Published online Nov 7, 2014. doi: 10.3748/wjg.v20.i41.15216
Revised: March 18, 2014
Accepted: June 14, 2014
Published online: November 7, 2014
Processing time: 285 Days and 0.5 Hours
The small and large intestine of the gastrointestinal tract (GIT) have evolved to have discrete functions with distinct anatomies and immune cell composition. The importance of these differences is underlined when considering that different pathogens have uniquely adapted to live in each region of the gut. Furthermore, different regions of the GIT are also associated with differences in susceptibility to diseases such as cancer and chronic inflammation. The large and small intestine, given their anatomical and functional differences, should be seen as two separate immunological sites. However, this distinction is often ignored with findings from one area of the GIT being inappropriately extrapolated to the other. Focussing largely on the murine small and large intestine, this review addresses the literature relating to the immunology and biology of the two sites, drawing comparisons between them and clarifying similarities and differences. We also highlight the gaps in our understanding and where further research is needed.
Core tip: The small and large intestine of the gastrointestinal tract (GIT) have evolved to have different functions and have a distinct anatomy, biology and immunology. Despite this, findings from the small intestine are often inappropriately attributed to large intestine and vice versa. The importance of these biological differences is underlined when considering that different regions of the GIT are associated with different infections and pathologies. This review addresses the literature relating to the small and large intestine – clarifying the similarities and differences between the two sites. We also highlight the gaps in our understanding where further research is needed.
- Citation: Bowcutt R, Forman R, Glymenaki M, Carding SR, Else KJ, Cruickshank SM. Heterogeneity across the murine small and large intestine. World J Gastroenterol 2014; 20(41): 15216-15232
- URL: https://www.wjgnet.com/1007-9327/full/v20/i41/15216.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i41.15216
The gastrointestinal tract (GIT) is the largest mucosal surface in the body; approximately 8.5 m long in humans and 30 cm long in mice, with approximately 80% of this compromising of the small intestine[1-3]. The duodenum, jejunum and ileum make up the anterior to posterior structure of the small intestine, with the caecum, colon and rectum comprising the large intestine. As a consequence of its main role in food processing and nutrient extraction, the mucosal surface of the intestine is at high risk from attack by pathogens that contaminate the host’s food. In addition to thwarting attack from pathogens, the intestine is also home to a large number of commensal microbes (the microbiome) to which tolerance must be maintained. Therefore, it is paramount that the local immune system strikes an effective balance between tolerance and immunity. The large and small intestine are anatomically and functionally different and should be considered as two separate immunological sites which is significant when considering the aetiology and pathogenesis of intestinal tract diseases. Regarding infection, the small and large intestine are susceptible to different pathogens; whipworm species such as Trichuris trichuria (human) or Trichuris muris (T. muris)(mouse) and Clostridium difficile colonise the large intestine whereas Ancylostoma duodenale (human), Ascaris lumbricoides (human), Heligmosomoides polygyrus (mouse), Trichinella spiralis (human and mouse) and Norovirus infect the small intestine. Similar distinctions are seen in allergy and autoimmunity; Coeliac disease is restricted to the small intestine and ulcerative colitis to the large intestine whereas Crohn’s Disease affects any region of the gastrointestinal tract (GIT). Moreover, cancer of the small intestine is rare whereas colon cancer is more common (2.4 patients per 100000 compared with 54 per 100000 respectively)[4]. Appreciating the immunological heterogeneity between the small and large intestine can provide answers to why this spectrum of disease along the GIT is apparent. Furthermore, a greater understanding of the cellular diversity within the GIT aids in the development of region-specific therapeutic targets for inflammatory bowel diseases and allergy. This review aims to highlight the differences and similarities between the small and large intestine of mice discussing both architectural and immune components.
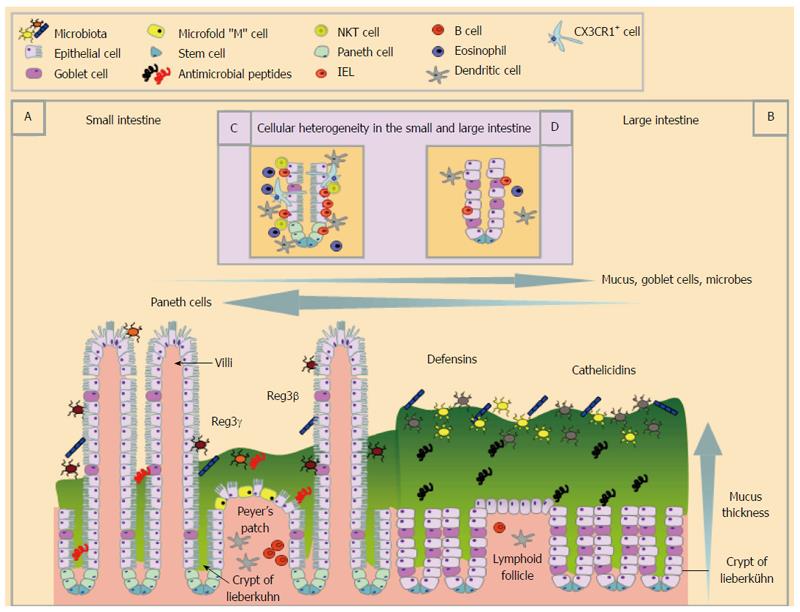
The large and small intestine are lined with a single layer of columnar epithelial cells that are linked via complexes of junctional proteins and desmosomes creating a sealed yet dynamic barrier[5]. Enterocytes and enteroendocrine cells are found throughout the GIT and are involved in nutrient and water absorption[6] and hormone production[7]. The base of the crypts in the GIT contain undifferentiated stem cells from which progenitor cells are derived[8-10] and develop into any of one of four major differentiated epithelial cell types; enterocytes, Paneth cells, goblet cells, enteroendocrine cells[6]. Leucine-rich repeat-containing G protein-coupled receptor 5 stem cells are intermingled with Paneth cells in the small intestine supplying necessary signals for the stem cell niche[11]. However, there are distinct cellular features between small and large intestinal epithelial cells (EC) that have evolved out of the need to perform different digestive processes. Small intestine EC play a critical role in nutrient absorption that is reflected in their unique structure. In addition to the small intestinal epithelium being folded in such a way to create the crypts of Lieberkühn, it is further adapted with finger-like projections called villi thus collectively increasing the surface area for maximal nutrient absorption (Figure 1). Furthermore, villous EC are striated with numerous actin-rich microvilli that further maximise their surface area. In contrast, the large intestine, which is primarily involved in water absorption and fermentation, has EC which lack villi.
Although the primary role of the villi is nutrient absorption, they are also intimately associated with immune cells (described below), highlighting their role in immunity. The different functions of the small and large intestine place different demands on the immune system. For example, most protein antigens are taken up in the small intestine by specialised cells including Microfold or “M” cells. M cells transcytose luminal particulate antigens, effectively passing them onto underlying immune cells. M cells are most abundant overlying the Peyer’s patches of the small intestine but have also been described in the villous epithelium[12]. In the large intestine, M cells are less numerous and are usually associated with isolated lymphoid follicles and colonic patches[13].
In addition to processing antigens, the intestinal epithelium plays a direct role in innate mucosal defence via production of anti-microbial peptides (AMPs). The requirement for different types of AMPs varies along the GIT reflecting the need to respond to different commensal and pathogenic microbes that colonise the GIT[14-17]. Paneth cells, a specialized lineage of cells unique to the small intestine, are a major source of AMPs including α-defensins (also called cryptdins in mice)[18], lysozyme, ribonucleases [such as angiogenin 4 (ANG4)], secretory phospholipase A2[14,16,17] and C-type lectins [such as the regenerating islet derived protein (REG3) family][19,20]. AMPs are thought to act as mediators in host defence against pathogens[21] and in shaping the microbiome composition for the maintenance of homeostasis[14,22]. AMPs such as REG3γ and REG3β promote spatial segregation of microbial populations from the epithelial surface of the small intestine[15,20]. AMPs also help shape microbial diversity and thus may contribute to the relatively low bacterial burden in the duodenum compared with the large intestine[23]. In addition to the production of AMPs, Paneth cells may directly recognise pathogens via pattern recognition receptors enabling them to initiate innate immune responses[17,20,23,24].
The unequal distribution of Paneth cells along the GIT, however, is not the sole factor participating in the differential arsenal of antimicrobial proteins between the small and the large intestine. Enterocytes along the entire GIT can be induced to express a distinct group of AMPs[16,17]. Enterocytes in the small intestine stimulate the production of REG3γ and REG3β[19,20], whereas those in the large intestine drive the synthesis of β-defensins[25] and cathelicidins[26]. As well as differences between the large and small intestine, within the large intestine there is also a different AMP profile between the proximal and distal gut which may be related to different microbial composition[27].
Goblet cells exhibit a non-homogenous distribution along the intestinal epithelium, and are more abundant in the large intestine. They release a panel of bioactive compounds including mucins, AMPs [such as resistin-like molecule β (Relmβ; also known as FIZZ2)[28], ANG4[29], REG3γ and REG3β[27]] and trefoil peptides[30] that further contribute to innate immune defence. REG3γ expression is minimal in the large intestine compared to the small intestine[15,20]. In contrast, Relmβ is expressed mainly in the large intestine and only a small amount is present in the normal terminal ileum and there is no Relmβ expression in the other areas of the small intestine[28]. Relmβ may exert a function analogous to REG3 proteins in the small intestine, keeping bacteria at a distance in the large intestine[15]. The prevalence of different AMPs at distinct sites of the GIT is due to both the distinctive primary cellular source but also due to different expression profiles of the same cells (enterocytes or goblet cells) in the different regions.
Alterations in microbial composition, due to physiologic[27,31] or pathologic states[32,33], as well as alterations in bacterial-epithelial interaction[27] can also change AMP expression patterns. The intricate cross talk between AMP expression and the microbiome generates a feedback loop, where AMPs can influence microbiome structure and microbes themselves can direct AMP synthesis[14,15]. Strikingly, short chain fatty acids, products of bacterial fermentation of dietary fibres and/or host mucin glycans can drive the expression of AMPs on colon epithelial cells[34], emphasizing the impact of bacterial metabolites on AMP site-specific expression. AMPs are concentrated within the mucus layer overlying the intestinal epithelium[35] (Figure 1).
Mucus functions as a physical, chemical and immunological barrier and is enriched in mucin glycoproteins[36], nonspecific AMPs[37] and secretory immunoglobulin A (sIgA) and G (IgG)[36]. Mucins are the major structural components of mucus, which comprises a family of 17 mucins[38] and are produced in the GIT as either membrane bound or secreted. In the small and large intestine, the mucus layer consists of secreted MUC2 and smaller amounts of transmembrane MUC1, 3A/B, 4, 12, 13, 15 and 16[36]. In the large intestine the mucus layer is approximately four-fold thicker than that of the duodenum and jejunum[39].
