Published online Aug 7, 2014. doi: 10.3748/wjg.v20.i29.9633
Revised: April 20, 2014
Accepted: May 19, 2014
Published online: August 7, 2014
Despite the great successes achieved in the fields of virology and diagnostics, several difficulties affect improvements in hepatitis C virus (HCV) infection control and eradication in the new era. New HCV infections still occur, especially in some of the poorest regions of the world, where HCV is endemic and long-term sequelae have a growing economic and health burden. An HCV vaccine is still no available, despite years of researches and discoveries about the natural history of infection and host-virus interactions: several HCV vaccine candidates have been developed in the last years, targeting different HCV antigens or using alternative delivery systems, but viral variability and adaption ability constitute major challenges for vaccine development. Many new antiviral drugs for HCV therapy are in preclinical or early clinical development, but different limitations affect treatment validity. Treatment predictors are important tools, as they provide some guidance for the management of therapy in patients with chronic HCV infection: in particular, the role of host genomics in HCV infection outcomes in the new era of direct-acting antivirals may evolve for new therapeutic targets, representing a chance for modulated and personalized treatment management, when also very potent therapies will be available. In the present review we discuss the most recent data about HCV epidemiology, the new perspectives for the prevention of HCV infection and the most recent evidence regarding HCV diagnosis, therapy and predictors of response to it.
Core tip: Challenges and opportunities will characterise the story of hepatitis C virus (HCV) infection also in the new era. Despite the great therapeutic advances, improvements in HCV surveillance, epidemiological mapping, testing, prevention and therapy are urgently needed. Currently HCV vaccine candidates have shown promising results in animal models and data from clinical phase 1/2 trials are expected. Predictors of response to therapy represent a chance for modulated and personalized treatment management, when also very potent therapies will be available. The ultimate goal of global HCV control will be achieved with the joint efforts of researchers and public health workers.
- Citation: Ansaldi F, Orsi A, Sticchi L, Bruzzone B, Icardi G. Hepatitis C virus in the new era: Perspectives in epidemiology, prevention, diagnostics and predictors of response to therapy. World J Gastroenterol 2014; 20(29): 9633-9652
- URL: https://www.wjgnet.com/1007-9327/full/v20/i29/9633.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i29.9633
The history of hepatitis C virus (HCV) has always been characterized by discoveries, challenges, opportunities and difficulties. Starting with the same virus name: a Lancet editorial in 1975 suggested the term non-A, non-B hepatitis to describe the hepatitis neither diagnosed as A nor B, underlining that the diagnosis was one of exclusion[1]. Fifteen years after, in 1989, Choo et al[2] successfully cloned a single cDNA clone derived from a new flavi-like virus, by using numerous molecular biological methods: the virus responsible for most post-transfusion hepatitis, also called type C hepatitis, parenterally transmitted non-A non-B hepatitis (PT-NANB), non-B transfusion-associated hepatitis, post-transfusion non-A non-B hepatitis, HC, was finally identified[3,4]. This discovery paved the way for the development of several diagnostic tests that have been developed over time, starting from the first-generation enzyme-linked immunosorbent assay (EIA-1) for the detection of antibodies to HCV epitopes, with low rates of sensitivity and specificity, until the introduction of molecular methods for the detection of acute infection, HCV RNA and genotyping analysis. Currently used molecular tests allow the detection, quantification and analysis of viral genomes and the identification of viral genotype or subtype, as well as detecting nucleotide or amino acid substitutions associated with resistance to antiviral drugs; new enzyme immunoassays can quantify hepatitis C core antigens, that can be used as alternatives to HCV RNA in patients with chronic HCV infection[5-12]. Despite the great successes achieved in the fields of virology and diagnostics, several difficulties affect improvements in HCV infection control and eradication. New HCV infections still occur, especially in some of the poorest regions of the world, where HCV is endemic and long-term sequelae such as cirrhosis and hepatocellular carcinoma (HCC) have a growing economic and health burden. In developed countries, the lack of recognition of infection is the main barrier to controlling existing infection and allowing an adequate therapy[13-15]. The development of an effective primary prevention measure is an unmet need: an HCV vaccine is still no available, despite years of researches and discoveries about the natural history of infection and host-virus interactions. Several HCV vaccine candidates have been developed in the last years, targeting different HCV antigens or using alternative delivery systems, but viral variability and adaption ability constitute major challenges for vaccine development[16]. Many new antiviral drugs for HCV therapy are in preclinical or early clinical development, but different limitations affect treatment validity, such as comorbidity and risk-conditions, drug-drug interactions, severe adverse effects, alternate genotypes and host immune response. Treatment predictors are important tools, as they provide some guidance for the management of therapy in patients with chronic HCV infection[17-19].
In this review we will discuss the most recent data about HCV epidemiology, the new perspectives for the prevention of HCV infection and the most recent evidence regarding HCV diagnosis, therapy and predictors of response to it.
HCV is a single-stranded RNA member of the Flaviviridae family, packed into a small (50 nm) enveloped viral particle. The single polyprotein precursor of approximately 3100 amino acids, originated by the translation of the single genomic open reading frame, is processed by cellular and viral proteases into 3 structural proteins (core, E1 and E2) and 7 non-structural (NS) polypeptides (p7, NS2, NS3, NS4A, NS4B, NS5A and NS5B). These proteins have different functional roles in the virus life cycle: the core protein constitutes the viral nucleocapsid; E1 and E2 are glycoproteins that form the functional envelope that facilitates viral entry into host cells and induces neutralizing antibody proliferation; the NS proteins are required for the constitution of the replicase complex, assembly, release of infectious particles and viral propagation[20,21]. The presence of 2 hypervariable regions (HVR) in the E2 envelope glycoprotein, the lack of proofreading ability and the high rate of generating new viral variants during infection allow HCV to continuously evolve, adapt and escape the host immune responses. Moreover, HCV has developed numerous strategies to impair immune responses and evade the host immune system, by delaying and reducing both the intrinsic and adaptive immune response arm[22-28]. All these immunological determinants partially explain HCV ability to persist in the infected organism and to establish a chronic infection, most often without production of striking symptoms, until the emergence of long-term complications such as hepatic fibrosis, cirrhosis and HCC. Approximately 75%-85% of people infected with HCV will develop chronic hepatitis, 60%-70% will develop hepatic steatosis or fibrosis, 5%-20% will develop cirrhosis and in 1%-5% disease will progress to life-threatening complications and HCC, within 20 years from acute infection[29,30].
It has been calculated that 130-170 million people are infected with HCV, with a global prevalence of infection estimated at 2%-3%[31-34]. HCV prevalence is characterized by a high variability between world’s regions, individual countries and between age and risk groups within countries: this can be partially explained by the characteristic of the analysed population and the primary mode of transmission. HCV prevalence is highest in Africa and the Middle East, where Egypt, Cameroon, Saudi Arabia, Iraq and Syria account for the majority of cases and prevalence ranges from 2% to 15%. North America, Australia, Japan and Northern and Western Europe report lower prevalence of HCV infection, with no country showing a rate > 2%. China, India, Egypt, Pakistan and Indonesia account for approximately half of the global HCV-infected subjects[33-36]. In general, developing countries present the major HCV-related burden but also the major limitations in surveillance: data from most African, Asian and South American countries are lacking. In Egypt, the country with the highest HCV prevalence, there is evidence for an age-related distribution of infection: HCV seroprevalence ranges from 19% in subjects < 18 years old to > 50% in the 30-year-old age group. In this country HCV is endemic and ongoing HCV transmission levels are high, mainly due to unsafe medical procedures and household contacts. The use of improper sterilization procedures during the eradication campaign of schistosomiasis carried out in Egypt from the 1950s to the 1980s has led to an extensive transmission of HCV among persons alive during that campaign, but blood transfusion and needle reuse still remain the principal risk factors. Although lower prevalence rates, other developing countries have a similar epidemiological pattern, with an age-related distribution of cases and a virus transmission linked to unsafe medical procedures and blood transfusions; however, recent data show the increasing role played by injection drug use in the spread of infection, especially in China and Iran[37-52]. HCV prevalence in the majority of developed countries is classified as low, but marked differences in the epidemiological picture exist among countries, principally related to temporal and transmission factors and resulted in diverse age-specific distribution of HCV cases[53]. Most recent survey on the number of HCV infected people in United States estimated a total of 5-7 million people seropositive, one third of which belonging to high-risk populations, such as incarcerated persons and homeless, and a general HCV prevalence of 1.6%-1.8%, with 75% of cases in subjects born between 1945 and 1965. The expanded consumption of illicit injection drugs, the use of unsafe medical procedures and contaminated blood transfusions are the most likely causes of the creation of the adult cohort of HCV cases, evidence confirmed by the decline in new infections recorded from the mid-1980s, due to improvements of healthcare practices and the more recent introduction of screening of blood and organ donors[54-58]. HCV prevalence and transmission routes in Australia are similar as in the United States, but age distribution of cases is quite different, with the peak prevalence recorded in people aged 30-39 years, probably related to an increase of parenteral drug use throughout the 1980s and 1990s[59,60]. Among developed countries with a low prevalence of HCV infection, Japan shows some distinctive features that differentiate it sharply from other countries: most of HCV cases are recorded in people aged 40-69 years, while HCV prevalence in younger people is very low, so reflecting the occurrence of infection in the distant past, linked to improper sterilization procedures and unsafe medical practice[61-63]. HCV epidemiological pattern in Europe is heterogeneous: Northern and Western European countries reported very low (< 1%) prevalence rates, while Southern and some Eastern countries reported intermediate-to-high prevalence rates (> 2.5%). Notably, completeness of collected information is limited in many countries of the Mediterranean and the Balkan area, particularly in high-risk groups. The observed differences reflect the variable modes of transmission among countries, strongly related to cultural practices, presence of safe and effective medical and screening procedures and prevalence of specific risk behaviours. In general, iatrogenic spread of HCV infection through blood transfusion occurred in the past is the main cause of the high HCV prevalence in the older population, observed in particular in Southern European countries. Improved blood supply safety from the 1990s limited HCV diffusion among younger cohorts, but sharing of injecting equipment among intravenous drug users has become the predominant route for HCV transmission. The expansion of intravenous drug use is recorded both in Western and in Eastern European countries. Nosocomial infections still occur in European countries, although the advances in medical procedures: 50%-70% of new HCV cases can be attributed to nosocomial exposure, according to recent estimates in Italy and Spain. Another important factor that substantially contributed to HCV epidemic in Europe is immigration from endemic areas, especially during the last 10-15 years in Northern and Western Europe[64-70]. It is important to highlight that a considerable proportion of HCV positive subjects are unaware of its status and many new infections are not diagnosed or reported: lack of recognition of infection affects epidemiology estimates and treatment opportunities, especially in high-risk groups, so hampering effective control of infection, even with treatments of high efficacy[15].
