratified guidelines. Lancet Oncol. 2013;14:e535-e547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 382] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 4. | Correa P. A human model of gastric carcinogenesis. Cancer Res. 1988;48:3554-3560. [PubMed] |
| 5. | de Vries AC, van Grieken NC, Looman CW, Casparie MK, de Vries E, Meijer GA, Kuipers EJ. Gastric cancer risk in patients with premalignant gastric lesions: a nationwide cohort study in the Netherlands. Gastroenterology. 2008;134:945-952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 483] [Cited by in RCA: 585] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 6. | Dinis-Ribeiro M, Areia M, de Vries AC, Marcos-Pinto R, Monteiro-Soares M, O’Connor A, Pereira C, Pimentel-Nunes P, Correia R, Ensari A. Management of precancerous conditions and lesions in the stomach (MAPS): guideline from the European Society of Gastrointestinal Endoscopy (ESGE), European Helicobacter Study Group (EHSG), European Society of Pathology (ESP), and the Sociedade Portuguesa de Endoscopia Digestiva (SPED). Endoscopy. 2012;44:74-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 442] [Cited by in RCA: 488] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 7. | Uchida M, Tsukamoto Y, Uchida T, Ishikawa Y, Nagai T, Hijiya N, Nguyen LT, Nakada C, Kuroda A, Okimoto T. Genomic profiling of gastric carcinoma in situ and adenomas by array-based comparative genomic hybridization. J Pathol. 2010;221:96-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 8. | World Medical Association. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310:2191-2194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16597] [Cited by in RCA: 18220] [Article Influence: 1518.3] [Reference Citation Analysis (0)] |
| 9. | Dixon MF. Gastrointestinal epithelial neoplasia: Vienna revisited. Gut. 2002;51:130-131. [PubMed] |
| 10. | Leja M, Wex T, Malfertheiner P. Markers for gastric cancer premalignant lesions: where do we go? Dig Dis. 2012;30:268-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 11. | Welch C, Santra MK, El-Assaad W, Zhu X, Huber WE, Keys RA, Teodoro JG, Green MR. Identification of a protein, G0S2, that lacks Bcl-2 homology domains and interacts with and antagonizes Bcl-2. Cancer Res. 2009;69:6782-6789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 114] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 12. | Fucci A, Colangelo T, Votino C, Pancione M, Sabatino L, Colantuoni V. The role of peroxisome proliferator-activated receptors in the esophageal, gastric, and colorectal cancer. PPAR Res. 2012;2012:242498. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 13. | Takahashi N, Okumura T, Motomura W, Fujimoto Y, Kawabata I, Kohgo Y. Activation of PPARgamma inhibits cell growth and induces apoptosis in human gastric cancer cells. FEBS Lett. 1999;455:135-139. [PubMed] |
| 14. | Schweiger M, Paar M, Eder C, Brandis J, Moser E, Gorkiewicz G, Grond S, Radner FP, Cerk I, Cornaciu I. G0/G1 switch gene-2 regulates human adipocyte lipolysis by affecting activity and localization of adipose triglyceride lipase. J Lipid Res. 2012;53:2307-2317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 87] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 15. | Das SK, Eder S, Schauer S, Diwoky C, Temmel H, Guertl B, Gorkiewicz G, Tamilarasan KP, Kumari P, Trauner M. Adipose triglyceride lipase contributes to cancer-associated cachexia. Science. 2011;333:233-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 478] [Cited by in RCA: 440] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 16. | Kusakabe M, Kutomi T, Watanabe K, Emoto N, Aki N, Kage H, Hamano E, Kitagawa H, Nagase T, Sano A. Identification of G0S2 as a gene frequently methylated in squamous lung cancer by combination of in silico and experimental approaches. Int J Cancer. 2010;126:1895-1902. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 50] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 17. | Kusakabe M, Watanabe K, Emoto N, Aki N, Kage H, Nagase T, Nakajima J, Yatomi Y, Ohishi N, Takai D. Impact of DNA demethylation of the G0S2 gene on the transcription of G0S2 in squamous lung cancer cell lines with or without nuclear receptor agonists. Biochem Biophys Res Commun. 2009;390:1283-1287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 18. | Kioka H, Kato H, Fujikawa M, Tsukamoto O, Suzuki T, Imamura H, Nakano A, Higo S, Yamazaki S, Matsuzaki T. Evaluation of intramitochondrial ATP levels identifies G0/G1 switch gene 2 as a positive regulator of oxidative phosphorylation. Proc Natl Acad Sci Unites States. 2014;111:273-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 19. | Yamada T, Lee PH, Burns A, Lacorazza D. G0S2, an early response gene, regulates quiescence in naive T cells. J Immunol. 2011;186:104-110. |