The mucus barrier consists of two layers; a thinner inner “firm” mucus layer, which is physically difficult to dislodge and is considered devoid of bacteria, and a thicker outer “loose” mucus layer, in which anaerobic microbiota may reside[40]. There is a debate in the literature regarding the presence of a firm mucus layer in the small intestine. Atuma et al[39] have shown the existence of a firm and loose mucus layer throughout the gut (including both the small and large intestine) with the mucus layer of the large intestine characterized by a more substantial layer of “firm” mucus. However, recent findings indicate that MUC2 forms a single soluble mucus gel in the small intestine, which is not attached to the epithelium and is penetrable to bacteria[41,42]. The absence of a firm organized mucus in the small intestine may allow direct antigen sampling by dendritic cells (DCs) conditioning them to a tolerogenic state, highlighting the importance of MUC2 in gut homeostasis[42]. The novel finding that MUC2 alone or coupled with bacterial antigens can deliver immunoregulatory signals both to underlying DCs and intestinal ECs advances the concept of the role of mucus as a barrier. Despite the recent advances on mucus research, there is much we do not know about the organization of mucus network in the small and large intestine and how AMPs may modulate it. Recent evidence that Reg3γ affects the distribution of mucus in the ileum but not in colon[43] raises important questions regarding the effects of AMPs on mucus structure given the unequal distribution of AMPs along the GIT.
A distinguishing feature of mucins is their heavy glycosylation, which is determined by the expression pattern of glycosyltransferases[36,41]. A disparity in mucin glycan content is evident between different sites of the GIT[44] and, in fact, in adjacent crypts or even in the same crypt[36]. Sialylated and sulfated glycans dominate the small intestine, whereas fucosylation is prevalent in the large intestine[44]. The mucin glycosylation profile may affect bacterial composition, providing potential adhesion sites[45] as well as bacterial energy sources[46]. Mice deficient in fucosylation of mucins exhibit an altered microbial structural configuration and expression profile in a manner dependent on the availability of diet-derived polysaccharides[47]. Thus, mucus functions not only as a barricade of enteric microorganisms, but also plays a role in the determination of bacterial composition alongside AMPs and sIgA.
The essential role of mucins in GIT health is demonstrated by reports showing the severe pathologies that develop in MUC-deficient animals[48-52]. Furthermore, de novo induction of MUC5AC in the mouse large intestine plays a key role in resistance to intestine nematode infection[53]. The mucin barrier responds dynamically to microbial signals and the immune response with an impact on mucin biosynthesis rate, secretion and glycosylation profiles[36,54,55]. Further research on the dynamic mucin changes during infection and inflammation is warranted to shed more light on mucus function and the potential for different roles in the small and large intestine.
The microbiota is the community of commensal, symbiotic, and pathogenic microorganisms that reside within the GIT with the combined genetic material of these microorganisms comprising the microbiome. The microbiota consists of approximately 500-1000 bacterial species[56-59] yet represents a small fraction of the known bacterial phyla. The most abundant phyla are Bacteroidetes and Firmicutes; however, species belonging to other phyla such as Proteobacteria and Actinobacteria are also encountered[22,60,61]. The GIT microbiota increases longitudinally along the GIT[59,62] with 1010-1011 bacteria per gram of intestinal contents in the large intestine but only 103-104 bacteria per gram of intestinal content in the duodenum and jejunum[63]. The bacterial density and abundance also increases transversely from the mucus to the lumen[62]. Additionally, the composition of the gut microbiome varies along the length of the gut depending on the intestinal site and specific niches[61,64]. For example, Firmicutes and, more specifically the families Lachnospiraceae and Ruminococcaceae are more prevalent at mucosal surfaces of the large intestine compared to the lumen[65]. The reasons for these differences include the slow transit time, changes in oxygen tension along the GIT, and the action of secreted factors including gastric acid and bile salts in the small intestine[66-68]. Moreover, the microbiota of the small intestine is adapted to process simple carbohydrates whereas large intestinal microbial communities efficiently degrade complex polysaccharides[69]. Among the multiple factors that can sculpt the GIT microbial architecture, diet and metabolic requirements have a major role in the microbiome differences throughout the GIT[70-72]. Interestingly, ileal microbiota display a relative instability over time in humans[73], whereas most bacterial strains of the large intestine reside for most or a lifetime[74]. Data in mice are lacking regarding the temporal variability of microbial populations in different sites.
The disparity in microbial communities inhabiting different niches of the gut is reflected by disparate mucosal immune responses, which are time and region dependent. Immune tolerance in response to the gut microbiome is established earlier in the large intestine compared with the small intestine[15]. Also, different bacteria strains can have different effects on host immune responses in different parts of the gut[75]. Administration of the probiotic Lactobacillus salivarius UCC118 elicits regulatory T cell responses in the small intestine, whereas, it activates inflammatory responses in the large intestine[75].
Differences in bacterial colonisation of the GIT impact on functionality in three ways: structural, protective, and metabolic. Due to the larger microbial load of the large intestine, there is likely to be a greater pressure on maintaining a structural barrier with a thicker mucus layer and abundant goblet cells[40]. As the presence of a firm mucus layer in the large intestine prevents direct contact between the microbiome and EC[40] then bacteria/epithelial cell crosstalk may be more prevalent in the small intestine. However, there may be non-cognate interactions with epithelial cells that are mediated via bacterial products or secretions which given the greater density of bacteria in the large intestine may be of greater significance in this region of the GIT. Isolated large intestine EC can distinguish between different commensal bacteria by secreting different chemokines[76] and thus may contribute directly to homeostasis. In terms of protection, the GIT microbiome can resist colonisation by pathogens by direct competition for nutrients and space and/or via production of microbicidal compounds[32]. The microbiome and its associated metabolites[77] play additional roles in resistance against infection by for example, priming immune cell function[78-80] and by promoting the development and/or regulation of innate lymphoid cells (ILCs)[62,81,82] and, specifically in the ileum, Th17 cells[83-85]. Conversely, the immune system also helps shape the composition of the microbiome[86]. It can be inferred, therefore, that changes in the microbiome along the GIT contribute to differences in the types and function of immune cells throughout the GIT.
The GIT contains the largest immune system in the body and is densely populated with many different immune cells. The greatest density of immune cells resides in the small intestine, which is perhaps not surprising given its absorptive function. Throughout the GIT differences in the make-up and functionality of immune cells are observed with many aspects of their function, particularly in the large intestine, not fully understood.
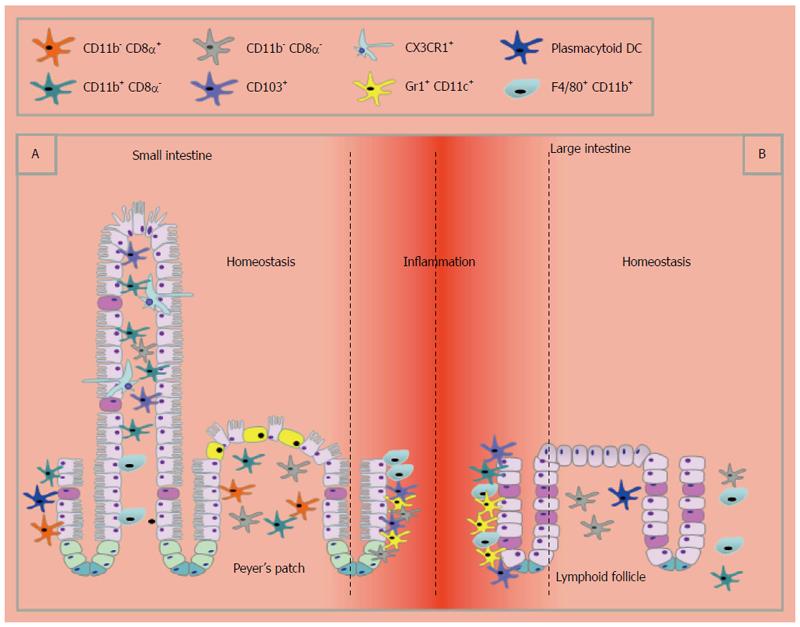
Mononuclear phagocytes can be subdivided into macrophages or dendritic cells, however, there has been extensive debate as to what distinguishes the two[87,88]. The extent of macrophage and DC heterogeneity within the large and small intestine is considerable (Table 1 and Figure 2), presumably reflecting the anatomical and physiological differences in the two sites. Data to date shows that DCs are more abundant in the small intestine. Nevertheless, DCs in the large intestine are functionally important to intestinal health as their absence or depletion can result in severe pathology and tissue destruction[89].
| Cell type | Small intestine | Large intestine | Ref. | ||
| Dendritic/macrophage Cells Conventional DCs (CD11chigh MHCIIhigh CD64-) | |||||
| CD11b- CD8α+ | Yes. Peyers Patches (PP) | Not readily detected but have been identified after the use of Flt3 ligand | [93] | ||
| CD11b+ CD8α- | Yes. PP, Lamina Propria (LP), and express CD103) | Yes. Isolated Lymphoid Follicles (ILF) and Colonic Patches. LP but scarce | [91,92] | ||
| CD11b- CD8α- | Yes. PP | Unknown | [91,92,94] | ||
| CD103+ | Yes | Yes. CD103+CD11b- are more abundant | [91,92] | ||
| Monocyte derived DCs | |||||
| Gr1+ CD11c+ plasmacytoid DCs | Yes. Upon inflammation | Yes Upon inflammation | [104,105] | ||
| CD11cintBB20+ plasmacytoid DC | Yes. Small populations in LP | Yes ILF but rare | [93,95] | ||
| CD11b+ F4/80+ | Yes. Intraepithelial lymphocytes (IEL) and lamina propria lymphocytes (LPL) | Mainly LPL, IEL are scarce | [110,193] | ||
| Macrophages | |||||
| CX3CR1+ | Yes Production of transepithelial identified. Higher frequencies in the jejunum. Fewer in ileum | Yes Production of transepithelial dendrites uncertain | [95,106-108,194,195] | ||
| Other innate cells | |||||
| Eosinophil | High numbers in steady state located below epithelium and in LP. Highest numbers are found in duodenum. High numbers of CD22+ eosinophils | High numbers in steady state located below epithelium and in LP. Low numbers of CD22+ eosinophils | [130-133,137,196] | ||
| Basophils | LP following infection, draining lymph node | Draining lymph node | [121-123] | ||
| Mast cells | Mucosal mast cells located close to the epithelium | Mucosal mast cells located close to the epithelium | [140] | ||
| NK cells | IEL and lymph nodes (LN) | IEL unknown, LN | [149,197] | ||
| NKT cells | IEL | IEL, LPL | [189,191,192] | ||
| Innate lymphoid cells | Nuocytes: Epithelium 0.2% of total cells and mesenteric lymph node (MLN) | Unknown | [151] | ||
| Innate type 2 helper (Ih2): MLN peritoneum | Unknown | [153] | |||
| Multipotent progenitor (MPP) type2: PP, MLN | Caecal patch | [155] | |||
| Natural helper cell: fat associated lymphoid cluster in helminth infection | Unknown | [198] | |||
| T cells IEL cells | 1 T cell per 5-10 epithelial cells | 1 T cell per 40 epithelial cells | [176] | ||
| Proximal | Mid | Distal | [199] | ||
| αβ T cells | 59% | 60% | 78% | 64% | |
| DN | 1% | 1% | 1% | 20% | |
| CD4 | 6% | 7% | 15% | 7% | |
| CD8αα | 43% | 32% | 20% | 58% | |
| CD8αβ | 46% | 54% | 57% | 12% | |
| gd T cells | 41% | 40% | 22% | 36% | |
| DN | 15% | 17% | 17% | 69% | |
| CD4 | 0.15% | 0.2% | 0.3% | 0.2% | |
| CD8αα | 83% | 82% | 82% | 30% | |
| CD8αβ | 0.3% | 0.5% | 0.4% | 0.2% | |
| LPL cells | |||||
| Th17 | 10%-15% of CD4+ cells | 2%-3% of CD4+ cells | [83] | ||
| Regulatory T cells | 50% of regulatory T cells are Tr1-like cells, 50% are FoxP3+ cells | Almost devoid of Tr1-like cells and 90% of regulatory T cells are Foxp3+ | [182] | ||
| B cells | |||||
| IgA+ | Yes LP and in Peyer’s patches | Found in small numbers | |||
DCs can be classified as conventional, inflammatory or plasmacytoid based upon the differential expression of key surface markers and their functional characteristics. There are clear regional differences of DC subtypes throughout the GIT, which may be of functional significance. Conventional lymphoid resident DC (CD11chigh, MHCIIhigh CD64-)[90] do not migrate or present antigen to T cells within their local environment and can be grouped into CD8α+, CD11b+ and CD8α-CD11b- DCs which within the small intestine are located in Peyer’s patches[91,92]. In the lamina propria (LP) of the small intestine CD11b+CD8α- DCs are the most abundant[93]. CD8α-CD11b- DCs have been identified in regions of the large intestine[94], although CD11chiCD11b-CD8α+ lymphoid DCs have not been identified in homeostasis[93,94]. Furthermore, whereas in the small intestine, DC are readily observed near the epithelium and are able to rapidly respond to damage or infection, in the large intestine DCs are rare in the LP and need to be recruited to the site of infection or injury[94,95].