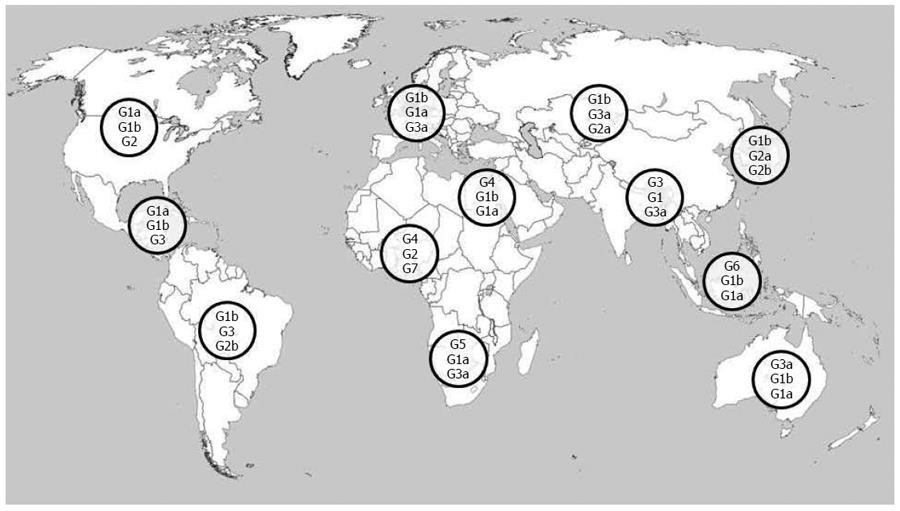
Also the genetic diversity of the virus contributes to complicate HCV epidemic picture and constitutes a severe challenge for effective therapy. HCV is characterized by an high genomic variability and is classified into 7 genotypes (1-7), which differ by more than 30% sequence diversity, and at least 67 subtypes, characterized by a about 20% sequence divergence, according to the last update to the previous consensus HCV classification[71-73]. Moreover, when HCV infects an individual, multiple closely related but distinct viruses, a “quasispecies” population, can be identified, with sequence variations up to 10%: as already observed, HCV polymerase is characterized by the absence of proofreading capacity, and this leads to an high mutational rate of 10-5-10-4 nucleotides per replication cycle[74]. Identification of HCV genotypes and subtypes is a crucial step for the definition of epidemiological patterns and effective treatment. Current commercially available methods allow the detection of nucleotide sequence disparity using direct or indirect approaches and new sequencing technologies can detect minor viral populations in complex quasispecies mixtures; however, improvements in specificity, reading sequence lengths and general clinical significance of generated sequences are required[75,76]. Global distribution of HCV genotypes is characterized by marked geographical differences, reflecting the evolving pattern of transmission modes and other influencing factors such as immigration and screening diffusion. In Figure 1 most common HCV genotypes across the world are shown. Genotype 1 is distributed worldwide and is responsible for the majority of cases in the Americas, Europe, Australia and Japan; subtype 1b is the commonest in Europe and Asia, while subtype 1a is distributed widely in Northern Europe and United States. Genotype 2 is more prevalent in industrialized countries as well as in South America and Asia, particularly in Japan and China, where the subtype 2a is commonly isolated; genotype 2b is widespread in Northern Europe and United States; genotype 2c is the most common subtype in Western and Southern Europe, Pakistan and India. Genotype 3, and in particular subtype 3a is prevalent in Europe, United States, Australia and Southern Asia; recent data indicate an increasing trend in isolation of genotype 3a and a general decrease of genotype 1b and 2 over time, probably related to the reduced iatrogenic transmission of infection and the diffusion of illicit parenteral drugs, especially in Eastern and Southern Europe[77-79]. Genotypes 4-7 are limited to distinct areas and/or countries. Genotypes 4 and 5 are mainly identified in Africa and the Middle East: genotype 4a is prevalent in Egypt, while genotype 4c is highly prevalent in Central Africa; genotype 5 is mostly isolated in South Africa. Genotype 6 and its numerous subtypes are found mainly in Southeast Asia, and in some countries such as Thailand, Vietnam, and Myanmar genotype 6 is the responsible for the majority of new HCV cases. Genotypes 4, 5 and 6 were believed to be confined in Northern Africa, Central Africa and Southeast Asia, respectively, but increasing migration waves and globalisation processes are causing a spread of these genotypes outside the cited areas, in nearby areas of Asia and as far as western countries such as United States, Canada and Northern Europe. Genotype 7 has a minor clinical relevance and was recently found in patients from Central Africa and Thailand[34,61,75,80-84].
Association between HCV genotype and fibrosis progression appears inconclusive, although increasing clinical and experimental data show that infection with genotype 3 is associated with a higher risk of severe hepatic steatosis, accelerated fibrosis progression rate and increased oncogenesis[82,85-87]. Conversely, each genotype has different response rates to antiviral therapies. While 80% of HCV genotype 2-and 3-infected patients reach sustained virological response (SVR) under pegylated-interferon with ribavirin (pegIFN/RBV) combination therapy, this regimen leads to a SVR only for about 50% of genotype 1- and 4-infections; genotypes 5 and 6 have intermediate response rates[87]. There is increasing evidence that SVR rates with dual pegIFN/RBV therapy for genotype 3 infected patients appear worse than for genotype 2 infections. The introduction of direct-acting antivirals (DAAs) into combined regimens markedly increased SVR rates for genotype 1 infected patients: the first-generation NS3-NS4A protease inhibitors telaprevir (TVR) and boceprevir (BOC), approved for treatment of only genotype 1, lead to SVR rates of 63%-75%, although with an increase of side effects. New combinations of DAAs have shown a favorable safety profile and an improvement of antiviral activities also against non-genotype 1 HCV, but risks of resistance, treatment failures and the well-known limitations of IFN-based regimens are issues that have still to be overcome[88-94].
The global burden of HCV disease is now fully recognised, thanks to several epidemiological and natural history studies performed during the two decades after virus’ discovery. Primary prevention of new infections and management of existing infections (secondary prevention) are the fundamental approaches to controlling HCV epidemic. Despite the great advances in the treatment of HCV infections, the still heavy public health burden and the limitations of current available therapies highlight the key role of primary prevention strategies to reducing worldwide disease diffusion. Among the different strategies to prevent infections of major public health relevance, vaccination has proved to be the most effective preventive approach to control infectious diseases and interrupt transmission chains. The history of the epidemic sustained by another hepatotropic virus, HBV, is the demonstration of the fundamental importance of the availability of an effective vaccine in preventing viral infections and virus associated disease[95-97]. The development of a preventive HCV vaccine constitutes an irreplaceable tool to control HCV spread, but several major hurdles hamper the achievement of this important purpose. As already observed, HCV is characterized by an high genetic diversity and variability, because of the lack of proof-reading activity of its polymerase: as such, infection is sustained by a quasispecies of multiple closely related but distinct viruses, with the ability to persist in infected people by escaping immune control of cytotoxic T lymphocytes (CTL) and antibodies against different regions of the viral envelope. Moreover, HCV is able to impair CD4+ T cell response at the beginning of infection and causes a rapid immune exhaustion of CD8+ T cells as the infection endures[98-102]. Strategies to develop a preventive HCV vaccine should consider these aspects: in particular, an effective HCV vaccine should elicit both a strong humoral immune response, by inducing neutralizing antibodies targeting multiple conserved B and T cell epitopes, and a cellular immune response, by stimulating a rapid activation of T-helper 1 lymphocytes as well as CTL. Moreover, fundamental steps for the development of an HCV vaccine will be the definition of suitable correlates of protection and cross-protection evaluations against the various HCV genotypes. The lack of convenient experimental model systems is another important challenge towards the full understanding of viral pathogenesis and immune response to HCV infection. Only recently the availability of a cell culture-derived HCV model (HCVcc), consisting of human hepatoma cell lines infected with the 2a strain of HCV, and of a cell-culture-based system that allows production of infectious HCV in physiologically relevant human hepatocytes (HCVpc) provide useful tools for the study of HCV interactions with host cell and for testing neutralizing and cross-protective potential of antibodies induced by various HCV vaccine candidates[103-110]. Nevertheless, these in vitro systems do not allow the study of T cell response to HCV infection, and a suitable animal model is still required to study innate and adaptive immune responses in vivo. Chimpanzees constitute the only acceptable animal model for HCV analysis, but ethical issues, high costs and scarce supply limit the use of these animals; moreover, chimpanzees have major differences in immunologic responses to infection from humans, so the results obtained with this model have to be interpreted with caution, especially those regarding protective immunity[111-115]. Another commonly used animal model for HCV research is a chimeric mouse model, in which engineered mice engrafted with human hepatocytes are able to be infected with HCV either from patient sera or produced in vitro: main limitations of this model are high mouse mortality rate and lack of adaptive immune response to HCV. Improved mouse models characterized by a partially reconstituted human immune system and human liver, susceptibility to HCV infection and ability to generate a specific response against the virus have been recently described[111,116,117]. Although very useful in viral pathogenesis understanding and vaccine development, animal models cannot substitute accurate evaluations in humans. The design of preventive vaccine trials presents several challenges, especially in the case of HCV: the number of enrolled subjects should be very high to ensure adequate power to the trial, study results may not be applicable to countries other than those where the trial has been performed, factors such as HCV prevalence, exposure frequency, infectivity and chronicity may affect significance of the trial, endpoints and correlates of protective immunity should be clearly defined[118-121]. Experience achieved in the field of HIV vaccine development, with several recent high-profile failures, highlights the need for accurate studies on HCV vaccine design.
Several approaches have been adopted to develop an effective preventive HCV vaccine: they can be classified on the basis of targeted immunity (humoral immunity, cell-mediated immunity or both) or employed strategy (recombinant protein or viral peptide vaccines, virus-like particles vaccines, DNA/recombinant vaccines, DNA/viral vector vaccines). HCV vaccine candidates combining recombinant envelope E1 and E2 proteins and adjuvants have been demonstrated to elicit humoral responses and production of neutralizing antibodies both in animal model and human phase I trials[122-124]. A subunit approach combining HCV core protein and ISCOMATRIX® adjuvant has been investigated to promote a broad and strong humoral and cellular immune response to HCV antigens in primates and healthy volunteers, with conflicting results[125,126]. Formulations combining several highly conserved CD4+ and CD8+ epitopes with different adjuvants, bacterial pore-forming toxoids, heat shock proteins or influenza-based virosomes are promising strategy for the induction of cross-protective humoral and cellular immunity[127]. A virus-like particles approach, based on insect cells infected with a recombinant baculovirus containing the cDNA of HCV structural proteins (core, E1, and E2), has been shown to induce both humoral and cellular immune responses in animal models, but protection against infection after HCV challenge has not been demonstrated. Similar recently described approaches include the use of engineered HBV S envelope protein, murine leukemia virus and vectored measles viruses[128-135]. DNA vaccines present the advantage of inducing cytotoxic lymphocyte responses; however, the induced immunity is often brief, weak and unlikely to be effective in infection prevention. Several strategies include a priming with a DNA vaccine followed by a protein-based vaccine to boost CD4+ and CD8+ T cells and humoral immune responses[127-138]. A potent T cell-mediated immunity can be obtained with the use of a defective or attenuated viral or bacterial vector expressing HCV structural and non-structural antigens: viral vectors include adenoviruses, vaccinia virus, modified vaccinia Ankara (MVA), pox virus and other viruses. Adenoviral vectors have shown the most promising results in inducing strong and broad CD4+ and CD8+ T cell responses. Vaccine strategies based on these vectors reduced peak viremia and induced protection against chronic infection in primates, but did not prevent HCV primary infection[139-143]. Currently, a phase 1/2 trial, designed to assess safety, immunogenicity and efficacy of a prime-boost vaccine based on an adenoviral vector and a MVA vector in active intravenous drug users aged 18 to 45 years in United States, is ongoing[144].
The diagnosis of hepatitis C infection is usually obtained by detection of anti-HCV antibodies. The anti-HCV reactivity by screening assays can indicate a past, acute or chronic hepatitis and despite the high specificity of the assays (> 99%) false positive results are not rare, especially in some clinical situation such as in pregnant women, in patients with immunologic or hematologic diseases and when testing is performed among population with low risk of infection. In all these circumstances the anti-HCV reactivity must be confirmed with a confirmatory test. Two major guidelines [European Association for the Study of the Liver (EASL) and Centers for Disease Control and Prevention (CDC)] currently recommend the detection of anti-HCV antibodies together with molecular determination of HCV-RNA for the diagnosis of HCV infection[145,146]. In addition, in the course of infection more and more often it is also advisable to assess the genotype of the virus, as well as its quantitative plasma load, also done by molecular tests. Particularly, these are useful, if not mandatory, in the phases of therapeutic decision, choice of treatment and control of efficacy. Thus, several viral markers, either serological or molecular, can be used in the course of HCV infection both for diagnostic and monitoring purposes.
Detection of anti-HCV antibodies: Several enzyme immunoassays (EIAs), microparticle EIAs and chemiluminescence immunoassays have been developed to detect anti-HCV antibodies[147]. First generation assays can identify as anti-HCV positive about 80% of patients with diagnosis of non-A non-B hepatitis[148]. Only after the characterization of conserved amino-acid sequences, in the NS5 region of the genome, throughout the genotypes, and the detection, within them, of those epitopes that shared T and B cell recognition, have the second and then the third generation of assays been produced[149]. Currently, third generation assays that include, in solid phase, antigens from the core and recombinant antigens from NS3, NS4 and NS5 regions are diffusely used[150]. The evolution from first, to second and finally to third generation has led to a progressive increase in sensitivity and specificity, as well as in a reduction of the window period (8-10 wk after exposure). In fact, this latter purpose has been reached, both in house-made and commercially standardized assays, by simultaneously detect antibody and antigens in the same assays, thus reaching the same sensitivity and specificity of the routinely used assays and also achieving the capability of detecting HCV infection about 21-50 d earlier than simple anti-HCV assays[151].
In the effort at having an assay capable to discriminate between acute and chronic HCV infection, anti-HCV IgM assays were produced. The attempts, however, have been frustrated by the presence of anti-HCV IgM antibodies both in the acute and in the chronic infection, although in different percentage, so that their significance is often unclear, and the assays are not used in clinical practice[152].
Avidity assays, used to distinguish primary from chronic or recurrent viral infection in many other diseases, have also been tested in the HCV infection. As reported by some authors, the avidity index has been found significantly lower in primary than in chronic, but also lower in past than in chronic infection. Even if these assays may sometimes be an useful help at assessing the timing of hepatitis C infection after the onset of symptoms, they nevertheless have had poor success in clinical practice as well[153,154].