| 20. | Heckmann BL, Zhang X, Xie X, Liu J. The G0/G1 switch gene 2 (G0S2): regulating metabolism and beyond. Biochim Biophys Acta. 2013;1831:276-281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 91] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 21. | Kim JY, Tillison K, Lee JH, Rearick DA, Smas CM. The adipose tissue triglyceride lipase ATGL/PNPLA2 is downregulated by insulin and TNF-alpha in 3T3-L1 adipocytes and is a target for transactivation by PPARgamma. Am J Physiol Endocrinol Metab. 2006;291:E115-E127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 161] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 22. | Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2:725-734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2972] [Cited by in RCA: 3211] [Article Influence: 139.6] [Reference Citation Analysis (0)] |
| 23. | Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5:749-759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2227] [Cited by in RCA: 2409] [Article Influence: 120.5] [Reference Citation Analysis (0)] |
| 24. | Naugler WE, Karin M. NF-kappaB and cancer-identifying targets and mechanisms. Curr Opin Genet Dev. 2008;18:19-26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 509] [Cited by in RCA: 494] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 25. | Bollrath J, Greten FR. IKK/NF-kappaB and STAT3 pathways: central signalling hubs in inflammation-mediated tumour promotion and metastasis. EMBO Rep. 2009;10:1314-1319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 314] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 26. | Baud V, Karin M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov. 2009;8:33-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 799] [Cited by in RCA: 857] [Article Influence: 53.6] [Reference Citation Analysis (0)] |
| 27. | Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883-899. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8437] [Cited by in RCA: 8164] [Article Influence: 544.3] [Reference Citation Analysis (0)] |
| 28. | Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2609] [Cited by in RCA: 2785] [Article Influence: 146.6] [Reference Citation Analysis (0)] |
| 29. | Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279:2610-2623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 839] [Cited by in RCA: 1009] [Article Influence: 77.6] [Reference Citation Analysis (0)] |
| 30. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51728] [Cited by in RCA: 47013] [Article Influence: 3358.1] [Reference Citation Analysis (5)] |
| 31. | Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3445] [Cited by in RCA: 3707] [Article Influence: 264.8] [Reference Citation Analysis (0)] |
| 32. | Muñoz-Pinedo C, El Mjiyad N, Ricci JE. Cancer metabolism: current perspectives and future directions. Cell Death Dis. 2012;3:e248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 276] [Cited by in RCA: 296] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 33. | Testino G. Gastric precancerous changes: carcinogenesis, clinical behaviour immunophenotype study and surveillance. Panminerva Med. 2006;48:109-118. [PubMed] |
| 34. | Lin LL, Huang HC, Juan HF. Discovery of biomarkers for gastric cancer: a proteomics approach. J Proteomics. 2012;75:3081-3097. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 71] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 35. | Zheng ZH, Sun XJ, Ma MC, Hao DM, Liu YH, Sun KL. [Studies of promoter methylation status and protein expression of E-cadherin gene in associated progression stages of gastric cancer]. Yichuan Xuebao. 2003;30:103-108. [PubMed] |
| 36. | Rugge M, Capelle LG, Cappellesso R, Nitti D, Kuipers EJ. Precancerous lesions in the stomach: from biology to clinical patient management. Best Pract Res Clin Gastroenterol. 2013;27:205-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 83] [Article Influence: 6.9] [Reference Citation Analysis (0)] |