Interestingly within the small intestine, approximately 80% of the CD11chi LP DC express CD103[93] which have been shown to induce FoxP3+ Treg development[96,97] and also the up-regulation of gut homing receptors on lymphocytes which is in part mediated by retinoic acid[98-101]. Although this function was originally thought to be unique to DCs of the small intestine, DCs in the large intestine can also induce expression of the GIT homing receptor α4β7 on T cells[102]. To add further complexity, CD103+ DCs can be divided into CD103+ CD11b+ and CD103+ CD11b- subtypes with the latter being more abundant in the large intestine[103]. Conventional CD103+ CD11b+CD8α- DC have a migratory phenotype and are thought to sample antigen in the small intestine under steady state conditions and contribute to immune tolerance. They are thought to lack expression of the macrophage markers CX3CR1 (Fracktalkine receptor) and F4/80 and are rare in the steady state large intestine[95].
The behaviour of monocyte-derived inflammatory DCs appears to be comparable in both the small and large intestine. They accumulate in the large and/or small intestine in infection[104] and inflammation[105], for example in T cell-mediated colitis and Toxoplasma gondii infection. In the small intestine plasmocytoid B220+ CD11cintermediate DCs are found in the LP in relatively low numbers but higher than that seen in the large intestine where they primarily reside in lymphoid follicles[94], representing less that 5% of DCs.
The identity of cells expressing CX3CR1 and their functional significance is again controversial. In the murine small intestine and in particular the jejunum, CX3CR1+ cells can produce transepithelial dendrites that extend through the epithelial barrier and directly sample luminal contents[106-108]. However, the phenomenon of transepithelial dendrites in the large intestine may be a rare or transient event[95]. It was originally thought that these cells were static in the epithelial layer although more recent data has suggested that these CX3CR1hi cells are also capable of migration[109]. Recently it has been demonstrated that CX3CR1+ cells are a heterogeneous population expressing varying levels of CX3CR1[90] calling for a re-evaluation of the data surrounding CX3CR1+ cells, in particular, analysis of the expression levels of CX3CR1 and CD64 to distinguish between macrophages and DC.
In addition to CX3CR1, macrophages can be further defined by expression of CD11b, F4/80 and CD64[90]. In the large intestine CD11b+F4/80+ macrophages predominantly reside within the LP and are rarely found associated with the epithelium in naïve animals[110] whereas, in the small intestine macrophages are found closely associated with overlaying enterocytes[87]. In the resting mouse colon, most F4/80+CD64+CD11b+ macrophages express unusually high levels of CX3CR1, but there is also a significant population of CX3CR1int cells that expands preferentially in inflammation. It has recently been determined that these cells represent stages in a single differentiation continuum from lymphocyte antigen 6C monocytes to mature CX3CR1hi macrophages[90,111]. Importantly, there are no known differences in macrophage subset proportions between the small and large intestines[111]. Macrophages exist in different activation states including classical and alternative, based upon the cytokines they are exposed to[112]. Alternatively activated macrophages have been shown to be important in small intestine pathogens such as the helminth Heligmosomoides polygyrus[113] but do not appear to play a direct role in the response to large intestine parasite T. muris[114]. Whether this contrast is due to the different helminths or a differential role in the small vs the large intestine is not known. It will be interesting to see if differences between the small and large intestinal monocyte/macrophage lineages become apparent as more research is done in this emerging area of research.
Neutrophils are an essential component of innate immunity and host defence. They initiate innate immunity as well as directly kill pathogens via the release of extracellular “traps”[115], contribute to inflammation by producing interleukin (IL)-17 and IL-23[116], and may act as professional antigen presenting cells in promoting Th1 and Th17 differentiation[115]. Neutrophils are rare in the intestine although their expansion in response to infection or injury is a hallmark feature of GIT diseases including inflammatory bowel disease (IBD)[117], colorectal carcinoma[118], coeliac disease and infection[119]. The microbiota may play a role in neutrophil expansion in ulcerative colitis and infection[117,120]. Data on regional differences along the GIT are lacking. Given the importance of neutrophils in effector immunity and their prominence in chronic inflammatory conditions it would be important to know if there are regional or functional differences in the neutrophils along the GIT that are of relevance to mucosal immunity and injury.
Basophils are uncommon or absent in the small intestinal mucosa, although they accumulate in high numbers in the LP of the small intestine following Nippostrongylus brasiliensis (N. brasiliensis) infection[121]. Furthermore, they are recruited to the draining lymph node during helminth infections and have a role in the development of type 2 responses[122,123]. The presence of basophils in the large intestine is not known, although one study suggests a role for basophils in the expulsion of the large intestinal parasite T. muris[123] implying the possibility of their presence or (transient) recruitment to the large intestine[123]. Several studies also suggest that basophils have the ability to present antigen[123,124]. Given the potential importance of basophils both in potentiating type 2 immunity it would be interesting to determine whether there are differences in basophil function along the GIT.
Eosinophils have multiple roles including supporting plasma cell function[125], recruitment of DCs[126] and regulation of obesity[127] with the GIT containing the largest number of eosinophils in the body under steady state conditions[128,129]. Within the GIT the duodenum contains the largest population of eosinophils[130]. In both the small and large intestine eosinophils reside primarily in the LP and submucosa with some being detected in the villi of the small intestine[130]. Eosinophilia is seen in response to infection and in inflammation of both the large[131,132] and small intestine[133]. Circulating eosinophils are recruited to the GIT by different mechanisms with steady state recruitment to the large intestine dependent on I-CAM1[134] and small intestine recruitment involving α4β7[135]. Additionally during inflammation, β7 and α4β1 integrins are important in recruitment to the small and large intestine, respectively[136]. Eosinophil populations within the GIT can be distinguished phenotypically with CD22+ eosinophils being most frequent in the jejunum and rarest in the large intestine[137]. The functional significance of these phenotypic variants is however not known although the increased frequency of eosinophils in the small vs the large intestine implies they may be of greater functional significance in this region of the GIT, at least in the steady state.
Mast cells are major effector cells in the immune response to infection, driving pro-inflammatory responses via the release of inflammatory mediators and recruitment of other immune cells[138,139]. They are long-lived cells and enriched at barrier surfaces including the skin and intestinal mucosa[138]. Mast cells are resident cells in both the large and small intestine and mastocytosis is often a feature of GIT infection and inflammation. Two types of mast cell have been identified in the murine small and large intestine based upon differential expression of mast cell proteases (MCP): Mucosal m-MCP1+ and m-MCP2+ cells that reside close to the epithelium and connective tissue mast cells that express mMCP4, mMCP5, mMCP6 and mMCP7 that are within the submucosa[140]. Mucosal mast cells produce lower levels of histamine and high amounts of cysteinyl leukotrienes compared with connective mast cells. Mast cells are critical cells in the expulsion of the small intestinal helminth Trichinella spiralis[141] whereas their role in resistance to the related large intestinal dwelling helminth T. muris infection is less clear[139,142] and presumably reflects functional differences between mast cells in the small and large intestine. Mast cells may also play a role in the pathophysiology of food allergy, cancer, irritable bowel syndrome and inflammatory bowel disease[143,144]. Mast cells may also contribute to altering intestinal permeability. It is known that mucosal mast cells can enhance epithelial barrier permeability in the small intestine[145] and mastocytosis is a feature of IBD, a hallmark feature of which is increased intestinal permeability. However, in the large intestine, which has a higher baseline epithelial resistance, a role for mast cells in mediating increased permeability is less likely[146]. Whether, these conflicting observations in permeability reflect the fact that mast cells are functionally different along the GIT or there are differences in mast cell subset function in homeostasis vs disease is not known, and is worthy of further investigation.
ILCs are a newly identified member of the lymphoid lineage that are capable of producing type 1, type 2 and Th17 cytokines yet do not express any of the cell surface markers associated with other immune cell lineages[147,148]. Three groups of ILCs have been described based primarily upon their cytokine profiles.
Group 1 ILCs produce Interferon (IFN)-γ[147] with the prototypical member being Natural killer (NK) cells that comprise about 2% of small intestinal epithelia-associated lymphocytes in both lymphocyte-replete and -deficient mice[149]. Although NK cells increase in the draining lymph nodes in response to infection in the large intestine[149] their exact number and distribution in the large intestine are currently unknown. As well as NK cells IFNγ-producing non-cytotoxic ILC1s have been described which are more prevalent in the small intestine than the large intestine[150].
Group 2 ILCs produce Th2 associated cytokines following stimulation and require IL-7 for their development[147] and include natural helper cells, nuocytes and IH2 cells. However, it is not clear if these populations represent one cell type or are distinct and stable subsets of group 2 ILCs[147]. All of these ILCs are scarce in naive mice[151] therefore differences between intestinal regions under steady state conditions are hard to analyse. The ILC populations are elicited after small intestinal helminth infection[151-153] with, nuocytes being the predominant IL-13 expressing cell type within the mesenteric lymph node and small intestine after N. brasiliensis infection[151]. Similarly, in the large intestine, nuocytes are the main IL-13 producing cell type during experimental colitis[154]. Differences in the infection models driving inflammation however make it difficult to compare the populations directly following infection in the small and large intestine.
A multipotent progenitor like cell cell has been described which gives rise to cells of the monocytes/macrophage and granulocyte lineage[155]. These cells expand under the influence of IL-25 and are found within Peyer’s patches of the small intestine and the caecal patch of the large intestine[156]. In the small intestine these cells expand in response to N. brasiliensis infection and in the large intestine they expand in response to T. muris[156], and have been shown to have important roles in promoting Th2 cytokine responses to helminth infections in both the small and large intestine.
Group 3 ILCs produce IL-17A and/or IL-22[147] with the prototypical members being LTi cells. Recently, another subset of group 3 ILCs has been identified that is crucial for the IL-22-mediated innate immune response against intestinal bacterial pathogens including Citrobacter rodentium[157]. Depletion of these IL-22 producing cells resulted in systemic inflammation and bacterial dissemination which could only be rescued by exogenous IL-22[143]. This population is primarily located in the LP of the intestine being three times more frequent in the small intestine than the large intestine[157].