Recently, rapid immuno-chromatographic assays for the detection of anti-HCV antibody, based on recombinant antigens from the core, NS3, NS4 and NS5 regions, were evaluated and shown to possess > 99% specificity and sensitivity ranging from 86% to 99%. As they are able to generate a result within an hour, they can be used as point-of-care testing[146,155,156].
Currently, CDC recommends the use of an approved screening test, either an EIA or a rapid test, and the use of another assay to confirm a positive result as a true positive one[146].
The recombinant immunoblot assay and other immunoblot assays are commonly used to confirm a reactive result at an anti-HCV screening test. The same antigens as in EIAs are used in these assays, but the antigens are separately coated on a membrane and the result depends on the number of bands present on the membrane. The immunoblot assays, more specific than EIAs, can confirm a true positive anti-HCV result but are unable to confirm an active HCV infection, which only a molecular test can reveal[148].
Detection of HCV antigen: In addition to the previously described tests that allow the simultaneous detection of antigens and antibodies, assays for the detection of HCV core antigen alone were also developed. It is now available an automated, quantitative chemiluminescence immunoassay which has been shown to have sensitivity and specificity ranging from 80% to 99% and from 96% to 99%, respectively[11,148]. Several studies demonstrated that the test can similarly detect and quantify all the genotypes and that the quantification of core HCV antigen shows a good correlation with the HCV-RNA levels[10,157]. On these bases, recently, Ottiger et al[158] proposed a new algorithm to confirm an anti-HCV reactive result and also a mathematical formula to extrapolate HCV-RNA levels by measuring HCV antigen. It must be noted, though, that slight differences across the genotypes and from one patient to another have been reported.
As this is an immunoassay, it is easy to use and less expensive compared to a molecular assay. Moreover, as it is able to detect core antigens it can be used to confirm acute infection and also to monitor HCV response to therapy. The lower limit of detection, varying according to the HCV genotype from 500 to 3000 IU/mL of HCV-RNA[159], represents the important limitation of the assay. However, even taking this restriction into account, HCV antigen assay can represent a useful diagnostic marker in those laboratories where HCV-RNA molecular tests cannot be performed[160], pending their hopefully fast conformation to international standards.
Detection of HCV-RNA: HCV-RNA is detectable in plasma and in serum 1 to 3 wk after infection, about 1 mo before the appearance of anti-HCV antibody, and is a hallmark of ongoing viral replication[152,161]. Nucleic acid testing (NAT) used for detecting and quantifying HCV-RNA characterizes the gold standard for HCV diagnosis and can be done by mean of polymerase chain reaction (PCR), branched DNA signal amplification (bDNA) and transcription mediated amplification. Currently, all NATs for detecting and quantifying HCV-RNA levels are standardized by the use of the WHO International Standard and the HCV-RNA results are expressed in Unit International (UI/mL)[162].
The development and the availability of the semi-automated or fully-automated real-time PCR that exhibit excellent sensitivity, specificity and high dynamic range will probably lead to the replacement of the qualitative assays. Several real-time assays are being commercialized which have been demonstrated to be accurate enough in detecting and quantifying HCV-RNA to be suitable for clinical practice. Differences have been reported with regard to the HCV-RNA quantification based on the genetic diversity of the viruses and, probably, due to the mismatching between primers or probes and HCV target sequences[163].
Detection of HCV genotype: As already said, 7 different HCV genotypes and several subtypes have been characterized so far[71]. The HCV genotype along with HCV-RNA baseline level is considered the major predictor of SVR to antiviral therapies. In clinical practice, HCV genotype can be assessed by commercially available techniques based on real-time PCR with genotype specific probes/primers, semi-automated sequencing and automated reverse hybridization that analyze the 5’ NC region of HCV genome, representing the most conserved one. However, analysis of 5’ NC region can lead to errors in subtype attribution, because it is not the most appropriate for discrimination among subtypes. For this reason a new version of automated reverse hybridization, the most commonly used method, analyzes both the 5’ NC and the core regions. The gold standard of genotyping is the sequencing of NS5B region, able to accurately assign the genotype, with the advantage that the obtained sequence can be used for phylogenetic analysis to epidemiological purposes[164-167].
Recently, hybridization on oligonucleotide microarray assay, containing genotype and subtype specific oligonucleotides on the corresponding sequences of the NS5B region, have been successfully developed for identifying HCV genotypes and subtypes[168].
To date and for many years, the peg-IFN/RBV combination, able to eradicate the virus in approximately 50% of treated patients[169], has characterized the standard of care (SOC) for chronic HCV infection. The recent development and availability of new molecules named DAAs are implementing the HCV therapeutic options[170,171].
These new DAAs include: NS3/NS4 protease inhibitors, divided into linear and macrocyclic, NS5a phosphoprotein inhibitors, NS5B polymerase nucleoside and non-nucleoside inhibitors and host-targeting antivirals[172].
Currently, only TVR and BOC, the first two NS3 protease inhibitors, are available and approved for use in Europe, in combination with SOC, in HCV, genotype 1, chronically infected patients[173,174]. Both are linear ketoamides molecules that target the catalytic site of the NS3/4A protease, blocking the release of HCV NS proteins required to assemble the viral replication complex. Moreover, they also work by stopping the release of immune modulating host proteins, thus promoting the innate immune response to HCV infection[175].
As HCV is a virus with high genetic heterogeneity, high rate of turnover and no proofreading activity, when used in mono-therapy DAAs cause the rapid emergence of resistant variants, so that they are approved for use in combination with peg-IFN/RBV[175].
The efficacy of TVR and BOC has been evaluated in phase III clinical trials. In summary, two trials have been performed for each: in naïve chronic HCV patients (advance for TVR and sprint 2 for BOC) and in treatment experienced SOC failed patients (realize for TVR and respond 2 for BOC)[93,94,176,177]. All these studies demonstrated significant improvement of SVR in DAAs arms compared to SOC. Several post-marketing studies are currently being performed confirming these favorable preliminary data. However, while representing new therapeutic chances for clinicians and patients, DAAs also entail new challenges and efforts to lab workers.
As already said, the use of TVR and BOC in HCV treatment leads to a substantial improvement in SVR rates, but the managing and monitoring of the patients in triple therapy has become more complicated than in SOC. Due to the differences in the “stopping rules”, also known as “futility rules”, between SOC and DAAs treatment, and between the two DAAs (TVR and BOC), HCV-RNA quantitative monitoring faces new challenges. Not only has it to be very accurate for several reasons: for understanding HCV-RNA kinetics, for establishing eligibility for response guided therapy (RGT) and for complying with stopping rules, it also has to be very frequently and rapidly performed[178,179] (Table 1). In fact, a prompt result from the laboratory is often important not only to avoid unnecessary side effects and the waste of ineffective expensive drug, but also to prevent the occurrence of resistant variants in patients for which it is impossible to achieve SVR. For these reasons, EASL Guidelines recommend the use of a real-time PCR-based assay with a lower limit of detection as low as 10-20 IU/mL for HCV-RNA detection and quantification[145].
| End-point | Boceprevir | Telaprevir |
| RGT | Non-cirrhotic treatment naïve patients, previously relapsers or partial responders | Non-cirrhotic treatment naïve patients previously relapsers |
| HCV-RNA undetectable at weeks 8 and 24 | HCV-RNA undetectable at weeks 4 and 24 | |
| Futility rules | HCV-RNA > 100 IU/mL at week 12 | HCV-RNA > 1000 IU/mL at weeks 4 and 12 |
| HCV-RNA detectable at week 24 | HCV-RNA detectable at week 24 |
Each real-time assay has its own linear range, with its own upper and lower limit of detection, but the terminology used to interpret the results is the same[178-182] (Table 2). It is possible to define: (1) lower limit of quantification (LLOQ): the lowest value of HCV-RNA that is possible to accurately quantify, HCV-RNA is detectable and quantifiable; (2) limit of detection (LOD): the lowest value of HCV-RNA that can be detected, always < LLOQ, HCV-RNA is detectable but not quantifiable; (3) < LLOQ: HCV-RNA is detectable but not quantifiable, the interpretation is the same as LOD; and (4) target no detected (TND): no HCV-RNA amplification, HCV-RNA is undetectable or not detected.
| Assay | Manufacturer | Method | LOD, IU/mL(dynamic range, IU/mL) |
| Qualitative | |||
| Amplicor HCV v2.0 | Roche molecular system | RT-PCR (manual) | 50 |
| Cobas Amplicor HCV v2.0 | Roche molecular system | Semi-automated RT-PCR | 50 |
| Ampliscreen1 | Roche molecular system | Semi-automated RT-PCR | < 50 |
| Versant HCV RNA | Siemens healthcare diagnostics | Semi-automated TMA | 10 |
| Procleix HIV-1/HCV1 | Chiron corporation | TMA (manual) | < 50 |
| Quantitative | |||
| Amplicor HCV Monitor | Roche molecular system | RT-PCR (manual) | N/A (600-700000) |
| Cobas Amplicor HCV Monitor v2.0 | Roche molecular system | Semi-automated RT-PCR | 600 (600-500000) |
| Versant HCV 1.0 Assay K-PCR | Siemens healthcare diagnostics | Semi-automated RT-PCR | 15 (15-100000000) |
| Abbott RealTime HCV Assay | Abbott diagnostics | Semi-automated RT-PCR | 12 (12-100000000) |
| LCx HCV RNA Quantitative Assay | Abbott diagnostics | Semi-automated RT-PCR | 25 (25-2630000) |
| Cobas Ampliprep/Taqman | Roche molecular system | Semi-automated RT-PCR | 15 (15-100000000) |
Timing of sample collection is also assessed by guidelines, depending on the specific futility rules of each drug, as otherwise it happens for SOC. However, HCV-RNA kinetics induced by DAA treatments seems to have a different trend in comparison to that observed by IFN-based therapies (typically biphasic)[183,184]. DAAs determine an initial shorter delay before the beginning of the biphasic decline, in addition the decline observed in each phase is faster. If no fast decrease in the HCV-RNA levels is observed within the first two days, that is at the end of the first phase of decline, resistant variants can be selected[185]. Due to the faster HCV-RNA kinetics induced by current DAAs, which will be at least equaled if not improved by the next generation ones, timing of sampling must be very strict during treatment with DAAs, and is likely to need a further restriction in a future revision of the guidelines.
In summary, for clinical purposes, every piece of information from HCV-RNA testing can be important. A detectable, though not quantifiable HCV-RNA (< LLOQ or LOD) may lead to different therapeutic decisions than undetectable HCV-RNA (TND or < LOD), thus all phases of HCV testing, including timing of sample collection, HCV-RNA measuring and result reporting, must be equally very accurate.
As already said, due to the virus HCV characteristics (high rate of turnover, no proof-reading activity with production of about 10-3-10-5 mutations per nucleotide per genomic replication), HCV exists as a whole of viral variants, called “quasi-species”. In other words, the viral population consists of a prevalent population, typically called “wild type” virus (the virus with the best fitness) and of the minority variants selected during HCV replication and favorable to the virus[74,171,186,187]. There are also differences in nucleotide sequences within genotype (greater) and subtypes (smaller)[71]. It is against this quite heterogeneous viral population that old and new drugs must work.
Resistant viral variants are quickly selected if the new DAAs, that have been shown to have low genetic barrier, are administered in mono-therapy[188]. The combination of the new DAAs with peg-IFN/RBV partially protects against the onset of resistance-associated mutations (RAMs). The function of peg-IFN/RBV is to suppress pre-existing resistant variants, therefore treatment failure occurs more easily in poor IFN responder patients, unfortunately those in greater need of DAAs and for whom DAAs are indicated. Factors associated with IFN response and tests to predict response are discussed elsewhere in this review.
So far, RAMs can be identified only with “in house” assays. Several methods for sequence analysis can be performed: TaqMAMA, hybridization assays, restriction enzyme assays, direct sequencing and next generation sequencing (NGS) techniques[188,189]. The well standardized and most commonly used in clinical practice direct sequencing can detect the prevalent population of the quasi-species (> 25%), whilst the NGS, used only in research laboratories, can also reveal minority species (> 5%). Currently, the NGS techniques present several problems for a routine use. Although they can produce a significant amount of information, they can generate sequence errors, unreadable sequences, false positive and false negative results for substitutions, insertions and deletions. For that reasons, sophisticated software and trained personnel are needed[9,188].
A list of primary RAMs to TVR and BOC has been drawn up and an overlapping of the resistance profile between the two drugs has been found. In vitro, a degree of resistance was established for each mutation by phenotypic analysis of the mutated strains and was divided into low, intermediate and high level of resistance (Table 3). If selective drug pressure continues, other compensatory mutations are likely to be induced, with an increase of replicative fitness[171,188,190,191]. However, while a high degree of cross resistance between these linear NS3/4 inhibitors was observed, cross resistance in the class of macrocyclic inhibitors seems to occur more rarely and in far less sites. Nevertheless, whereas mutations at R155 amino acid position were potentially selected by all the NS3/4 protease inhibitors, the mutations at D168 amino acid position were selected only by the new macrocyclic NS3/4 inhibitors[192-195] (Table 4).
| Degree of resistance | Mutations | Fold increase |
| Low level | V36A/M/C | 3.5-7 |
| T54A/S | 6-12 | |
| R155K/T/Q | 8.5-11 | |
| Intermediate | V36A/M + R155K/T | 57-71 |
| High level | A156V/T | 74-410 |
| V36A/M + A156V/T | > 700 |
Differences between subtypes have been described both in terms of response to treatment and in terms of selection of resistant variants. Indeed, with both protease inhibitors SVR rates are higher in subtype 1b than in 1a. In fact, not only are the mutations subtype specific, but they also depend on the genetic barrier of the subtype. Typically, when treatment failure occurs R155K/T and V36M mutations are selected in genotype 1a, whereas A156S/T, V36A and T54A in genotype 1b. R155K mutation is selected faster in subtype 1a than in subtype 1b, because the change of a single nucleotide is sufficient to cause an amino-acid substitution in the first, whilst two changes are required in the latter[196,197].
Unlike HIV, which integrates the viral genome into host cells and HBV whose viral genome is present in the nucleus of hepatocytes as cccDNA, HCV has no latent reservoir. While warranting the possibility of viral eradication, this implies that the virus is not stored and the RAMs, selected by treatment, tend to be replaced by wild-type virus after the end of drug pressure. Although data from literature indicate an extremely high variability from one patient to another in the time required for the disappearance of the HCV resistant variants, probably depending on the viral fitness of the respective variant, this aspect must be taken into account when sequencing HCV in search for RAMs[171,196].
In DAAs failing patients HCV sequencing for individuation of RAMs can yield useful information at an investigative level, but it is unclear its role as a clinical tool and it is not currently recommended in this setting. Likewise, at the moment screening for resistance is not indicated in naïve patients before initiation of treatment with TVR or BOC, mainly because the circulation of mutated strains in the general population has to be very low. However, an entire new generation of DAAs is currently being developed, some of which with potential use in IFN-free settings and some of which very close to commercial release. One of the emerging aspects in terms of viral susceptibility to these newest drugs is that some polymorphisms, detected in certain viral subtypes, can selectively reduce susceptibility to one or more of them. In fact, for instance, the protease inhibitor simeprevir has most recently been approved by FDA with the mandatory condition that patients harboring a polymorphic Q80K variant strain of genotype 1a-HCV have to be excluded from treatment. Thus, a viral genotype has to be acquired in potential candidates for simeprevir treatment. It is likely that similar limitations will be issued for other new drugs in the nearest future. Therefore, resistance testing in naïve patients, which seemed a mere research exercise so far for the lacking of data indicating a clinical utility, may soon become a pretreatment requirement at least in selected cases.
The primary goal of therapy against HCV is to eradicate infection and its target is to achieve SVR, defined as having undetectable serum HCV RNA 24 wk after cessation of treatment. The main antiviral tools are unsuccessful in about 50% of cases[198-201], in particular in HCV/HIV co-infected. Given the variability of response and in order to reduce several side effects and to avoid the heavy medical cost, baseline viral and host parameters to predict an individual response before the treatment would be quite useful[202]. Several studies have proven the role of both viral factors (such as HCV genotype, quasispecies diversity, baseline viremia) and host factors (i.e., age, sex, ethnicity, grade of liver fibrosis, body mass index, co-morbidities) in predicting the natural course of hepatitis C and response to therapy.
However, the observations that the treatment response rate in African/Americans is less than half of that observed in Caucasians[203] suggested that additional factors associated with patient genetic background are related to likelihood of an SVR and may influence tailored treatment duration.
More recently, a number of independent genome-wide association studies (GWAS) have shown associations between single nucleotide polymorphisms (SNPs) near the interleukin 28B (IL28B) gene and viral clearance or SVR to peg-IFN/RBV[204,205] as well as inosine triphosphatase (ITPA) and RBV-anemia[203]. Other SNPs have also been identified to correlate with treatment outcomes or drug-related toxicities.
The role of host genomics in HCV infection outcomes in the new era of DAAs may evolve thanks to new therapeutic targets, especially when more potent and numerous new DAAs will be available and a chance of modulating and personalizing treatment and patient management will become real.
Several studies have demonstrated the role of SNPs upstream of the IL28B gene on chromosome 19 coding for IFN-λ3 (rs12979860, rs8099917, rs12980275, and rs8103142)[204-211] in predicting treatment induced and spontaneous clearance from HCV infection. Based on both GWAS and genetic mapping data, patients with the SNPs haplotypes associated with HCV clearance are more likely to achieve SVR and this could be explained by the biology of these genetic variations. The IL28B gene encodes for IFN-λ3, belonging to the IFN-λ family which plays an important role in antiviral immunity. Recent findings described the association between the IL28B SNPs and the expression of intrahepatic interferon-stimulated genes (ISG) in liver[203]. Low ISG expression prior to treatment has been already correlated with high response to IFN-based therapy and the protective IL28B genotype is associated to lower levels of ISG expression, suggesting that ISG induction in part segregates according to IL28B haplotype[203,212].
The first and largest GWAS was conducted in the IDEAL trial, in which about 1600 HCV genotype 1- infected patients of European, African-American or Hispanic ancestry were enrolled: although more than 500000 SNPs were studied, rs12979860 with its favorable bi-allelic variant CC represented the best predictor of SVR in these ethnic groups. Specifically, carriers of the protective genotype achieved an SVR in 82% in comparison of an SVR of 38% in the CT or TT-genotype[207].
The C allele distribution shows a geographical pattern with a lower frequency among African individuals than European descents and is more prevalent in East Asians, where the highest rates of SVR were observed. This might account for more than half of the ethnic difference in treatment response rates[206].
Other SNPs of IL28B (rs8105790, rs11881222, rs28416813, rs4803219 and rs7248668) have been identified in HCV genotype 1 infected patients[204,213,214], although SNPs rs12979860 and rs8099917 were found to be predominant in different populations and strongly associated with treatment outcome: in particular the favorable variants, homozygosis for the SNPs rs12979860 (CC) and rs8099917 (TT), are significantly associated with SVR in HCV-genotype 1-infected patients treated with peg-IFN/RBV. A recent meta-analysis confirmed the strength of these genotypes as independent predictors[215]; however, in consideration of the high prevalence of rs12979860 in different populations and its relevant effect on treatment outcome, the determination of this SNP seemed sufficient for predicting response to therapy[216] and the current consensus considered it as the strongest predictor[217], suggesting the possibility of personalized therapy with its significant clinical and pharmacoeconomic implications.
Thus, carriers of rs1297860CC infected with HCV genotypes 1/4 may be treated with peg-IFN/RBV, whereas carriers of the allele T with HCV genotype 1 and no advanced liver fibrosis may delay therapy and wait for new DAAs[218]. On this question the EASL Clinical Practice Guidelines for the management of HCV infection indicate the determination of IL28B polymorphisms as an useful tool to identify patient’s likelihood of response to therapy, but with a low predictive value, and for this reason it should not be used to defer therapy in carriers of unfavorable genotype, but mostly in need of treatment (i.e., patients with fibrosis). Moreover, IL28 genotyping is less clinically significant in patients with more IFN-responsive 2/3 HCV genotype infections[145].
In the new era of DAAs therapy an important question is whether and how the determination of IL28B polymorphisms may be useful in predicting patient’s likelihood of response and the potential implications for treatment decision making: data from SPRINT-2 trial in naïve chronic HCV patients and from RESPOND-2 trial in those treatment experienced indicated that rs1297860 is associated with SVR to triple therapy with boceprevir in combination with peg-IFN/RBV (rs1297860 CC vs non-CC; OR 2.6 and 2.1, respectively)[94]. Likewise SVR rates to triple therapy with TVR are 87%-90% in carriers of favorable in comparison to 59%-72% in carriers of non-favorable IL28B genotype[203,219]. In consideration to the expected high SVR rates associated to DAAs therapy, in the future it is reasonable that IL28B variations will be useful for identifying patients appropriate for simpler peg-IFN/RBV regimen and allocating the more expensive triple therapy to patients with unfavorable IL28 genotypes.
However, further cost-effectiveness analyses of tailored therapies are needed for better understanding how IL28 genotyping may help to evaluate the risk/benefit antiviral treatment profile.
Hemolytic anemia is a common side effect of RBV-based HCV therapy, although reversible and dose-related. However, in more than 15% of cases it is cause of ribavirin dose reduction or premature withdrawal from therapy[218].
In a recent GWAS, Fellay et al[220] identified two variants (rs1127354 and rs7270101) in the ITPA gene functionally responsible for ITPA deficiency and correlated to the risk of RBV-induced anemia in European-American population. Later it was confirmed that rs1127354 is strongly associated with protection against anemia in a Japanese cohort of 474 patients[221] and similar data were shown by Naggie et al[222] in HCV/HIV co-infected patients with all HCV genotypes.
Tanaka et al[223] also reported the association of two SNPs, rs11697186 and rs6139030, which were within and around DDRGK1 gene on chromosome 20, with reduced platelet counts in response to peg-IFN/RBV treatment in 303 Japanese HCV patients. The SNP rs11697186 was in strong linkage disequilibrium with rs 1127354 ITPA variants[224].
Overall, SNP ITPA was not associated to treatment outcomes and does not predict SVR[225]. These findings further support the perspective of pharmacogenetic diagnostic tools tailoring therapy to minimize drug-induced adverse events, even though in the future DAAs likely will progressively replace RBV-based treatment.
The low-density lipoprotein receptor (LDLR) promotes HCV endocytosis and thus particle entry to the cell. Association of polymorphisms within the LDLR gene with the pathogenesis of familial hypercholesterolemia, atherosclerosis and obesity is known[226]. LDLR gene variant was associated to SVR to peg-IFN/RBV[227] and in HIV/HCV co-infected higher levels of LDLR are observed in carriers of rs1297860 CC than in non-CC carriers[228], probably due to the reduced LDLR gene expression induced by a lower activity of endogenous IFN-α in CC carriers[229]. Most likely for this reason LDLR can improve the predictive value of IL28B variants for HCV treatment response[228].
Previous studies supported the importance of 25-OH vitamin D deficiency {defined as 25-hydroxyvitamin D [25(OH)D3] serum concentration < 20 ng/mL} in influencing HCV therapy outcome[230,231] and this is relevant because vitamin D status might represent a potentially modifiable determinant of treatment outcome[232]. Very interestingly, some studies showed the promising possibility of adding vitamin D to conventional peg-α-2b/ribavirin therapy for treatment-naïve patients with chronic HCV genotype 1 and 2/3 infection significantly improves the viral response[231,233].
In recent times, the interesting findings were that three SNPs identified in two large GWAS (GC, encoding the vitamin D binding protein; DHCR7, encoding 7-dehydrocholesterol reductase; and CYP2R1, encoding a liver 25-hydroxylase) were genetic determinants of reduced 25(OH)D3 serum levels[234,235]. More recently, Lange et al[236] investigated the association between these SNPs and the risk of HCV-induced HCC development in 1279 chronic HCV infected patients with HCC and 4325 without HCC, respectively. The results of this study indicated a mild association of genetic variations in CYP2R1, GC and DHCR7 with progression to HCC in patients with chronic hepatitis C and confirmed the functional role of vitamin D in the prevention of HCV-related HCC.
Some studies have shown the usefulness of combined measurement of serum bile acids and ferritin as prognostic markers to predict failure of SVR to antiviral treatment in chronic hepatitis C[237]. A recent study identified a common polymorphism in the ABCB11 gene as host factor affecting antiviral treatment response in chronic hepatitis C, investigating the influence of plasma bile acids concentrations and an ABCB11 polymorphism associated with lower transporter expression on viral load and SVR in 441 Caucasian patients treated with peg-IFN/RBV. The results supported the role for bile acids as host factor affecting therapy response in HCV-2/3 patients, whereas a weaker association was found for HCV-1[238].
In recent years several studies identified genetic variants associated with fibrosis progression besides the above mentioned IL28B, VDR and ABCB11: Patatin-like phospholipase 3, ring finger protein 7, receptor tyrosine kinase MerTK, Tubby like protein 1[224]. These data were replicated in different studies[203].
Challenges and opportunities will characterise the story of HCV infection also in the new era. Despite the great therapeutic advances, improvements in HCV surveillance, epidemiological mapping, testing, prevention and therapy are urgently needed. Currently HCV vaccine candidates have shown promising results in animal models and data from clinical phase 1/2 trials are expected. Predictors of response to therapy represent a chance for modulated and personalized treatment management, when also very potent therapies will be available. The ultimate goal of global HCV control will be achieved with the joint efforts of researchers and public health workers.