Based upon the experimental evidence obtained to date, ILCs may play an important role in GIT barrier defence and perhaps intestinal inflammation; however more work needs to be done in this newly identified population of cells and differences between the small and large intestine are still emerging.
B cells play a key role in adaptive mucosal immune responses via antibody production. In the GIT, IgA secreting plasma cells represent the major pathway to blocking pathogen invasion. The GIT contains the largest number of IgA-producing plasma cells, which collectively produce and secrete 3-5 mg of antibody into the lumen each day[158]. IgA producing plasma cells are enriched in the LP of the small intestine but are found in smaller numbers in the large intestine LP (our unpublished observations). There are regional differences in the distribution of B cell follicles that contain IgA producing B cells. In the small intestine they reside within the Peyer’s patches whereas in the large intestine they reside in isolated lymphoid follicles, although they are rarely seen, further emphasising differences in antigen presentation across the GIT. The mechanisms driving IgA production in the small and large intestine may overlap, particularly with regard to the role of the intestinal microbiota. In the small intestine the microbiota elicits IgA production, which leads to the development of gut-associated lymphoid tissue (GALT)[159,160]. Conversely, IgA can supress the growth of commensal bacteria[161]. In the large intestine, Bacteroides acidifaciens, a member of the microbiota has been shown to promote IgA production[162] but its not known whether this commensal drives GALT development.
In the GIT T cells are compartmentalised into the GALT, the LP, and the epithelium where they reside as epithelia-associated lymphocytes or intestinal intraepithelial lymphocytes (iIEL)[163]. Different mucosal T cell subsets reside in the different compartments of the GIT and there are striking differences in the distribution and function of iIEL subsets in the small vs large intestine iIEL, which has been extensively reviewed elsewhere[164-167] and will not be discussed in any detail here.
iIELs that express the integrin CD103[168-172] are in close contact with the intestinal epithelium and contribute to the maintenance of barrier integrity[170,173-175]. They are highly heterogeneous, comprising many different subsets that exhibit distinct regional differences throughout the GIT (Table 1). iIELs can be divided into two main groups, induced IELs (type A) that arise from conventional T cells and become activated in the periphery, and natural iIELs (type B) that develop in the thymus[164,165,176]. Both types of iIELs can be further divided based on the expression of the αβ or γδ T Cell Receptor (TCR) and on expression of CD4 and CD8 (Table 1). The distribution of iIELs in the small intestine is approximately one IEL per 5-10 epithelial cells vs one IEL per 40 epithelial cells in the large intestine[177,178]. A key difference between populations of iIELs throughout the GIT is the ratio between naïve and activated to memory IELs, with the small intestine containing a smaller proportion of naïve cells than the large intestine[179]. The function of iIELS are not fully elucidated however in addition to immune and cytotoxic roles, they play a vital role in small intestinal epithelial barrier maintenance[168] and defence by promoting epithelial tight junctions and Paneth cell anti-microbial production[180]. It is not known whether colonic iIEL have analogous functions to these small intestinal populations.
Compared to iIELs, the distribution of LP T cell subsets throughout the GIT is less well characterised. TCRαβ+ CD4+ cells are found in greater numbers in the LP of the both the small and large intestine in comparison to the epithelium[163]. In addition, classical CD4+ Th subsets are found in the LP with Th17 and Treg cells present in high numbers[83]. Indeed the intestine has the highest levels of Th17, Th22 and FoxP3+ cells in the body[181]. This enrichment of Th17 cells is particularly evident in the small intestine where approximately 10%-15% of CD4+ cells in healthy mice are IL-17+ compared to 2-3% in the large intestine[182]. The GIT is also enriched in regulatory T cells (Treg) that can be subdivided into Foxp3- Tr1-like cells that produce large amounts of IL-10, and “natural” Foxp3+ Treg. In the LP of the small intestine approximately 50% of regulatory T cells have a Tr1-like phenotype, in stark contrast to the large intestine, which is relatively devoid of Tr1-like cells. Instead, virtually all Treg cells in the large intestine are Foxp3+ (89%) in comparison to 54%-58% of the Treg cells in the small intestine[183]. Indeed, it has become increasingly clear that mechanisms of tolerance differ greatly between the small and large intestine with differential induction of Foxp3+ Tregs and Tr1 cells. Nevertheless, more detailed studies are needed to understand the contribution of dietary and bacterial antigens, antigen presenting cells and tissue resident cells in the induction of Tr1 and Treg cells and the differences between the small and large intestine[184].
In addition to differences in T cell composition throughout the GIT there are differences in the mechanisms of T cell homing to the gut. This is believed to be one of the key ways the tissue-specificity of T cells are maintained[181]. The adhesion molecules expressed by small intestine homing T cells are well characterised with LFA-1, α4β7, αEβ7 (CD103) and CCR9 and retinoic acid playing an important role[100,101]. In contrast, the mechanisms involved in the recruitment of T cells to the large intestine are less well defined. It has been demonstrated that migration of T cells to the large intestine is dependent on α4β7 but independent of CCR9[185,186] and retinoic acid[187]. Recent work has identified a key role for transforming growth factor-β, combined with unknown factors, in induction of GPR15 expression to drive Treg cell homing to the large but not small intestine[188]. Furthermore, CD4 T cell recruitment to the large intestine has been shown to be chemokine dependent although the chemokines involved have yet to be identified[93]. The differences in homing to the large and small intestine still needs to be further clarified but research in this area has been hampered by a lot of overlap between the chemokines and their receptors.
Natural killer T (NKT) cells are a subset of T cells expressing the TCR/CD3 complex, NK.1 and Ly-49[189] that are CD1d-restricted T cells that respond to lipid rather than protein antigens. NKT cells have both regulatory and effector functions and play critical roles in the regulation of immune responses in many disease settings including cancer[190]. NKT subsets do differ in frequency between the large and small intestine and have been extensively reviewed by van Dieren et al[191]. Higher numbers of NKT cells are found in the large intestine (up to 11% are epithelia-associated and 7% LP associated) compared to the small intestine (2%-6% of epithelia-associated lymphocytes)[191,192].
We have reviewed, for the first time, the broad regional diversity of the cells, barrier and immune system of the small and large intestine. From the external environment and the microbiome to the cells of the innate and adaptive immune system the small and large intestine have clear differences that have marked effects on functionality and responses to commensal organisms as well as pathogens. Given the significance of the small and large intestine not only functionally but also in terms of disease it is important to understand how these organs and the cells within them function, and how and why they are different. A greater understanding of the immune system-microbiome interactions in the small and large intestine should reveal answers to important unanswered questions about disease susceptibility and mechanisms of inflammation that are specific to the small or large intestine. Specifically, questions to be addressed include: identifying mechanisms of epithelial crosstalk with the microbiome and whether it is the same in small and large intestine, how the microbiome promotes immune function when access to the LP is limited. What the role of innate cells like eosinophils, ILCs and mast cells are in the regions of the gut in homeostasis and inflammation and how do they differ across the GIT, e.g., is CD22 a functional marker on small intestinal eosinophils? How do colonic and small intestinal macrophage and dendritic cell subsets differ functionally across the GIT in homeostasis and infection. A widening awareness of the differences between the small and large intestine can also influence the development of therapeutics, e.g., in instances where probiotics may be beneficial for the treatment of diseases of the small intestinal but not those of the large intestine. In addition, in Crohn’s disease which can affect both the small and large intestine we may want to look at using specialised combination therapy in order to tackle the disease in both sites instead of the use of one drug which may be efficacious in one gut region. Although this review has focused on murine studies, some analogous data has been generated for humans but there is even more which remains unknown, especially in homeostasis, in the human GIT. It is clear that we can no longer make assumptions about similarities in function of our gut cells along the GIT.
P- Reviewer: Servin AL, Zouiten-Mekki L S- Editor: Gou SX L- Editor: A E- Editor: Wang CH
| 1. | Standring S, Gray HA. Gray’s anatomy: the anatomical basis of clinical practice. 40th ed. Edinburgh: Churchill Livingstone 2008; . |
| 2. | Axelsson LG, Landström E, Goldschmidt TJ, Grönberg A, Bylund-Fellenius AC. Dextran sulfate sodium (DSS) induced experimental colitis in immunodeficient mice: effects in CD4(+) -cell depleted, athymic and NK-cell depleted SCID mice. Inflamm Res. 1996;45:181-191. [PubMed] |
| 3. | Foong JP, Nguyen TV, Furness JB, Bornstein JC, Young HM. Myenteric neurons of the mouse small intestine undergo significant electrophysiological and morphological changes during postnatal development. J Physiol. 2012;590:2375-2390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 62] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 4. | Howlader NNA, Krapcho M, Neyman N, Aminou R, Altekruse SF, Kosary CL, Ruhl J, Tatalovich Z, Cho H, Mariotto A. EER Cancer Statistics Review. Surveillance Epidemiology and End Results 2009. Available from: http://seer.cancer.gov/csr/1975_2009_pops09/. |
| 5. | Shen L, Turner JR. Role of epithelial cells in initiation and propagation of intestinal inflammation. Eliminating the static: tight junction dynamics exposed. Am J Physiol Gastrointest Liver Physiol. 2006;290:G577-G582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 127] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 6. | Rescigno M. The intestinal epithelial barrier in the control of homeostasis and immunity. Trends Immunol. 2011;32:256-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 226] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 7. | Palazzo M, Balsari A, Rossini A, Selleri S, Calcaterra C, Gariboldi S, Zanobbio L, Arnaboldi F, Shirai YF, Serrao G. Activation of enteroendocrine cells via TLRs induces hormone, chemokine, and defensin secretion. J Immunol. 2007;178:4296-4303. [PubMed] |
| 8. | Leedham SJ, Brittan M, McDonald SA, Wright NA. Intestinal stem cells. J Cell Mol Med. 2005;9:11-24. [PubMed] |
| 9. | Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003-1007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3854] [Cited by in RCA: 4324] [Article Influence: 240.2] [Reference Citation Analysis (0)] |
| 10. | Marshman E, Booth C, Potten CS. The intestinal epithelial stem cell. Bioessays. 2002;24:91-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 426] [Cited by in RCA: 413] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 11. | Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415-418. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2119] [Cited by in RCA: 1928] [Article Influence: 137.7] [Reference Citation Analysis (0)] |
| 12. | Jang MH, Kweon MN, Iwatani K, Yamamoto M, Terahara K, Sasakawa C, Suzuki T, Nochi T, Yokota Y, Rennert PD. Intestinal villous M cells: an antigen entry site in the mucosal epithelium. Proc Natl Acad Sci USA. 2004;101:6110-6115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 345] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 13. | Chang SY, Cha HR, Uematsu S, Akira S, Igarashi O, Kiyono H, Kweon MN. Colonic patches direct the cross-talk between systemic compartments and large intestine independently of innate immunity. J Immunol. 2008;180:1609-1618. [PubMed] |
| 14. | Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9:356-368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 725] [Cited by in RCA: 875] [Article Influence: 62.5] [Reference Citation Analysis (0)] |