P- Reviewer: Conti B, Svicher V S- Editor: Qi Y L- Editor: A E- Editor: Wang CH
| 2. | Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359-362. [PubMed] [Cited in This Article: ] |
| 3. | Houghton M. Discovery of the hepatitis C virus. Liver Int. 2009;29 Suppl 1:82-88. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 75] [Article Influence: 5.0] [Reference Citation Analysis (1)] |
| 4. | Alter MJ, Margolis HS, Krawczynski K, Judson FN, Mares A, Alexander WJ, Hu PY, Miller JK, Gerber MA, Sampliner RE. The natural history of community-acquired hepatitis C in the United States. The Sentinel Counties Chronic non-A, non-B Hepatitis Study Team. N Engl J Med. 1992;327:1899-1905. [PubMed] [Cited in This Article: ] |
| 5. | Alter HJ, Purcell RH, Shih JW, Melpolder JC, Houghton M, Choo QL, Kuo G. Detection of antibody to hepatitis C virus in prospectively followed transfusion recipients with acute and chronic non-A, non-B hepatitis. N Engl J Med. 1989;321:1494-1500. [PubMed] [Cited in This Article: ] |
| 6. | van der Poel CL, Reesink HW, Lelie PN, Leentvaar-Kuypers A, Choo QL, Kuo G, Houghton M. Anti-hepatitis C antibodies and non-A, non-B post-transfusion hepatitis in The Netherlands. Lancet. 1989;2:297-298. [PubMed] [Cited in This Article: ] |
| 7. | Bukh J, Purcell RH, Miller RH. Importance of primer selection for the detection of hepatitis C virus RNA with the polymerase chain reaction assay. Proc Natl Acad Sci USA. 1992;89:187-191. [PubMed] [Cited in This Article: ] |
| 8. | Cha TA, Kolberg J, Irvine B, Stempien M, Beall E, Yano M, Choo QL, Houghton M, Kuo G, Han JH. Use of a signature nucleotide sequence of hepatitis C virus for detection of viral RNA in human serum and plasma. J Clin Microbiol. 1991;29:2528-2534. [PubMed] [Cited in This Article: ] |
| 9. | Chevaliez S, Rodriguez C, Pawlotsky JM. New virologic tools for management of chronic hepatitis B and C. Gastroenterology. 2012;142:1303-1313.e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 101] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 10. | Park Y, Lee JH, Kim BS, Kim do Y, Han KH, Kim HS. New automated hepatitis C virus (HCV) core antigen assay as an alternative to real-time PCR for HCV RNA quantification. J Clin Microbiol. 2010;48:2253-2256. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 11. | Hosseini-Moghaddam SM, Iran-Pour E, Rotstein C, Husain S, Lilly L, Renner E, Mazzulli T. Hepatitis C core Ag and its clinical applicability: potential advantages and disadvantages for diagnosis and follow-up? Rev Med Virol. 2012;22:156-165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 12. | Descamps V, Op de Beeck A, Plassart C, Brochot E, François C, Helle F, Adler M, Bourgeois N, Degré D, Duverlie G. Strong correlation between liver and serum levels of hepatitis C virus core antigen and RNA in chronically infected patients. J Clin Microbiol. 2012;50:465-468. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 13. | Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57:1333-1342. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1770] [Cited by in F6Publishing: 1796] [Article Influence: 163.3] [Reference Citation Analysis (0)] |
| 14. | Grebely J, Dore GJ. What is killing people with hepatitis C virus infection? Semin Liver Dis. 2011;31:331-339. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 110] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 15. | Denniston MM, Klevens RM, McQuillan GM, Jiles RB. Awareness of infection, knowledge of hepatitis C, and medical follow-up among individuals testing positive for hepatitis C: National Health and Nutrition Examination Survey 2001-2008. Hepatology. 2012;55:1652-1661. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 270] [Cited by in F6Publishing: 287] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 16. | Di Lorenzo C, Angus AG, Patel AH. Hepatitis C virus evasion mechanisms from neutralizing antibodies. Viruses. 2011;3:2280-2300. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 50] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 17. | Barritt AS, Fried MW. Maximizing opportunities and avoiding mistakes in triple therapy for hepatitis C virus. Gastroenterology. 2012;142:1314-1323.e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 18. | FDA Drug Safety Communication. Serious skin reactions after combination treatment with the Hepatitis C drugs Incivek (telaprevir), peginterferon alfa, and ribavirin. 19 December 2012. Available from: http://www.fda.gov/Drugs/DrugSafety/ucm332731.htm. [Cited in This Article: ] |
| 19. | Schaefer EA, Chung RT. Anti-hepatitis C virus drugs in development. Gastroenterology. 2012;142:1340-1350.e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 72] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 20. | Khaliq S, Jahan S, Pervaiz A. Sequence variability of HCV Core region: important predictors of HCV induced pathogenesis and viral production. Infect Genet Evol. 2011;11:543-556. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 33] [Article Influence: 2.5] [Reference Citation Analysis (2)] |
| 21. | Vieyres G, Thomas X, Descamps V, Duverlie G, Patel AH, Dubuisson J. Characterization of the envelope glycoproteins associated with infectious hepatitis C virus. J Virol. 2010;84:10159-10168. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 162] [Cited by in F6Publishing: 169] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 22. | Thimme R, Binder M, Bartenschlager R. Failure of innate and adaptive immune responses in controlling hepatitis C virus infection. FEMS Microbiol Rev. 2012;36:663-683. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 99] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 23. | Gale M, Foy EM. Evasion of intracellular host defence by hepatitis C virus. Nature. 2005;436:939-945. [PubMed] [Cited in This Article: ] |
| 24. | Lambotin M, Raghuraman S, Stoll-Keller F, Baumert TF, Barth H. A look behind closed doors: interaction of persistent viruses with dendritic cells. Nat Rev Microbiol. 2010;8:350-360. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 25. | Cheent K, Khakoo SI. Natural killer cells and hepatitis C: action and reaction. Gut. 2011;60:268-278. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 75] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 26. | Rehermann B. Hepatitis C virus versus innate and adaptive immune responses: a tale of coevolution and coexistence. J Clin Invest. 2009;119:1745-1754. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 394] [Cited by in F6Publishing: 405] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 27. | Lin W, Kim SS, Yeung E, Kamegaya Y, Blackard JT, Kim KA, Holtzman MJ, Chung RT. Hepatitis C virus core protein blocks interferon signaling by interaction with the STAT1 SH2 domain. J Virol. 2006;80:9226-9235. [PubMed] [Cited in This Article: ] |
| 28. | Kumthip K, Chusri P, Jilg N, Zhao L, Fusco DN, Zhao H, Goto K, Cheng D, Schaefer EA, Zhang L. Hepatitis C virus NS5A disrupts STAT1 phosphorylation and suppresses type I interferon signaling. J Virol. 2012;86:8581-8591. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 67] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 29. | World Health Organization Hepatitis C Fact Sheet 2012. Available from: http://www.who.int/mediacentre/factsheets/fs164/en/. [Cited in This Article: ] |
| 30. | Wise M, Bialek S, Finelli L, Bell BP, Sorvillo F. Changing trends in hepatitis C-related mortality in the United States, 1995-2004. Hepatology. 2008;47:1128-1135. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 157] [Cited by in F6Publishing: 170] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 31. | Lavanchy D. The global burden of hepatitis C. Liver Int. 2009;29 Suppl 1:74-81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 933] [Cited by in F6Publishing: 1020] [Article Influence: 68.0] [Reference Citation Analysis (0)] |
| 32. | Global Burden Of Hepatitis C Working Group. Global burden of disease (GBD) for hepatitis C. J Clin Pharmacol. 2004;44:20-29. [PubMed] [Cited in This Article: ] |
| 33. | Lavanchy D. Evolving epidemiology of hepatitis C virus. Clin Microbiol Infect. 2011;17:107-115. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 919] [Cited by in F6Publishing: 929] [Article Influence: 71.5] [Reference Citation Analysis (2)] |
| 34. | Hepatitis C--global prevalence (update). Wkly Epidemiol Rec. 1999;74:425-427. [PubMed] [Cited in This Article: ] |
| 35. | Nerrienet E, Pouillot R, Lachenal G, Njouom R, Mfoupouendoun J, Bilong C, Mauclere P, Pasquier C, Ayouba A. Hepatitis C virus infection in cameroon: A cohort-effect. J Med Virol. 2005;76:208-214. [PubMed] [Cited in This Article: ] |
| 36. | Guerra J, Garenne M, Mohamed MK, Fontanet A. HCV burden of infection in Egypt: results from a nationwide survey. J Viral Hepat. 2012;19:560-567. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 131] [Cited by in F6Publishing: 135] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 37. | Miller FD, Abu-Raddad LJ. Evidence of intense ongoing endemic transmission of hepatitis C virus in Egypt. Proc Natl Acad Sci USA. 2010;107:14757-14762. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 137] [Cited by in F6Publishing: 146] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 38. | Centers for Disease Control and Prevention (CDC). Progress toward prevention and control of hepatitis C virus infection--Egypt, 2001-2012. MMWR Morb Mortal Wkly Rep. 2012;61:545-549. [PubMed] [Cited in This Article: ] |
| 39. | Frank C, Mohamed MK, Strickland GT, Lavanchy D, Arthur RR, Magder LS, El Khoby T, Abdel-Wahab Y, Aly Ohn ES, Anwar W. The role of parenteral antischistosomal therapy in the spread of hepatitis C virus in Egypt. Lancet. 2000;355:887-891. [PubMed] [Cited in This Article: ] |
| 40. | Paez Jimenez A, Sharaf Eldin N, Rimlinger F, El-Daly M, El-Hariri H, El-Hoseiny M, Mohsen A, Mostafa A, Delarocque-Astagneau E, Abdel-Hamid M. HCV iatrogenic and intrafamilial transmission in Greater Cairo, Egypt. Gut. 2010;59:1554-1560. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 41. | Plancoulaine S, Mohamed MK, Arafa N, Bakr I, Rekacewicz C, Trégouët DA, Obach D, El Daly M, Thiers V, Féray C. Dissection of familial correlations in hepatitis C virus (HCV) seroprevalence suggests intrafamilial viral transmission and genetic predisposition to infection. Gut. 2008;57:1268-1274. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 36] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 42. | Mohamed MK, Abdel-Hamid M, Mikhail NN, Abdel-Aziz F, Medhat A, Magder LS, Fix AD, Strickland GT. Intrafamilial transmission of hepatitis C in Egypt. Hepatology. 2005;42:683-687. [PubMed] [Cited in This Article: ] |
| 43. | Ali SA, Donahue RM, Qureshi H, Vermund SH. Hepatitis B and hepatitis C in Pakistan: prevalence and risk factors. Int J Infect Dis. 2009;13:9-19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 145] [Cited by in F6Publishing: 170] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 44. | Waheed Y, Shafi T, Safi SZ, Qadri I. Hepatitis C virus in Pakistan: a systematic review of prevalence, genotypes and risk factors. World J Gastroenterol. 2009;15:5647-5653. [PubMed] [Cited in This Article: ] |
| 45. | Merat S, Rezvan H, Nouraie M, Jafari E, Abolghasemi H, Radmard AR, Zaer-rezaii H, Amini-Kafiabad S, Maghsudlu M, Pourshams A. Seroprevalence of hepatitis C virus: the first population-based study from Iran. Int J Infect Dis. 2010;14 Suppl 3:e113-e116. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 85] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 46. | Alavian SM. Hepatitis C infection in Iran: A review article. Iranian J Clin Infect Dis. 2009;4:47-59. [Cited in This Article: ] |
| 47. | Xia GL, Liu CB, Cao HL, Bi SL, Zhan MY, Su CA, Nan JH, Qi XQ. Prevalence of hepatitis B and C virus infections in the general Chinese population. Results from a nationwide cross-sectional seroepidemiologic study of hepatitis A, B, C, D, and E virus infections in China, 1992. Int Hep Comm. 1996;5:62-73. [Cited in This Article: ] |
| 48. | Lu J, Zhou Y, Lin X, Jiang Y, Tian R, Zhang Y, Wu J, Zhang F, Zhang Y, Wang Y. General epidemiological parameters of viral hepatitis A, B, C, and E in six regions of China: a cross-sectional study in 2007. PLoS One. 2009;4:e8467. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 89] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 49. | Liu F, Chen K, He Z, Ning T, Pan Y, Cai H, Ke Y. Hepatitis C seroprevalence and associated risk factors, Anyang, China. Emerg Infect Dis. 2009;15:1819-1822. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 50. | Yan Z, Fan K, Wang Y, Fan Y, Tan Z, Deng G. Changing pattern of clinical epidemiology on hepatitis C virus infection in southwest china. Hepat Mon. 2012;12:196-204. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 19] [Reference Citation Analysis (0)] |
| 51. | Paez Jimenez A, Mohamed MK, Eldin NS, Seif HA, El Aidi S, Sultan Y, Elsaid N, Rekacewicz C, El-Hoseiny M, El-Daly M. Injection drug use is a risk factor for HCV infection in urban Egypt. PLoS One. 2009;4:e7193. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 52. | Thaikruea L, Thongsawat S, Maneekarn N, Netski D, Thomas DL, Nelson KE. Risk factors for hepatitis C virus infection among blood donors in northern Thailand. Transfusion. 2004;44:1433-1440. [PubMed] [Cited in This Article: ] |
| 53. | Alter MJ. Epidemiology of hepatitis C virus infection. World J Gastroenterol. 2007;13:2436-2441. [PubMed] [Cited in This Article: ] |
| 54. | Alter MJ, Kruszon-Moran D, Nainan OV, McQuillan GM, Gao F, Moyer LA, Kaslow RA, Margolis HS. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N Engl J Med. 1999;341:556-562. [PubMed] [Cited in This Article: ] |
| 55. | Chak E, Talal AH, Sherman KE, Schiff ER, Saab S. Hepatitis C virus infection in USA: an estimate of true prevalence. Liver Int. 2011;31:1090-1101. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 309] [Cited by in F6Publishing: 320] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 56. | Armstrong GL, Alter MJ, McQuillan GM, Margolis HS. The past incidence of hepatitis C virus infection: implications for the future burden of chronic liver disease in the United States. Hepatology. 2000;31:777-782. [PubMed] [Cited in This Article: ] |
| 57. | Williams I. Epidemiology of hepatitis C in the United States. Am J Med. 1999;107:2S-9S. [PubMed] [Cited in This Article: ] |
| 58. | Williams IT, Bell BP, Kuhnert W, Alter MJ. Incidence and transmission patterns of acute hepatitis C in the United States, 1982-2006. Arch Intern Med. 2011;171:242-248. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 119] [Cited by in F6Publishing: 126] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 59. | The Kirby Institute. HIV, Viral Hepatitis and Sexually Transmissible Infections in Australia. Annual Surveillance Report 2012. Available from: http://www.kirby.unsw.edu.au/sites/default/files/hiv/resources/2012AnnualSurvAnnual.pdf. [Cited in This Article: ] |
| 60. | Razali K, Thein HH, Bell J, Cooper-Stanbury M, Dolan K, Dore G, George J, Kaldor J, Karvelas M, Li J. Modelling the hepatitis C virus epidemic in Australia. Drug Alcohol Depend. 2007;91:228-235. [PubMed] [Cited in This Article: ] |
| 61. | Sievert W, Altraif I, Razavi HA, Abdo A, Ahmed EA, Alomair A, Amarapurkar D, Chen CH, Dou X, El Khayat H. A systematic review of hepatitis C virus epidemiology in Asia, Australia and Egypt. Liver Int. 2011;31 Suppl 2:61-80. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 385] [Cited by in F6Publishing: 417] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 62. | Yoshizawa H. Hepatocellular carcinoma associated with hepatitis C virus infection in Japan: projection to other countries in the foreseeable future. Oncology. 2002;62 Suppl 1:8-17. [PubMed] [Cited in This Article: ] |
| 63. | Tanaka Y, Hanada K, Orito E, Akahane Y, Chayama K, Yoshizawa H, Sata M, Ohta N, Miyakawa Y, Gojobori T. Molecular evolutionary analyses implicate injection treatment for schistosomiasis in the initial hepatitis C epidemics in Japan. J Hepatol. 2005;42:47-53. [PubMed] [Cited in This Article: ] |
| 64. | Dalgard O, Jeansson S, Skaug K, Raknerud N, Bell H. Hepatitis C in the general adult population of Oslo: prevalence and clinical spectrum. Scand J Gastroenterol. 2003;38:864-870. [PubMed] [Cited in This Article: ] |
| 65. | Harris RJ, Ramsay M, Hope VD, Brant L, Hickman M, Foster GR, De Angelis D. Hepatitis C prevalence in England remains low and varies by ethnicity: an updated evidence synthesis. Eur J Public Health. 2012;22:187-192. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 72] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 66. | Delarocque-Astagneau E, Pillonel J, De Valk H, Perra A, Laperche S, Desenclos JC. An incident case-control study of modes of hepatitis C virus transmission in France. Ann Epidemiol. 2007;17:755-762. [PubMed] [Cited in This Article: ] |
| 67. | Duberg A, Janzon R, Bäck E, Ekdahl K, Blaxhult A. The epidemiology of hepatitis C virus infection in Sweden. Euro Surveill. 2008;13. [PubMed] [Cited in This Article: ] |
| 68. | Prasad L, Spicher VM, Zwahlen M, Rickenbach M, Helbling B, Negro F. Cohort Profile: the Swiss Hepatitis C Cohort Study (SCCS). Int J Epidemiol. 2007;36:731-737. [PubMed] [Cited in This Article: ] |
| 69. | European Centre for Disease Prevention and Control (ECDC). Annual epidemiological report on communicable diseases in Europe 2008. Available from: http://ecdc.europa.eu/en/files/pdf/Publications/081215_AER_long_2008.pdf. [Cited in This Article: ] |
| 70. | Esteban JI, Sauleda S, Quer J. The changing epidemiology of hepatitis C virus infection in Europe. J Hepatol. 2008;48:148-162. [PubMed] [Cited in This Article: ] |
| 71. | Simmonds P, Bukh J, Combet C, Deléage G, Enomoto N, Feinstone S, Halfon P, Inchauspé G, Kuiken C, Maertens G. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962-973. [PubMed] [Cited in This Article: ] |
| 72. | Gottwein JM, Bukh J. Cutting the gordian knot-development and biological relevance of hepatitis C virus cell culture systems. Adv Virus Res. 2008;71:51-133. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 48] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 73. | Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology. 2014;59:318-327. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 965] [Cited by in F6Publishing: 924] [Article Influence: 92.4] [Reference Citation Analysis (0)] |
| 74. | Duffy S, Shackelton LA, Holmes EC. Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet. 2008;9:267-276. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1015] [Cited by in F6Publishing: 1001] [Article Influence: 62.6] [Reference Citation Analysis (0)] |
| 75. | Chao DT, Abe K, Nguyen MH. Systematic review: epidemiology of hepatitis C genotype 6 and its management. Aliment Pharmacol Ther. 2011;34:286-296. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 76. | Poordad F, Dieterich D. Treating hepatitis C: current standard of care and emerging direct-acting antiviral agents. J Viral Hepat. 2012;19:449-464. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 77. | Chlabicz S, Flisiak R, Kowalczuk O, Grzeszczuk A, Pytel-Krolczuk B, Prokopowicz D, Chyczewski L. Changing HCV genotypes distribution in Poland--relation to source and time of infection. J Clin Virol. 2008;42:156-159. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 43] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 78. | Tallo T, Norder H, Tefanova V, Krispin T, Schmidt J, Ilmoja M, Orgulas K, Pruunsild K, Priimägi L, Magnius LO. Genetic characterization of hepatitis C virus strains in Estonia: fluctuations in the predominating subtype with time. J Med Virol. 2007;79:374-382. [PubMed] [Cited in This Article: ] |
| 79. | Katsoulidou A, Sypsa V, Tassopoulos NC, Boletis J, Karafoulidou A, Ketikoglou I, Tsantoulas D, Vafiadi I, Hatzis G, Skoutelis A. Molecular epidemiology of hepatitis C virus (HCV) in Greece: temporal trends in HCV genotype-specific incidence and molecular characterization of genotype 4 isolates. J Viral Hepat. 2006;13:19-27. [PubMed] [Cited in This Article: ] |
| 80. | Nguyen MH, Keeffe EB. Prevalence and treatment of hepatitis C virus genotypes 4, 5, and 6. Clin Gastroenterol Hepatol. 2005;3:S97-S101. [PubMed] [Cited in This Article: ] |
| 81. | Mauss S, Berger F, Vogel M, Pfeiffer-Vornkahl H, Alshuth U, Rockstroh JK, Niederau C, Hüppe D. Treatment results of chronic hepatitis C genotype 5 and 6 infections in Germany. Z Gastroenterol. 2012;50:441-444. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 82. | Bostan N, Mahmood T. An overview about hepatitis C: a devastating virus. Crit Rev Microbiol. 2010;36:91-133. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 81] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 83. | Bunchorntavakul C, Chavalitdhamrong D, Tanwandee T. Hepatitis C genotype 6: A concise review and response-guided therapy proposal. World J Hepatol. 2013;5:496-504. [PubMed] [Cited in This Article: ] |
| 84. | Cornberg M, Razavi HA, Alberti A, Bernasconi E, Buti M, Cooper C, Dalgard O, Dillion JF, Flisiak R, Forns X. A systematic review of hepatitis C virus epidemiology in Europe, Canada and Israel. Liver Int. 2011;31 Suppl 2:30-60. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 287] [Cited by in F6Publishing: 304] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 85. | Negro F. Steatosis and insulin resistance in response to treatment of chronic hepatitis C. J Viral Hepat. 2012;19 Suppl 1:42-47. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 86. | van der Meer AJ, Veldt BJ, Feld JJ, Wedemeyer H, Dufour JF, Lammert F, Duarte-Rojo A, Heathcote EJ, Manns MP, Kuske L. Association between sustained virological response and all-cause mortality among patients with chronic hepatitis C and advanced hepatic fibrosis. JAMA. 2012;308:2584-2593. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1165] [Cited by in F6Publishing: 1128] [Article Influence: 94.0] [Reference Citation Analysis (0)] |
| 87. | Goossens N, Negro F. Is genotype 3 of the hepatitis C virus the new villain? Hepatology. 2014;59:2403-2412. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 106] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 88. | Manns MP, Wedemeyer H, Cornberg M. Treating viral hepatitis C: efficacy, side effects, and complications. Gut. 2006;55:1350-1359. [PubMed] [Cited in This Article: ] |
| 89. | Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med. 2013;19:837-849. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 416] [Cited by in F6Publishing: 408] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 90. | Khattab MA, Ferenci P, Hadziyannis SJ, Colombo M, Manns MP, Almasio PL, Esteban R, Abdo AA, Harrison SA, Ibrahim N. Management of hepatitis C virus genotype 4: recommendations of an international expert panel. J Hepatol. 2011;54:1250-1262. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 95] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 91. | Lee LY, Tong CY, Wong T, Wilkinson M. New therapies for chronic hepatitis C infection: a systematic review of evidence from clinical trials. Int J Clin Pract. 2012;66:342-355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 92. | Zeuzem S. Interferon-based therapy for chronic hepatitis C: current and future perspectives. Nat Clin Pract Gastroenterol Hepatol. 2008;5:610-622. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 77] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 93. | Jacobson IM, McHutchison JG, Dusheiko G, Di Bisceglie AM, Reddy KR, Bzowej NH, Marcellin P, Muir AJ, Ferenci P, Flisiak R. Telaprevir for previously untreated chronic hepatitis C virus infection. N Engl J Med. 2011;364:2405-2416. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1866] [Cited by in F6Publishing: 1835] [Article Influence: 141.2] [Reference Citation Analysis (0)] |
| 94. | Poordad F, McCone J, Bacon BR, Bruno S, Manns MP, Sulkowski MS, Jacobson IM, Reddy KR, Goodman ZD, Boparai N. Boceprevir for untreated chronic HCV genotype 1 infection. N Engl J Med. 2011;364:1195-1206. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1948] [Cited by in F6Publishing: 1951] [Article Influence: 150.1] [Reference Citation Analysis (0)] |
| 95. | World Health Organization. Prevention and control of viral hepatitis infection: framework for global action. Available from: http://www.who.int/entity/csr/disease/hepatitis/GHP_framework.pdf. [Cited in This Article: ] |
| 96. | Chang MH, You SL, Chen CJ, Liu CJ, Lee CM, Lin SM, Chu HC, Wu TC, Yang SS, Kuo HS. Decreased incidence of hepatocellular carcinoma in hepatitis B vaccinees: a 20-year follow-up study. J Natl Cancer Inst. 2009;101:1348-1355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 433] [Cited by in F6Publishing: 393] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 97. | Ni YH, Huang LM, Chang MH, Yen CJ, Lu CY, You SL, Kao JH, Lin YC, Chen HL, Hsu HY. Two decades of universal hepatitis B vaccination in taiwan: impact and implication for future strategies. Gastroenterology. 2007;132:1287-1293. [PubMed] [Cited in This Article: ] |
| 98. | Lindenbach BD, Thiel HJ, Rice CM. Flaviviridae: the viruses and their replication. Fields Virology. 5th ed. Philadelphia: Lippincott-Raven Publishers 2007; . [Cited in This Article: ] |
| 99. | Weiner AJ, Brauer MJ, Rosenblatt J, Richman KH, Tung J, Crawford K, Bonino F, Saracco G, Choo QL, Houghton M. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology. 1991;180:842-848. [PubMed] [Cited in This Article: ] |