| 15. | El Aidy S, van Baarlen P, Derrien M, Lindenbergh-Kortleve DJ, Hooiveld G, Levenez F, Doré J, Dekker J, Samsom JN, Nieuwenhuis EE. Temporal and spatial interplay of microbiota and intestinal mucosa drive establishment of immune homeostasis in conventionalized mice. Mucosal Immunol. 2012;5:567-579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 162] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 16. | Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012;12:503-516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 627] [Cited by in RCA: 662] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 17. | Muniz LR, Knosp C, Yeretssian G. Intestinal antimicrobial peptides during homeostasis, infection, and disease. Front Immunol. 2012;3:310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 171] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 18. | Shi J. Defensins and Paneth cells in inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:1284-1292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 40] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 19. | Cash HL, Whitham CV, Hooper LV. Refolding, purification, and characterization of human and murine RegIII proteins expressed in Escherichia coli. Protein Expr Purif. 2006;48:151-159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 51] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 20. | Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1124] [Cited by in RCA: 1072] [Article Influence: 76.6] [Reference Citation Analysis (0)] |
| 21. | Ayabe T, Satchell DP, Wilson CL, Parks WC, Selsted ME, Ouellette AJ. Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nat Immunol. 2000;1:113-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 776] [Cited by in RCA: 780] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 22. | Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjöberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76-83. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 835] [Cited by in RCA: 908] [Article Influence: 56.8] [Reference Citation Analysis (0)] |
| 23. | Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci USA. 2008;105:20858-20863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 829] [Cited by in RCA: 763] [Article Influence: 44.9] [Reference Citation Analysis (0)] |
| 24. | Petnicki-Ocwieja T, Hrncir T, Liu YJ, Biswas A, Hudcovic T, Tlaskalova-Hogenova H, Kobayashi KS. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc Natl Acad Sci USA. 2009;106:15813-15818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 421] [Cited by in RCA: 455] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 25. | O‘Neil DA, Porter EM, Elewaut D, Anderson GM, Eckmann L, Ganz T, Kagnoff MF. Expression and regulation of the human beta-defensins hBD-1 and hBD-2 in intestinal epithelium. J Immunol. 1999;163:6718-6724. [PubMed] |
| 26. | Koon HW, Shih DQ, Chen J, Bakirtzi K, Hing TC, Law I, Ho S, Ichikawa R, Zhao D, Xu H. Cathelicidin signaling via the Toll-like receptor protects against colitis in mice. Gastroenterology. 2011;141:1852-1863.e1-3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 93] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 27. | Burger-van Paassen N, Loonen LM, Witte-Bouma J, Korteland-van Male AM, de Bruijn AC, van der Sluis M, Lu P, Van Goudoever JB, Wells JM, Dekker J. Mucin Muc2 deficiency and weaning influences the expression of the innate defense genes Reg3β, Reg3γ and angiogenin-4. PLoS One. 2012;7:e38798. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 65] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 28. | He W, Wang ML, Jiang HQ, Steppan CM, Shin ME, Thurnheer MC, Cebra JJ, Lazar MA, Wu GD. Bacterial colonization leads to the colonic secretion of RELMbeta/FIZZ2, a novel goblet cell-specific protein. Gastroenterology. 2003;125:1388-1397. [PubMed] |
| 29. | Forman RA, deSchoolmeester ML, Hurst RJ, Wright SH, Pemberton AD, Else KJ. The goblet cell is the cellular source of the anti-microbial angiogenin 4 in the large intestine post Trichuris muris infection. PLoS One. 2012;7:e42248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 50] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 30. | Artis D, Grencis RK. The intestinal epithelium: sensors to effectors in nematode infection. Mucosal Immunol. 2008;1:252-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 89] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 31. | Ménard S, Förster V, Lotz M, Gütle D, Duerr CU, Gallo RL, Henriques-Normark B, Pütsep K, Andersson M, Glocker EO. Developmental switch of intestinal antimicrobial peptide expression. J Exp Med. 2008;205:183-193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 112] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 32. | Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14:685-690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1062] [Cited by in RCA: 1082] [Article Influence: 90.2] [Reference Citation Analysis (0)] |
| 33. | Salzman NH. Microbiota-immune system interaction: an uneasy alliance. Curr Opin Microbiol. 2011;14:99-105. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 70] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 34. | Schauber J, Svanholm C, Termén S, Iffland K, Menzel T, Scheppach W, Melcher R, Agerberth B, Lührs H, Gudmundsson GH. Expression of the cathelicidin LL-37 is modulated by short chain fatty acids in colonocytes: relevance of signalling pathways. Gut. 2003;52:735-741. [PubMed] |
| 35. | Meyer-Hoffert U, Hornef MW, Henriques-Normark B, Axelsson LG, Midtvedt T, Pütsep K, Andersson M. Secreted enteric antimicrobial activity localises to the mucus surface layer. Gut. 2008;57:764-771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 221] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 36. | McGuckin MA, Lindén SK, Sutton P, Florin TH. Mucin dynamics and enteric pathogens. Nat Rev Microbiol. 2011;9:265-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 907] [Cited by in RCA: 1033] [Article Influence: 73.8] [Reference Citation Analysis (0)] |
| 37. | Stappenbeck TS. Paneth cell development, differentiation, and function: new molecular cues. Gastroenterology. 2009;137:30-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 38. | Cani PD, Possemiers S, Van de Wiele T, Guiot Y, Everard A, Rottier O, Geurts L, Naslain D, Neyrinck A, Lambert DM. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58:1091-1103. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1735] [Cited by in RCA: 1884] [Article Influence: 117.8] [Reference Citation Analysis (1)] |
| 39. | Atuma C, Strugala V, Allen A, Holm L. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am J Physiol Gastrointest Liver Physiol. 2001;280:G922-G929. [PubMed] |
| 40. | Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA. 2008;105:15064-15069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1326] [Cited by in RCA: 1526] [Article Influence: 89.8] [Reference Citation Analysis (1)] |
| 41. | Hansson GC. Role of mucus layers in gut infection and inflammation. Curr Opin Microbiol. 2012;15:57-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 337] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 42. | Shan M, Gentile M, Yeiser JR, Walland AC, Bornstein VU, Chen K, He B, Cassis L, Bigas A, Cols M. Mucus enhances gut homeostasis and oral tolerance by delivering immunoregulatory signals. Science. 2013;342:447-453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 408] [Cited by in RCA: 481] [Article Influence: 40.1] [Reference Citation Analysis (0)] |
| 43. | Loonen LM, Stolte EH, Jaklofsky MT, Meijerink M, Dekker J, van Baarlen P, Wells JM. REG3γ-deficient mice have altered mucus distribution and increased mucosal inflammatory responses to the microbiota and enteric pathogens in the ileum. Mucosal Immunol. 2014;7:939-947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 139] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 44. | Holmén Larsson JM, Thomsson KA, Rodríguez-Piñeiro AM, Karlsson H, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. III. Gastrointestinal Muc5ac and Muc2 mucin O-glycan patterns reveal a regiospecific distribution. Am J Physiol Gastrointest Liver Physiol. 2013;305:G357-G363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 154] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 45. | Juge N. Microbial adhesins to gastrointestinal mucus. Trends Microbiol. 2012;20:30-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 202] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 46. | Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955-1959. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 994] [Cited by in RCA: 879] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 47. | Kashyap PC, Marcobal A, Ursell LK, Smits SA, Sonnenburg ED, Costello EK, Higginbottom SK, Domino SE, Holmes SP, Relman DA. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc Natl Acad Sci USA. 2013;110:17059-17064. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 209] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 48. | Bergstrom KS, Kissoon-Singh V, Gibson DL, Ma C, Montero M, Sham HP, Ryz N, Huang T, Velcich A, Finlay BB. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog. 2010;6:e1000902. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 397] [Cited by in RCA: 471] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 49. | Hasnain SZ, Wang H, Ghia JE, Haq N, Deng Y, Velcich A, Grencis RK, Thornton DJ, Khan WI. Mucin gene deficiency in mice impairs host resistance to an enteric parasitic infection. Gastroenterology. 2010;138:1763-1771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 159] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 50. | Van der Sluis M, De Koning BAE, De Bruijn ACJM, Velcich A, Meijerink JPP, Van Goudoever JB, Baller HA, Dekker J, Van Seuningen I, Renes IB. Muc2-Deficient Mice Spontaneously Develop Colitis, Indicating That MUC2 Is Critical for Colonic Protection. Gastroenterology. 2006;131:117-129. |
| 51. | McAuley JL, Linden SK, Png CW, King RM, Pennington HL, Gendler SJ, Florin TH, Hill GR, Korolik V, McGuckin MA. MUC1 cell surface mucin is a critical element of the mucosal barrier to infection. J Clin Invest. 2007;117:2313-2324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 323] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 52. | McGuckin MA, Every AL, Skene CD, Linden SK, Chionh YT, Swierczak A, McAuley J, Harbour S, Kaparakis M, Ferrero R. Muc1 mucin limits both Helicobacter pylori colonization of the murine gastric mucosa and associated gastritis. Gastroenterology. 2007;133:1210-1218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 153] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 53. | Hasnain SZ, Evans CM, Roy M, Gallagher AL, Kindrachuk KN, Barron L, Dickey BF, Wilson MS, Wynn TA, Grencis RK. Muc5ac: a critical component mediating the rejection of enteric nematodes. J Exp Med. 2011;208:893-900. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 245] [Cited by in RCA: 236] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 54. | Turner JE, Stockinger B, Helmby H. IL-22 mediates goblet cell hyperplasia and worm expulsion in intestinal helminth infection. PLoS Pathog. 2013;9:e1003698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 121] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 55. | Artis D, Humphreys NE, Bancroft AJ, Rothwell NJ, Potten CS, Grencis RK. Tumor necrosis factor alpha is a critical component of interleukin 13-mediated protective T helper cell type 2 responses during helminth infection. J Exp Med. 1999;190:953-962. [PubMed] |
| 56. | Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207-214. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9292] [Cited by in RCA: 8039] [Article Influence: 618.4] [Reference Citation Analysis (2)] |
| 57. | Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59-65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9101] [Cited by in RCA: 7819] [Article Influence: 521.3] [Reference Citation Analysis (4)] |
| 58. | Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915-1920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3394] [Cited by in RCA: 3537] [Article Influence: 176.9] [Reference Citation Analysis (5)] |
| 59. | Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355-1359. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3709] [Cited by in RCA: 3187] [Article Influence: 167.7] [Reference Citation Analysis (0)] |
| 60. | Sina C, Lipinski S, Gavrilova O, Aden K, Rehman A, Till A, Rittger A, Podschun R, Meyer-Hoffert U, Haesler R. Extracellular cathepsin K exerts antimicrobial activity and is protective against chronic intestinal inflammation in mice. Gut. 2013;62:520-530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 61. | Swidsinski A, Loening-Baucke V, Lochs H, Hale LP. Spatial organization of bacterial flora in normal and inflamed intestine: a fluorescence in situ hybridization study in mice. World J Gastroenterol. 2005;11:1131-1140. [PubMed] |
| 62. | Sommer F, Bäckhed F. The gut microbiota--masters of host development and physiology. Nat Rev Microbiol. 2013;11:227-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2011] [Cited by in RCA: 2390] [Article Influence: 199.2] [Reference Citation Analysis (0)] |
| 63. | Berg RD. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4:430-435. [PubMed] |