| 100. | Kato N, Sekiya H, Ootsuyama Y, Nakazawa T, Hijikata M, Ohkoshi S, Shimotohno K. Humoral immune response to hypervariable region 1 of the putative envelope glycoprotein (gp70) of hepatitis C virus. J Virol. 1993;67:3923-3930. [PubMed] [Cited in This Article: ] |
| 101. | Farci P, Shimoda A, Coiana A, Diaz G, Peddis G, Melpolder JC, Strazzera A, Chien DY, Munoz SJ, Balestrieri A. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000;288:339-344. [PubMed] [Cited in This Article: ] |
| 102. | Bowen DG, Walker CM. Mutational escape from CD8+ T cell immunity: HCV evolution, from chimpanzees to man. J Exp Med. 2005;201:1709-1714. [PubMed] [Cited in This Article: ] |
| 103. | Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436:933-938. [PubMed] [Cited in This Article: ] |
| 104. | Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich HG, Mizokami M. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791-796. [PubMed] [Cited in This Article: ] |
| 105. | Zhong J, Gastaminza P, Chung J, Stamataki Z, Isogawa M, Cheng G, McKeating JA, Chisari FV. Persistent hepatitis C virus infection in vitro: coevolution of virus and host. J Virol. 2006;80:11082-11093. [PubMed] [Cited in This Article: ] |
| 106. | Lohmann V, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110-113. [PubMed] [Cited in This Article: ] |
| 107. | Bartenschlager R. Hepatitis C virus molecular clones: from cDNA to infectious virus particles in cell culture. Curr Opin Microbiol. 2006;9:416-422. [PubMed] [Cited in This Article: ] |
| 108. | Yi M, Villanueva RA, Thomas DL, Wakita T, Lemon SM. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc Natl Acad Sci USA. 2006;103:2310-2315. [PubMed] [Cited in This Article: ] |
| 109. | Thomas E, Gonzalez VD, Li Q, Modi AA, Chen W, Noureddin M, Rotman Y, Liang TJ. HCV infection induces a unique hepatic innate immune response associated with robust production of type III interferons. Gastroenterology. 2012;142:978-988. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 222] [Cited by in F6Publishing: 217] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 110. | Podevin P, Carpentier A, Pène V, Aoudjehane L, Carrière M, Zaïdi S, Hernandez C, Calle V, Méritet JF, Scatton O. Production of infectious hepatitis C virus in primary cultures of human adult hepatocytes. Gastroenterology. 2010;139:1355-1364. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 125] [Cited by in F6Publishing: 124] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 111. | Bukh J. Animal models for the study of hepatitis C virus infection and related liver disease. Gastroenterology. 2012;142:1279-1287.e3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 104] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 112. | Farci P, Alter HJ, Wong DC, Miller RH, Govindarajan S, Engle R, Shapiro M, Purcell RH. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci USA. 1994;91:7792-7796. [PubMed] [Cited in This Article: ] |
| 113. | Zhang P, Zhong L, Struble EB, Watanabe H, Kachko A, Mihalik K, Virata-Theimer ML, Alter HJ, Feinstone S, Major M. Depletion of interfering antibodies in chronic hepatitis C patients and vaccinated chimpanzees reveals broad cross-genotype neutralizing activity. Proc Natl Acad Sci USA. 2009;106:7537-7541. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 119] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 114. | Barth H, Rybczynska J, Patient R, Choi Y, Sapp RK, Baumert TF, Krawczynski K, Liang TJ. Both innate and adaptive immunity mediate protective immunity against hepatitis C virus infection in chimpanzees. Hepatology. 2011;54:1135-1148. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 115. | Meunier JC, Gottwein JM, Houghton M, Russell RS, Emerson SU, Bukh J, Purcell RH. Vaccine-induced cross-genotype reactive neutralizing antibodies against hepatitis C virus. J Infect Dis. 2011;204:1186-1190. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 82] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 116. | Washburn ML, Bility MT, Zhang L, Kovalev GI, Buntzman A, Frelinger JA, Barry W, Ploss A, Rice CM, Su L. A humanized mouse model to study hepatitis C virus infection, immune response, and liver disease. Gastroenterology. 2011;140:1334-1344. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 238] [Cited by in F6Publishing: 226] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 117. | Dorner M, Horwitz JA, Donovan BM, Labitt RN, Budell WC, Friling T, Vogt A, Catanese MT, Satoh T, Kawai T. Completion of the entire hepatitis C virus life cycle in genetically humanized mice. Nature. 2013;501:237-241. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 178] [Cited by in F6Publishing: 175] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 118. | Strickland GT, El-Kamary SS, Klenerman P, Nicosia A. Hepatitis C vaccine: supply and demand. Lancet Infect Dis. 2008;8:379-386. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 67] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 119. | Heller T, Rehermann B. Acute hepatitis C: a multifaceted disease. Semin Liver Dis. 2005;25:7-17. [PubMed] [Cited in This Article: ] |
| 120. | Zou S, Dodd RY, Stramer SL, Strong DM. Probability of viremia with HBV, HCV, HIV, and HTLV among tissue donors in the United States. N Engl J Med. 2004;351:751-759. [PubMed] [Cited in This Article: ] |
| 121. | Ghany MG, Strader DB, Thomas DL, Seeff LB. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology. 2009;49:1335-1374. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2252] [Cited by in F6Publishing: 2192] [Article Influence: 146.1] [Reference Citation Analysis (1)] |
| 122. | Choo QL, Kuo G, Ralston R, Weiner A, Chien D, Van Nest G, Han J, Berger K, Thudium K, Kuo C. Vaccination of chimpanzees against infection by the hepatitis C virus. Proc Natl Acad Sci USA. 1994;91:1294-1298. [PubMed] [Cited in This Article: ] |
| 123. | Frey SE, Houghton M, Coates S, Abrignani S, Chien D, Rosa D, Pileri P, Ray R, Di Bisceglie AM, Rinella P. Safety and immunogenicity of HCV E1E2 vaccine adjuvanted with MF59 administered to healthy adults. Vaccine. 2010;28:6367-6373. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 177] [Cited by in F6Publishing: 182] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 124. | Houghton M. Prospects for prophylactic and therapeutic vaccines against the hepatitis C viruses. Immunol Rev. 2011;239:99-108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 120] [Cited by in F6Publishing: 119] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 125. | Drane D, Maraskovsky E, Gibson R, Mitchell S, Barnden M, Moskwa A, Shaw D, Gervase B, Coates S, Houghton M. Priming of CD4+ and CD8+ T cell responses using a HCV core ISCOMATRIX vaccine: a phase I study in healthy volunteers. Hum Vaccin. 2009;5:151-157. [PubMed] [Cited in This Article: ] |
| 126. | Polakos NK, Drane D, Cox J, Ng P, Selby MJ, Chien D, O’Hagan DT, Houghton M, Paliard X. Characterization of hepatitis C virus core-specific immune responses primed in rhesus macaques by a nonclassical ISCOM vaccine. J Immunol. 2001;166:3589-3598. [PubMed] [Cited in This Article: ] |
| 127. | Reali E, Houghton M, Abrignani S. Hepatitis C virus vaccines. Vaccines. 5th ed. Netherlands: Saunders Elsevier 2008; 1187-1199. [Cited in This Article: ] |
| 128. | Beaumont E, Patient R, Hourioux C, Dimier-Poisson I, Roingeard P. Chimeric hepatitis B virus/hepatitis C virus envelope proteins elicit broadly neutralizing antibodies and constitute a potential bivalent prophylactic vaccine. Hepatology. 2013;57:1303-1313. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 129. | Baumert TF, Ito S, Wong DT, Liang TJ. Hepatitis C virus structural proteins assemble into viruslike particles in insect cells. J Virol. 1998;72:3827-3836. [PubMed] [Cited in This Article: ] |
| 130. | Murata K, Lechmann M, Qiao M, Gunji T, Alter HJ, Liang TJ. Immunization with hepatitis C virus-like particles protects mice from recombinant hepatitis C virus-vaccinia infection. Proc Natl Acad Sci USA. 2003;100:6753-6758. [PubMed] [Cited in This Article: ] |
| 131. | Jeong SH, Qiao M, Nascimbeni M, Hu Z, Rehermann B, Murthy K, Liang TJ. Immunization with hepatitis C virus-like particles induces humoral and cellular immune responses in nonhuman primates. J Virol. 2004;78:6995-7003. [PubMed] [Cited in This Article: ] |
| 132. | Elmowalid GA, Qiao M, Jeong SH, Borg BB, Baumert TF, Sapp RK, Hu Z, Murthy K, Liang TJ. Immunization with hepatitis C virus-like particles results in control of hepatitis C virus infection in chimpanzees. Proc Natl Acad Sci USA. 2007;104:8427-8432. [PubMed] [Cited in This Article: ] |
| 133. | Chua BY, Johnson D, Tan A, Earnest-Silveira L, Sekiya T, Chin R, Torresi J, Jackson DC. Hepatitis C VLPs delivered to dendritic cells by a TLR2 targeting lipopeptide results in enhanced antibody and cell-mediated responses. PLoS One. 2012;7:e47492. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 134. | Garrone P, Fluckiger AC, Mangeot PE, Gauthier E, Dupeyrot-Lacas P, Mancip J, Cangialosi A, Du Chéné I, LeGrand R, Mangeot I. A prime-boost strategy using virus-like particles pseudotyped for HCV proteins triggers broadly neutralizing antibodies in macaques. Sci Transl Med. 2011;3:94ra71. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 109] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 135. | Reyes-del Valle J, de la Fuente C, Turner MA, Springfeld C, Apte-Sengupta S, Frenzke ME, Forest A, Whidby J, Marcotrigiano J, Rice CM. Broadly neutralizing immune responses against hepatitis C virus induced by vectored measles viruses and a recombinant envelope protein booster. J Virol. 2012;86:11558-11566. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 34] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 136. | Chiarella P, Fazio VM, Signori E. Application of electroporation in DNA vaccination protocols. Curr Gene Ther. 2010;10:281-286. [PubMed] [Cited in This Article: ] |
| 137. | Youn JW, Park SH, Lavillette D, Cosset FL, Yang SH, Lee CG, Jin HT, Kim CM, Shata MT, Lee DH. Sustained E2 antibody response correlates with reduced peak viremia after hepatitis C virus infection in the chimpanzee. Hepatology. 2005;42:1429-1436. [PubMed] [Cited in This Article: ] |
| 138. | Klade CS, Wedemeyer H, Berg T, Hinrichsen H, Cholewinska G, Zeuzem S, Blum H, Buschle M, Jelovcan S, Buerger V. Therapeutic vaccination of chronic hepatitis C nonresponder patients with the peptide vaccine IC41. Gastroenterology. 2008;134:1385-1395. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 118] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 139. | Youn JW, Hu YW, Tricoche N, Pfahler W, Shata MT, Dreux M, Cosset FL, Folgori A, Lee DH, Brotman B. Evidence for protection against chronic hepatitis C virus infection in chimpanzees by immunization with replicating recombinant vaccinia virus. J Virol. 2008;82:10896-10905. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 48] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 140. | Rollier CS, Paranhos-Baccala G, Verschoor EJ, Verstrepen BE, Drexhage JA, Fagrouch Z, Berland JL, Komurian-Pradel F, Duverger B, Himoudi N. Vaccine-induced early control of hepatitis C virus infection in chimpanzees fails to impact on hepatic PD-1 and chronicity. Hepatology. 2007;45:602-613. [PubMed] [Cited in This Article: ] |
| 141. | Haller AA, Lauer GM, King TH, Kemmler C, Fiolkoski V, Lu Y, Bellgrau D, Rodell TC, Apelian D, Franzusoff A. Whole recombinant yeast-based immunotherapy induces potent T cell responses targeting HCV NS3 and Core proteins. Vaccine. 2007;25:1452-1463. [PubMed] [Cited in This Article: ] |
| 142. | Habersetzer F, Honnet G, Bain C, Maynard-Muet M, Leroy V, Zarski JP, Feray C, Baumert TF, Bronowicki JP, Doffoël M. A poxvirus vaccine is safe, induces T-cell responses, and decreases viral load in patients with chronic hepatitis C. Gastroenterology. 2011;141:890-899.e1-4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 65] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 143. | Rollier CS, Reyes-Sandoval A, Cottingham MG, Ewer K, Hill AV. Viral vectors as vaccine platforms: deployment in sight. Curr Opin Immunol. 2011;23:377-382. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 154] [Cited by in F6Publishing: 144] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 144. | Liang TJ. Current progress in development of hepatitis C virus vaccines. Nat Med. 2013;19:869-878. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 129] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 145. | European Association for the Study of the Liver. EASL Clinical Practice Guidelines: management of hepatitis C virus infection. J Hepatol. 2011;55:245-264. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 889] [Cited by in F6Publishing: 968] [Article Influence: 74.5] [Reference Citation Analysis (0)] |