| 64. | Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426-429. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 381] [Cited by in RCA: 459] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 65. | Nava GM, Friedrichsen HJ, Stappenbeck TS. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 2011;5:627-638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 201] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 66. | Schwarz R, Kaspar A, Seelig J, Künnecke B. Gastrointestinal transit times in mice and humans measured with 27Al and 19F nuclear magnetic resonance. Magn Reson Med. 2002;48:255-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 78] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 67. | Kashyap PC, Marcobal A, Ursell LK, Larauche M, Duboc H, Earle KA, Sonnenburg ED, Ferreyra JA, Higginbottom SK, Million M. Complex interactions among diet, gastrointestinal transit, and gut microbiota in humanized mice. Gastroenterology. 2013;144:967-977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 318] [Cited by in RCA: 341] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 68. | Musso G, Gambino R, Cassader M. Interactions between gut microbiota and host metabolism predisposing to obesity and diabetes. Annu Rev Med. 2011;62:361-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 426] [Cited by in RCA: 443] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 69. | Zoetendal EG, Raes J, van den Bogert B, Arumugam M, Booijink CC, Troost FJ, Bork P, Wels M, de Vos WM, Kleerebezem M. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012;6:1415-1426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 436] [Cited by in RCA: 511] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 70. | Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213-223. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2421] [Cited by in RCA: 2187] [Article Influence: 128.6] [Reference Citation Analysis (0)] |
| 71. | Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1496] [Cited by in RCA: 1587] [Article Influence: 105.8] [Reference Citation Analysis (0)] |
| 72. | Murphy EF, Cotter PD, Healy S, Marques TM, O’Sullivan O, Fouhy F, Clarke SF, O’Toole PW, Quigley EM, Stanton C. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut. 2010;59:1635-1642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 647] [Cited by in RCA: 708] [Article Influence: 47.2] [Reference Citation Analysis (0)] |
| 73. | Booijink CC, El-Aidy S, Rajilić-Stojanović M, Heilig HG, Troost FJ, Smidt H, Kleerebezem M, De Vos WM, Zoetendal EG. High temporal and inter-individual variation detected in the human ileal microbiota. Environ Microbiol. 2010;12:3213-3227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 226] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 74. | Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1329] [Cited by in RCA: 1444] [Article Influence: 120.3] [Reference Citation Analysis (0)] |
| 75. | Smelt MJ, de Haan BJ, Bron PA, van Swam I, Meijerink M, Wells JM, Faas MM, de Vos P. Probiotics can generate FoxP3 T-cell responses in the small intestine and simultaneously inducing CD4 and CD8 T cell activation in the large intestine. PLoS One. 2013;8:e68952. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 42] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 76. | Lan JG, Cruickshank SM, Singh JC, Farrar M, Lodge JP, Felsburg PJ, Carding SR. Different cytokine response of primary colonic epithelial cells to commensal bacteria. World J Gastroenterol. 2005;11:3375-3384. [PubMed] |
| 77. | Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14:676-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 589] [Cited by in RCA: 669] [Article Influence: 55.8] [Reference Citation Analysis (0)] |
| 78. | Reading NC, Kasper DL. The starting lineup: key microbial players in intestinal immunity and homeostasis. Front Microbiol. 2011;2:148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 79. | Hansen CH, Nielsen DS, Kverka M, Zakostelska Z, Klimesova K, Hudcovic T, Tlaskalova-Hogenova H, Hansen AK. Patterns of early gut colonization shape future immune responses of the host. PLoS One. 2012;7:e34043. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 225] [Cited by in RCA: 215] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 80. | Mulder IE, Schmidt B, Lewis M, Delday M, Stokes CR, Bailey M, Aminov RI, Gill BP, Pluske JR, Mayer CD. Restricting microbial exposure in early life negates the immune benefits associated with gut colonization in environments of high microbial diversity. PLoS One. 2011;6:e28279. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 103] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 81. | Kamada N, Núñez G. Role of the gut microbiota in the development and function of lymphoid cells. J Immunol. 2013;190:1389-1395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 109] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 82. | Brown EM, Sadarangani M, Finlay BB. The role of the immune system in governing host-microbe interactions in the intestine. Nat Immunol. 2013;14:660-667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 284] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 83. | Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121-1133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3769] [Cited by in RCA: 4121] [Article Influence: 216.9] [Reference Citation Analysis (0)] |
| 84. | Gaboriau-Routhiau V, Rakotobe S, Lécuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677-689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1045] [Cited by in RCA: 1127] [Article Influence: 70.4] [Reference Citation Analysis (0)] |
| 85. | Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, Yagita H, Ishii N, Evans R, Honda K. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808-812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 815] [Cited by in RCA: 864] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 86. | Sonnenberg GF, Artis D. Innate lymphoid cell interactions with microbiota: implications for intestinal health and disease. Immunity. 2012;37:601-610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 212] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 87. | Persson EK, Jaensson E, Agace WW. The diverse ontogeny and function of murine small intestinal dendritic cell/macrophage subsets. Immunobiology. 2010;215:692-697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 38] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 88. | Pabst O, Bernhardt G. The puzzle of intestinal lamina propria dendritic cells and macrophages. Eur J Immunol. 2010;40:2107-2111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 77] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 89. | Qualls JE, Kaplan AM, van Rooijen N, Cohen DA. Suppression of experimental colitis by intestinal mononuclear phagocytes. J Leukoc Biol. 2006;80:802-815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 198] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 90. | Tamoutounour S, Henri S, Lelouard H, de Bovis B, de Haar C, van der Woude CJ, Woltman AM, Reyal Y, Bonnet D, Sichien D. CD64 distinguishes macrophages from dendritic cells in the gut and reveals the Th1-inducing role of mesenteric lymph node macrophages during colitis. Eur J Immunol. 2012;42:3150-3166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 411] [Cited by in RCA: 425] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 91. | Iwasaki A, Kelsall BL. Localization of distinct Peyer’s patch dendritic cell subsets and their recruitment by chemokines macrophage inflammatory protein (MIP)-3alpha, MIP-3beta, and secondary lymphoid organ chemokine. J Exp Med. 2000;191:1381-1394. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 450] [Cited by in RCA: 445] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 92. | Kelsall BL, Strober W. Distinct populations of dendritic cells are present in the subepithelial dome and T cell regions of the murine Peyer’s patch. J Exp Med. 1996;183:237-247. [PubMed] |
| 93. | Chirdo FG, Millington OR, Beacock-Sharp H, Mowat AM. Immunomodulatory dendritic cells in intestinal lamina propria. Eur J Immunol. 2005;35:1831-1840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 179] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 94. | Cruickshank SM, English NR, Felsburg PJ, Carding SR. Characterization of colonic dendritic cells in normal and colitic mice. World J Gastroenterol. 2005;11:6338-6347. [PubMed] |
| 95. | Cruickshank SM, Deschoolmeester ML, Svensson M, Howell G, Bazakou A, Logunova L, Little MC, English N, Mack M, Grencis RK. Rapid dendritic cell mobilization to the large intestinal epithelium is associated with resistance to Trichuris muris infection. J Immunol. 2009;182:3055-3062. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 37] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 96. | Coombes JL, Siddiqui KR, Arancibia-Cárcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757-1764. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2056] [Cited by in RCA: 2199] [Article Influence: 122.2] [Reference Citation Analysis (0)] |
| 97. | Worthington JJ, Czajkowska BI, Melton AC, Travis MA. Intestinal dendritic cells specialize to activate transforming growth factor-β and induce Foxp3+ regulatory T cells via integrin αvβ8. Gastroenterology. 2011;141:1802-1812. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 143] [Cited by in RCA: 138] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 98. | Johansson-Lindbom B, Svensson M, Wurbel MA, Malissen B, Márquez G, Agace W. Selective generation of gut tropic T cells in gut-associated lymphoid tissue (GALT): requirement for GALT dendritic cells and adjuvant. J Exp Med. 2003;198:963-969. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 366] [Cited by in RCA: 366] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 99. | Svensson M, Marsal J, Uronen-Hansson H, Cheng M, Jenkinson W, Cilio C, Jacobsen SE, Sitnicka E, Anderson G, Agace WW. Involvement of CCR9 at multiple stages of adult T lymphopoiesis. J Leukoc Biol. 2008;83:156-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 100. | Mora JR, Bono MR, Manjunath N, Weninger W, Cavanagh LL, Rosemblatt M, Von Andrian UH. Selective imprinting of gut-homing T cells by Peyer’s patch dendritic cells. Nature. 2003;424:88-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 849] [Cited by in RCA: 845] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 101. | Iwata M, Hirakiyama A, Eshima Y, Kagechika H, Kato C, Song SY. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 2004;21:527-538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1148] [Cited by in RCA: 1218] [Article Influence: 58.0] [Reference Citation Analysis (0)] |
| 102. | Jaensson E, Uronen-Hansson H, Pabst O, Eksteen B, Tian J, Coombes JL, Berg PL, Davidsson T, Powrie F, Johansson-Lindbom B. Small intestinal CD103+ dendritic cells display unique functional properties that are conserved between mice and humans. J Exp Med. 2008;205:2139-2149. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 489] [Cited by in RCA: 468] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 103. | Denning TL, Norris BA, Medina-Contreras O, Manicassamy S, Geem D, Madan R, Karp CL, Pulendran B. Functional specializations of intestinal dendritic cell and macrophage subsets that control Th17 and regulatory T cell responses are dependent on the T cell/APC ratio, source of mouse strain, and regional localization. J Immunol. 2011;187:733-747. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 272] [Cited by in RCA: 273] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 104. | Dunay IR, Damatta RA, Fux B, Presti R, Greco S, Colonna M, Sibley LD. Gr1(+) inflammatory monocytes are required for mucosal resistance to the pathogen Toxoplasma gondii. Immunity. 2008;29:306-317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 356] [Cited by in RCA: 343] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 105. | Siddiqui KR, Laffont S, Powrie F. E-cadherin marks a subset of inflammatory dendritic cells that promote T cell-mediated colitis. Immunity. 2010;32:557-567. [PubMed] |
| 106. | Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, Granucci F, Kraehenbuhl JP, Ricciardi-Castagnoli P. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001;2:361-367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1870] [Cited by in RCA: 1821] [Article Influence: 75.9] [Reference Citation Analysis (0)] |
| 107. | Niess JH, Brand S, Gu X, Landsman L, Jung S, McCormick BA, Vyas JM, Boes M, Ploegh HL, Fox JG. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1209] [Cited by in RCA: 1230] [Article Influence: 61.5] [Reference Citation Analysis (0)] |
| 108. | Chieppa M, Rescigno M, Huang AY, Germain RN. Dynamic imaging of dendritic cell extension into the small bowel lumen in response to epithelial cell TLR engagement. J Exp Med. 2006;203:2841-2852. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 541] [Cited by in RCA: 546] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 109. | Diehl GE, Longman RS, Zhang JX, Breart B, Galan C, Cuesta A, Schwab SR, Littman DR. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. 2013;494:116-120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 322] [Cited by in RCA: 374] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 110. | Little MC, Bell LV, Cliffe LJ, Else KJ. The characterization of intraepithelial lymphocytes, lamina propria leukocytes, and isolated lymphoid follicles in the large intestine of mice infected with the intestinal nematode parasite Trichuris muris. J Immunol. 2005;175:6713-6722. [PubMed] |
| 111. | Bain CC, Scott CL, Uronen-Hansson H, Gudjonsson S, Jansson O, Grip O, Guilliams M, Malissen B, Agace WW, Mowat AM. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol. 2013;6:498-510. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 603] [Cited by in RCA: 705] [Article Influence: 58.8] [Reference Citation Analysis (0)] |
| 112. | Mosser DM. The many faces of macrophage activation. J Leukoc Biol. 2003;73:209-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1340] [Cited by in RCA: 1359] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 113. | Anthony RM, Urban JF, Alem F, Hamed HA, Rozo CT, Boucher JL, Van Rooijen N, Gause WC. Memory T(H)2 cells induce alternatively activated macrophages to mediate protection against nematode parasites. Nat Med. 2006;12:955-960. [PubMed] |
| 114. | Bowcutt R, Bell LV, Little M, Wilson J, Booth C, Murray PJ, Else KJ, Cruickshank SM. Arginase-1-expressing macrophages are dispensable for resistance to infection with the gastrointestinal helminth Trichuris muris. Parasite Immunol. 2011;33:411-420. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 115. | Abi Abdallah DS, Denkers EY. Neutrophils cast extracellular traps in response to protozoan parasites. Front Immunol. 2012;3:382. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 116. | Mei J, Liu Y, Dai N, Hoffmann C, Hudock KM, Zhang P, Guttentag SH, Kolls JK, Oliver PM, Bushman FD. Cxcr2 and Cxcl5 regulate the IL-17/G-CSF axis and neutrophil homeostasis in mice. J Clin Invest. 2012;122:974-986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 156] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 117. | Robinson CE, Kottapalli V, D’Astice M, Fields JZ, Winship D, Keshavarzian A. Regulation of neutrophils in ulcerative colitis by colonic factors: a possible mechanism of neutrophil activation and tissue damage. J Lab Clin Med. 1997;130:590-602. [PubMed] |
| 118. | Rao HL, Chen JW, Li M, Xiao YB, Fu J, Zeng YX, Cai MY, Xie D. Increased intratumoral neutrophil in colorectal carcinomas correlates closely with malignant phenotype and predicts patients’ adverse prognosis. PLoS One. 2012;7:e30806. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 158] [Cited by in RCA: 214] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 119. | Bennouna S, Bliss SK, Curiel TJ, Denkers EY. Cross-talk in the innate immune system: neutrophils instruct recruitment and activation of dendritic cells during microbial infection. J Immunol. 2003;171:6052-6058. [PubMed] |
| 120. | Milo LA, Correa-Matos NJ, Donovan SM, Tappenden KA. Neutrophil and small intestinal lymphocyte migration after Salmonella typhimurium infection: impact of fermentable fiber. J Pediatr Gastroenterol Nutr. 2004;39:73-79. [PubMed] |
| 121. | Ohnmacht C, Voehringer D. Basophil effector function and homeostasis during helminth infection. Blood. 2009;113:2816-2825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 143] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 122. | Kim S, Prout M, Ramshaw H, Lopez AF, LeGros G, Min B. Cutting edge: basophils are transiently recruited into the draining lymph nodes during helminth infection via IL-3, but infection-induced Th2 immunity can develop without basophil lymph node recruitment or IL-3. J Immunol. 2010;184:1143-1147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 116] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 123. | Perrigoue JG, Saenz SA, Siracusa MC, Allenspach EJ, Taylor BC, Giacomin PR, Nair MG, Du Y, Zaph C, van Rooijen N. MHC class II-dependent basophil-CD4+ T cell interactions promote T(H)2 cytokine-dependent immunity. Nat Immunol. 2009;10:697-705. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 500] [Cited by in RCA: 473] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 124. | Kim S, Shen T, Min B. Basophils can directly present or cross-present antigen to CD8 lymphocytes and alter CD8 T cell differentiation into IL-10-producing phenotypes. J Immunol. 2009;183:3033-3039. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 125. | Chu VT, Fröhlich A, Steinhauser G, Scheel T, Roch T, Fillatreau S, Lee JJ, Löhning M, Berek C. Eosinophils are required for the maintenance of plasma cells in the bone marrow. Nat Immunol. 2011;12:151-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 382] [Cited by in RCA: 389] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 126. | Jacobsen EA, Zellner KR, Colbert D, Lee NA, Lee JJ. Eosinophils regulate dendritic cells and Th2 pulmonary immune responses following allergen provocation. J Immunol. 2011;187:6059-6068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 105] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 127. | Wu D, Molofsky AB, Liang HE, Ricardo-Gonzalez RR, Jouihan HA, Bando JK, Chawla A, Locksley RM. Eosinophils sustain adipose alternatively activated macrophages associated with glucose homeostasis. Science. 2011;332:243-247. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1139] [Cited by in RCA: 1060] [Article Influence: 75.7] [Reference Citation Analysis (0)] |
| 128. | Powell N, Walker MM, Talley NJ. Gastrointestinal eosinophils in health, disease and functional disorders. Nat Rev Gastroenterol Hepatol. 2010;7:146-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 78] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 129. | Zuo L, Rothenberg ME. Gastrointestinal eosinophilia. Immunol Allergy Clin North Am. 2007;27:443-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 190] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 130. | Mishra A, Hogan SP, Lee JJ, Foster PS, Rothenberg ME. Fundamental signals that regulate eosinophil homing to the gastrointestinal tract. J Clin Invest. 1999;103:1719-1727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 297] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 131. | Svensson M, Bell L, Little MC, DeSchoolmeester M, Locksley RM, Else KJ. Accumulation of eosinophils in intestine-draining mesenteric lymph nodes occurs after Trichuris muris infection. Parasite Immunol. 2011;33:1-11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 31] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 132. | Woodruff SA, Masterson JC, Fillon S, Robinson ZD, Furuta GT. Role of eosinophils in inflammatory bowel and gastrointestinal diseases. J Pediatr Gastroenterol Nutr. 2011;52:650-661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 70] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 133. | Hogan SP, Mishra A, Brandt EB, Foster PS, Rothenberg ME. A critical role for eotaxin in experimental oral antigen-induced eosinophilic gastrointestinal allergy. Proc Natl Acad Sci USA. 2000;97:6681-6686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 128] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 134. | Forbes E, Hulett M, Ahrens R, Wagner N, Smart V, Matthaei KI, Brandt EB, Dent LA, Rothenberg ME, Tang M. ICAM-1-dependent pathways regulate colonic eosinophilic inflammation. J Leukoc Biol. 2006;80:330-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 44] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 135. | Brandt EB, Zimmermann N, Muntel EE, Yamada Y, Pope SM, Mishra A, Hogan SP, Rothenberg ME. The alpha4bbeta7-integrin is dynamically expressed on murine eosinophils and involved in eosinophil trafficking to the intestine. Clin Exp Allergy. 2006;36:543-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 50] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 136. | Svensson-Frej M. Immunobiology of intestinal eosinophils - a dogma in the changing? J Innate Immun. 2011;3:565-576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 137. | Wen T, Mingler MK, Blanchard C, Wahl B, Pabst O, Rothenberg ME. The pan-B cell marker CD22 is expressed on gastrointestinal eosinophils and negatively regulates tissue eosinophilia. J Immunol. 2012;188:1075-1082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 138. | Galli SJ, Tsai M. Mast cells in allergy and infection: versatile effector and regulatory cells in innate and adaptive immunity. Eur J Immunol. 2010;40:1843-1851. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 310] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 139. | Hepworth MR, Daniłowicz-Luebert E, Rausch S, Metz M, Klotz C, Maurer M, Hartmann S. Mast cells orchestrate type 2 immunity to helminths through regulation of tissue-derived cytokines. Proc Natl Acad Sci USA. 2012;109:6644-6649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 132] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 140. | Collington SJ, Williams TJ, Weller CL. Mechanisms underlying the localisation of mast cells in tissues. Trends Immunol. 2011;32:478-485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 93] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 141. | Donaldson LE, Schmitt E, Huntley JF, Newlands GF, Grencis RK. A critical role for stem cell factor and c-kit in host protective immunity to an intestinal helminth. Int Immunol. 1996;8:559-567. [PubMed] |
| 142. | Betts CJ, Else KJ. Mast cells, eosinophils and antibody-mediated cellular cytotoxicity are not critical in resistance to Trichuris muris. Parasite Immunol. 1999;21:45-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 76] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 143. | Crowe SE, Perdue MH. Gastrointestinal food hypersensitivity: basic mechanisms of pathophysiology. Gastroenterology. 1992;103:1075-1095. [PubMed] |
| 144. | Sander LE, Lorentz A, Sellge G, Coëffier M, Neipp M, Veres T, Frieling T, Meier PN, Manns MP, Bischoff SC. Selective expression of histamine receptors H1R, H2R, and H4R, but not H3R, in the human intestinal tract. Gut. 2006;55:498-504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 118] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 145. | Groschwitz KR, Ahrens R, Osterfeld H, Gurish MF, Han X, Abrink M, Finkelman FD, Pejler G, Hogan SP. Mast cells regulate homeostatic intestinal epithelial migration and barrier function by a chymase/Mcpt4-dependent mechanism. Proc Natl Acad Sci USA. 2009;106:22381-22386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 150] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 146. | Shea-Donohue T, Stiltz J, Zhao A, Notari L. Mast cells. Curr Gastroenterol Rep. 2010;12:349-357. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 52] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 147. | Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie AN, Mebius RE. Innate lymphoid cells--a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1678] [Cited by in RCA: 1858] [Article Influence: 154.8] [Reference Citation Analysis (0)] |
| 148. | Walker JA, Barlow JL, McKenzie AN. Innate lymphoid cells--how did we miss them? Nat Rev Immunol. 2013;13:75-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 519] [Cited by in RCA: 536] [Article Influence: 44.7] [Reference Citation Analysis (0)] |
| 149. | Hepworth MR, Grencis RK. Disruption of Th2 immunity results in a gender-specific expansion of IL-13 producing accessory NK cells during helminth infection. J Immunol. 2009;183:3906-3914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 150. | Vonarbourg C, Mortha A, Bui VL, Hernandez PP, Kiss EA, Hoyler T, Flach M, Bengsch B, Thimme R, Hölscher C. Regulated expression of nuclear receptor RORγt confers distinct functional fates to NK cell receptor-expressing RORγt(+) innate lymphocytes. Immunity. 2010;33:736-751. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 595] [Cited by in RCA: 579] [Article Influence: 38.6] [Reference Citation Analysis (0)] |
| 151. | Neill DR, Wong SH, Bellosi A, Flynn RJ, Daly M, Langford TK, Bucks C, Kane CM, Fallon PG, Pannell R. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature. 2010;464:1367-1370. [PubMed] |
| 152. | Koyasu S, Moro K, Tanabe M, Takeuchi T. Natural helper cells: a new player in the innate immune response against helminth infection. Adv Immunol. 2010;108:21-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 153. | Price AE, Liang HE, Sullivan BM, Reinhardt RL, Eisley CJ, Erle DJ, Locksley RM. Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proc Natl Acad Sci USA. 2010;107:11489-11494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 851] [Cited by in RCA: 924] [Article Influence: 61.6] [Reference Citation Analysis (0)] |
| 154. | Camelo A, Barlow JL, Drynan LF, Neill DR, Ballantyne SJ, Wong SH, Pannell R, Gao W, Wrigley K, Sprenkle J. Blocking IL-25 signalling protects against gut inflammation in a type-2 model of colitis by suppressing nuocyte and NKT derived IL-13. J Gastroenterol. 2012;47:1198-1211. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 103] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 155. | Saenz SA, Siracusa MC, Monticelli LA, Ziegler CG, Kim BS, Brestoff JR, Peterson LW, Wherry EJ, Goldrath AW, Bhandoola A. IL-25 simultaneously elicits distinct populations of innate lymphoid cells and multipotent progenitor type 2 (MPPtype2) cells. J Exp Med. 2013;210:1823-1837. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 112] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 156. | Saenz SA, Noti M, Artis D. Innate immune cell populations function as initiators and effectors in Th2 cytokine responses. Trends Immunol. 2010;31:407-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 122] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 157. | Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, Mention JJ, Thiam K, Cerf-Bensussan N, Mandelboim O. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958-970. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 895] [Cited by in RCA: 915] [Article Influence: 53.8] [Reference Citation Analysis (0)] |
| 158. | Brandtzaeg P, Pabst R. Let’s go mucosal: communication on slippery ground. Trends Immunol. 2004;25:570-577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 219] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 159. | Shroff KE, Cebra JJ. Development of mucosal humoral immune responses in germ-free (GF) mice. Adv Exp Med Biol. 1995;371A:441-446. [PubMed] |
| 160. | Cebra JJ, Periwal SB, Lee G, Lee F, Shroff KE. Development and maintenance of the gut-associated lymphoid tissue (GALT): the roles of enteric bacteria and viruses. Dev Immunol. 1998;6:13-18. [PubMed] |
| 161. | Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, Fagarasan S. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci USA. 2004;101:1981-1986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 521] [Cited by in RCA: 554] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 162. | Yanagibashi T, Hosono A, Oyama A, Tsuda M, Suzuki A, Hachimura S, Takahashi Y, Momose Y, Itoh K, Hirayama K. IgA production in the large intestine is modulated by a different mechanism than in the small intestine: Bacteroides acidifaciens promotes IgA production in the large intestine by inducing germinal center formation and increasing the number of IgA+ B cells. Immunobiology. 2013;218:645-651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 125] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 163. | van Wijk F, Cheroutre H. Intestinal T cells: facing the mucosal immune dilemma with synergy and diversity. Semin Immunol. 2009;21:130-138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 60] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 164. | Hayday A, Theodoridis E, Ramsburg E, Shires J. Intraepithelial lymphocytes: exploring the Third Way in immunology. Nat Immunol. 2001;2:997-1003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 376] [Cited by in RCA: 367] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 165. | Cheroutre H, Lambolez F, Mucida D. The light and dark sides of intestinal intraepithelial lymphocytes. Nat Rev Immunol. 2011;11:445-456. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 551] [Cited by in RCA: 495] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 166. | Cepek KL, Shaw SK, Parker CM, Russell GJ, Morrow JS, Rimm DL, Brenner MB. Adhesion between epithelial cells and T lymphocytes mediated by E-cadherin and the alpha E beta 7 integrin. Nature. 1994;372:190-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 875] [Cited by in RCA: 885] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 167. | Kilshaw PJ, Murant SJ. A new surface antigen on intraepithelial lymphocytes in the intestine. Eur J Immunol. 1990;20:2201-2207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 158] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 168. | Dalton JE, Cruickshank SM, Egan CE, Mears R, Newton DJ, Andrew EM, Lawrence B, Howell G, Else KJ, Gubbels MJ. Intraepithelial gammadelta+ lymphocytes maintain the integrity of intestinal epithelial tight junctions in response to infection. Gastroenterology. 2006;131:818-829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 110] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 169. | Matsumoto S, Nanno M, Watanabe N, Miyashita M, Amasaki H, Suzuki K, Umesaki Y. Physiological roles of gammadelta T-cell receptor intraepithelial lymphocytes in cytoproliferation and differentiation of mouse intestinal epithelial cells. Immunology. 1999;97:18-25. [PubMed] |
| 170. | Inagaki-Ohara K, Dewi FN, Hisaeda H, Smith AL, Jimi F, Miyahira M, Abdel-Aleem AS, Horii Y, Nawa Y. Intestinal intraepithelial lymphocytes sustain the epithelial barrier function against Eimeria vermiformis infection. Infect Immun. 2006;74:5292-5301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 78] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 171. | Komano H, Fujiura Y, Kawaguchi M, Matsumoto S, Hashimoto Y, Obana S, Mombaerts P, Tonegawa S, Yamamoto H, Itohara S. Homeostatic regulation of intestinal epithelia by intraepithelial gamma delta T cells. Proc Natl Acad Sci USA. 1995;92:6147-6151. [PubMed] |
| 172. | Guy-Grand D, DiSanto JP, Henchoz P, Malassis-Séris M, Vassalli P. Small bowel enteropathy: role of intraepithelial lymphocytes and of cytokines (IL-12, IFN-gamma, TNF) in the induction of epithelial cell death and renewal. Eur J Immunol. 1998;28:730-744. [PubMed] |
| 173. | Culshaw RJ, Bancroft GJ, McDonald V. Gut intraepithelial lymphocytes induce immunity against Cryptosporidium infection through a mechanism involving gamma interferon production. Infect Immun. 1997;65:3074-3079. [PubMed] |
| 174. | Mennechet FJ, Kasper LH, Rachinel N, Minns LA, Luangsay S, Vandewalle A, Buzoni-Gatel D. Intestinal intraepithelial lymphocytes prevent pathogen-driven inflammation and regulate the Smad/T-bet pathway of lamina propria CD4+ T cells. Eur J Immunol. 2004;34:1059-1067. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 53] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 175. | Bozić F, Marinculić A, Duraković E. Analysis of intestinal intraepithelial lymphocyte populations in experimental Trichinella spiralis infection of mice. Folia Parasitol (Praha). 2000;47:55-59. [PubMed] |
| 176. | Beagley KW, Fujihashi K, Lagoo AS, Lagoo-Deenadaylan S, Black CA, Murray AM, Sharmanov AT, Yamamoto M, McGhee JR, Elson CO. Differences in intraepithelial lymphocyte T cell subsets isolated from murine small versus large intestine. J Immunol. 1995;154:5611-5619. [PubMed] |
| 177. | Bonneville M, Janeway CA, Ito K, Haser W, Ishida I, Nakanishi N, Tonegawa S. Intestinal intraepithelial lymphocytes are a distinct set of gamma delta T cells. Nature. 1988;336:479-481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 307] [Cited by in RCA: 309] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 178. | Goodman T, Lefrançois L. Expression of the gamma-delta T-cell receptor on intestinal CD8+ intraepithelial lymphocytes. Nature. 1988;333:855-858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 493] [Cited by in RCA: 510] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 179. | Kunisawa J, Takahashi I, Kiyono H. Intraepithelial lymphocytes: their shared and divergent immunological behaviors in the small and large intestine. Immunol Rev. 2007;215:136-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 76] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 180. | Walker CR, Hautefort I, Dalton JE, Overweg K, Egan CE, Bongaerts RJ, Newton DJ, Cruickshank SM, Andrew EM, Carding SR. Intestinal intraepithelial lymphocyte-enterocyte crosstalk regulates production of bactericidal angiogenin 4 by Paneth cells upon microbial challenge. PLoS One. 2013;8:e84553. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 51] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 181. | Kim CH. Host and microbial factors in regulation of T cells in the intestine. Front Immunol. 2013;4:141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 182. | Maynard CL, Harrington LE, Janowski KM, Oliver JR, Zindl CL, Rudensky AY, Weaver CT. Regulatory T cells expressing interleukin 10 develop from Foxp3+ and Foxp3- precursor cells in the absence of interleukin 10. Nat Immunol. 2007;8:931-941. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 437] [Cited by in RCA: 487] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 183. | Agace W. Generation of gut-homing T cells and their localization to the small intestinal mucosa. Immunol Lett. 2010;128:21-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 32] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 184. | Mayne CG, Williams CB. Induced and natural regulatory T cells in the development of inflammatory bowel disease. Inflamm Bowel Dis. 2013;19:1772-1788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 140] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 185. | McDermott MR, Mark DA, Befus AD, Baliga BS, Suskind RM, Bienenstock J. Impaired intestinal localization of mesenteric lymphoblasts associated with vitamin A deficiency and protein-calorie malnutrition. Immunology. 1982;45:1-5. [PubMed] |
| 186. | Koboziev I, Karlsson F, Grisham MB. Gut-associated lymphoid tissue, T cell trafficking, and chronic intestinal inflammation. Ann N Y Acad Sci. 2010;1207 Suppl 1:E86-E93. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 87] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 187. | Svensson M, Russell K, Mack M, Else KJ. CD4+ T-cell localization to the large intestinal mucosa during Trichuris muris infection is mediated by G alpha i-coupled receptors but is CCR6- and CXCR3-independent. Immunology. 2010;129:257-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 188. | Kim SV, Xiang WV, Kwak C, Yang Y, Lin XW, Ota M, Sarpel U, Rifkin DB, Xu R, Littman DR. GPR15-mediated homing controls immune homeostasis in the large intestine mucosa. Science. 2013;340:1456-1459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 221] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 189. | Zeissig S, Kaser A, Dougan SK, Nieuwenhuis EE, Blumberg RS. Role of NKT cells in the digestive system. III. Role of NKT cells in intestinal immunity. Am J Physiol Gastrointest Liver Physiol. 2007;293:G1101-G1105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 190. | Terabe M, Berzofsky JA. The role of NKT cells in tumor immunity. Adv Cancer Res. 2008;101:277-348. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 242] [Cited by in RCA: 228] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 191. | van Dieren JM, van der Woude CJ, Kuipers EJ, Escher JC, Samsom JN, Blumberg RS, Nieuwenhuis EE. Roles of CD1d-restricted NKT cells in the intestine. Inflamm Bowel Dis. 2007;13:1146-1152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 36] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 192. | Bannai M, Kawamura T, Naito T, Kameyama H, Abe T, Kawamura H, Tsukada C, Watanabe H, Hatakeyama K, Hamada H. Abundance of unconventional CD8(+) natural killer T cells in the large intestine. Eur J Immunol. 2001;31:3361-3369. [PubMed] |
| 193. | Medina-Contreras O, Geem D, Laur O, Williams IR, Lira SA, Nusrat A, Parkos CA, Denning TL. CX3CR1 regulates intestinal macrophage homeostasis, bacterial translocation, and colitogenic Th17 responses in mice. J Clin Invest. 2011;121:4787-4795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 249] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 194. | Hapfelmeier S, Müller AJ, Stecher B, Kaiser P, Barthel M, Endt K, Eberhard M, Robbiani R, Jacobi CA, Heikenwalder M. Microbe sampling by mucosal dendritic cells is a discrete, MyD88-independent step in DeltainvG S. Typhimurium colitis. J Exp Med. 2008;205:437-450. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 147] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 195. | Schulz O, Jaensson E, Persson EK, Liu X, Worbs T, Agace WW, Pabst O. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J Exp Med. 2009;206:3101-3114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 515] [Cited by in RCA: 535] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 196. | Voehringer D, van Rooijen N, Locksley RM. Eosinophils develop in distinct stages and are recruited to peripheral sites by alternatively activated macrophages. J Leukoc Biol. 2007;81:1434-1444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 130] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 197. | McDermott JR, Humphreys NE, Forman SP, Donaldson DD, Grencis RK. Intraepithelial NK cell-derived IL-13 induces intestinal pathology associated with nematode infection. J Immunol. 2005;175:3207-3213. [PubMed] |
| 198. | Moro K, Yamada T, Tanabe M, Takeuchi T, Ikawa T, Kawamoto H, Furusawa J, Ohtani M, Fujii H, Koyasu S. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature. 2010;463:540-544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1453] [Cited by in RCA: 1567] [Article Influence: 97.9] [Reference Citation Analysis (0)] |
| 199. | Suzuki H. Differences in intraepithelial lymphocytes in the proximal, middle, distal parts of small intestine, cecum, and colon of mice. Immunol Invest. 2009;38:780-796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.0] [Reference Citation Analysis (0)] |