| 146. | Centers for Disease Control and Prevention (CDC). Testing for HCV infection: an update of guidance for clinicians and laboratorians. MMWR Morb Mortal Wkly Rep. 2013;62:362-365. [PubMed] [Cited in This Article: ] |
| 147. | de Leuw P, Sarrazin C, Zeuzem S. How to use virological tools for the optimal management of chronic hepatitis C. Liver Int. 2011;31 Suppl 1:3-12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 148. | Kamili S, Drobeniuc J, Araujo AC, Hayden TM. Laboratory diagnostics for hepatitis C virus infection. Clin Infect Dis. 2012;55 Suppl 1:S43-S48. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 119] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 149. | Lin HJ, Lau JY, Lauder IJ, Shi N, Lai CL, Hollinger FB. The hepatitis C virus genome: a guide to its conserved sequences and candidate epitopes. Virus Res. 1993;30:27-41. [PubMed] [Cited in This Article: ] |
| 150. | Barrera JM, Francis B, Ercilla G, Nelles M, Achord D, Darner J, Lee SR. Improved detection of anti-HCV in post-transfusion hepatitis by a third-generation ELISA. Vox Sang. 1995;68:15-18. [PubMed] [Cited in This Article: ] |
| 151. | Ansaldi F, Icardi G. Simultaneous detection of anti-HCV antibody and HCV core antigen. Methods Mol Biol. 2009;510:15-23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 152. | Chevaliez S. Virological tools to diagnose and monitor hepatitis C virus infection. Clin Microbiol Infect. 2011;17:116-121. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 45] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 153. | Coppola N, Pisapia R, Tonziello G, Masiello A, Martini S, Pisaturo M, Messina V, Sagnelli C, Macera M, Signoriello G. Improvement in the aetiological diagnosis of acute hepatitis C: a diagnostic protocol based on the anti-HCV-IgM titre and IgG Avidity Index. J Clin Virol. 2009;46:222-229. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 32] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 154. | Kanno A, Kazuyama Y. Immunoglobulin G antibody avidity assay for serodiagnosis of hepatitis C virus infection. J Med Virol. 2002;68:229-233. [PubMed] [Cited in This Article: ] |
| 155. | Smith BD, Teshale E, Jewett A, Weinbaum CM, Neaigus A, Hagan H, Jenness SM, Melville SK, Burt R, Thiede H. Performance of premarket rapid hepatitis C virus antibody assays in 4 national human immunodeficiency virus behavioral surveillance system sites. Clin Infect Dis. 2011;53:780-786. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 68] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 156. | Smith BD, Drobeniuc J, Jewett A, Branson BM, Garfein RS, Teshale E, Kamili S, Weinbaum CM. Evaluation of three rapid screening assays for detection of antibodies to hepatitis C virus. J Infect Dis. 2011;204:825-831. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 75] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 157. | Bouvier-Alias M, Patel K, Dahari H, Beaucourt S, Larderie P, Blatt L, Hezode C, Picchio G, Dhumeaux D, Neumann AU. Clinical utility of total HCV core antigen quantification: a new indirect marker of HCV replication. Hepatology. 2002;36:211-218. [PubMed] [Cited in This Article: ] |
| 158. | Ottiger C, Gygli N, Huber AR. Detection limit of architect hepatitis C core antigen assay in correlation with HCV RNA, and renewed confirmation algorithm for reactive anti-HCV samples. J Clin Virol. 2013;58:535-540. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 159. | Ross RS, Viazov S, Salloum S, Hilgard P, Gerken G, Roggendorf M. Analytical performance characteristics and clinical utility of a novel assay for total hepatitis C virus core antigen quantification. J Clin Microbiol. 2010;48:1161-1168. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 74] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 160. | Moscato GA, Giannelli G, Grandi B, Pieri D, Marsi O, Guarducci I, Batini I, Altomare E, Antonaci S, Capria A. Quantitative determination of hepatitis C core antigen in therapy monitoring for chronic hepatitis C. Intervirology. 2011;54:61-65. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 161. | Pawlotsky JM. Use and interpretation of hepatitis C virus diagnostic assays. Clin Liver Dis. 2003;7:127-137. [PubMed] [Cited in This Article: ] |
| 162. | Saldanha J, Heath A, Aberham C, Albrecht J, Gentili G, Gessner M, Pisani G. World Health Organization collaborative study to establish a replacement WHO international standard for hepatitis C virus RNA nucleic acid amplification technology assays. Vox Sang. 2005;88:202-204. [PubMed] [Cited in This Article: ] |
| 163. | Kessler HH, Hübner M, Konrad PM, Stelzl E, Stübler MM, Baser MH, Santner BI. Genotype impact on HCV RNA levels determined with the VERSANT HCV RNA 1.0 assay (kPCR). J Clin Virol. 2013;58:522-527. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 164. | Bouchardeau F, Cantaloube JF, Chevaliez S, Portal C, Razer A, Lefrère JJ, Pawlotsky JM, De Micco P, Laperche S. Improvement of hepatitis C virus (HCV) genotype determination with the new version of the INNO-LiPA HCV assay. J Clin Microbiol. 2007;45:1140-1145. [PubMed] [Cited in This Article: ] |
| 165. | Chevaliez S, Bouvier-Alias M, Vandervenet C, Pawlotsky JM. HCV genotype determination in clinical practice: weaknesses of assays based on the 5’noncoding region and improvement with the core-coding region. Hepatology. 2007;46 Suppl 1:839A. [Cited in This Article: ] |
| 166. | Stelzl E, van der Meer C, Gouw R, Beld M, Grahovac M, Marth E, Kessler HH. Determination of the hepatitis C virus subtype: comparison of sequencing and reverse hybridization assays. Clin Chem Lab Med. 2007;45:167-170. [PubMed] [Cited in This Article: ] |
| 167. | Verbeeck J, Stanley MJ, Shieh J, Celis L, Huyck E, Wollants E, Morimoto J, Farrior A, Sablon E, Jankowski-Hennig M. Evaluation of Versant hepatitis C virus genotype assay (LiPA) 2.0. J Clin Microbiol. 2008;46:1901-1906. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 99] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 168. | Gryadunov D, Nicot F, Dubois M, Mikhailovich V, Zasedatelev A, Izopet J. Hepatitis C virus genotyping using an oligonucleotide microarray based on the NS5B sequence. J Clin Microbiol. 2010;48:3910-3917. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 169. | Zeuzem S, Berg T, Moeller B, Hinrichsen H, Mauss S, Wedemeyer H, Sarrazin C, Hueppe D, Zehnter E, Manns MP. Expert opinion on the treatment of patients with chronic hepatitis C. J Viral Hepat. 2009;16:75-90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 181] [Cited by in F6Publishing: 188] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 170. | Asselah T, Benhamou Y, Marcellin P. Protease and polymerase inhibitors for the treatment of hepatitis C. Liver Int. 2009;29 Suppl 1:57-67. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 57] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 171. | Thompson AJ, McHutchison JG. Antiviral resistance and specifically targeted therapy for HCV (STAT-C). J Viral Hepat. 2009;16:377-387. [PubMed] [Cited in This Article: ] |
| 172. | Susser S, Welsch C, Wang Y, Zettler M, Domingues FS, Karey U, Hughes E, Ralston R, Tong X, Herrmann E. Characterization of resistance to the protease inhibitor boceprevir in hepatitis C virus-infected patients. Hepatology. 2009;50:1709-1718. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 243] [Cited by in F6Publishing: 261] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 173. | Victrelis TM (boceprevir). Summary of Products Characteristics, 2012. Available from: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Product_Information/human/ 002332/WC500109786.pdf. [Cited in This Article: ] |
| 174. | Incivo TM (telaprevir). Summary of Product Characteristics, 2012. Available from: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Product_Information/human/ 002313/ WC500115529.pdf. [Cited in This Article: ] |
| 175. | Thompson AJ, Locarnini SA, Beard MR. Resistance to anti-HCV protease inhibitors. Curr Opin Virol. 2011;1:599-606. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 176. | Bacon BR, Gordon SC, Lawitz E, Marcellin P, Vierling JM, Zeuzem S, Poordad F, Goodman ZD, Sings HL, Boparai N. Boceprevir for previously treated chronic HCV genotype 1 infection. N Engl J Med. 2011;364:1207-1217. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1287] [Cited by in F6Publishing: 1288] [Article Influence: 99.1] [Reference Citation Analysis (0)] |
| 177. | Zeuzem S, Andreone P, Pol S, Lawitz E, Diago M, Roberts S, Focaccia R, Younossi Z, Foster GR, Horban A. Telaprevir for retreatment of HCV infection. N Engl J Med. 2011;364:2417-2428. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1205] [Cited by in F6Publishing: 1194] [Article Influence: 91.8] [Reference Citation Analysis (0)] |
| 178. | Harrington PR, Zeng W, Naeger LK. Clinical relevance of detectable but not quantifiable hepatitis C virus RNA during boceprevir or telaprevir treatment. Hepatology. 2012;55:1048-1057. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 53] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 179. | Vierling JM. Hepatitis C virus viral assays in the direct-acting antiviral era. Clin Liver Dis. 2013;17:27-45. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 180. | Pawlotsky JM. Treatment of hepatitis C: how will we use viral kinetics, response-guided therapy? Curr Gastroenterol Rep. 2013;15:309. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 181. | Rosenberg WM, Tanwar S, Trembling P. Complexities of HCV management in the new era of direct-acting antiviral agents. QJM. 2014;107:17-19. [PubMed] [Cited in This Article: ] |
| 182. | Cobb B, Pockros PJ, Vilchez RA, Vierling JM. HCV RNA viral load assessments in the era of direct-acting antivirals. Am J Gastroenterol. 2013;108:471-475. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 183. | Neumann AU, Lam NP, Dahari H, Gretch DR, Wiley TE, Layden TJ, Perelson AS. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282:103-107. [PubMed] [Cited in This Article: ] |
| 184. | Lam NP, Neumann AU, Gretch DR, Wiley TE, Perelson AS, Layden TJ. Dose-dependent acute clearance of hepatitis C genotype 1 virus with interferon alfa. Hepatology. 1997;26:226-231. [PubMed] [Cited in This Article: ] |
| 185. | Dahari H, Guedj J, Perelson AS, Layden TJ. Hepatitis C Viral Kinetics in the Era of Direct Acting Antiviral Agents and IL28B. Curr Hepat Rep. 2011;10:214-227. [PubMed] [Cited in This Article: ] |
| 186. | Bartenschlager R, Lohmann V. Replication of hepatitis C virus. J Gen Virol. 2000;81:1631-1648. [PubMed] [Cited in This Article: ] |
| 187. | Bukh J, Miller RH, Purcell RH. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin Liver Dis. 1995;15:41-63. [PubMed] [Cited in This Article: ] |
| 188. | Sarrazin C, Zeuzem S. Resistance to direct antiviral agents in patients with hepatitis C virus infection. Gastroenterology. 2010;138:447-462. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 418] [Cited by in F6Publishing: 436] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 189. | Jacka B, Lamoury F, Simmonds P, Dore GJ, Grebely J, Applegate T. Sequencing of the Hepatitis C Virus: A Systematic Review. PLoS One. 2013;8:e67073. [PubMed] [Cited in This Article: ] |
| 190. | Kieffer TL, Sarrazin C, Miller JS, Welker MW, Forestier N, Reesink HW, Kwong AD, Zeuzem S. Telaprevir and pegylated interferon-alpha-2a inhibit wild-type and resistant genotype 1 hepatitis C virus replication in patients. Hepatology. 2007;46:631-639. [PubMed] [Cited in This Article: ] |
| 191. | Sarrazin C, Kieffer TL, Bartels D, Hanzelka B, Müh U, Welker M, Wincheringer D, Zhou Y, Chu HM, Lin C. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology. 2007;132:1767-1777. [PubMed] [Cited in This Article: ] |
| 192. | Berger KL, Triki I, Cartier M, Marquis M, Massariol MJ, Böcher WO, Datsenko Y, Steinmann G, Scherer J, Stern JO. Baseline hepatitis C virus (HCV) NS3 polymorphisms and their impact on treatment response in clinical studies of the HCV NS3 protease inhibitor faldaprevir. Antimicrob Agents Chemother. 2014;58:698-705. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 32] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 193. | Vermehren J, Sarrazin C. The role of resistance in HCV treatment. Best Pract Res Clin Gastroenterol. 2012;26:487-503. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 99] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 194. | Phenotype Working Group, Drug development Advisory Group. Clinically relevant HCV drug resistance mutations figures and tables (updated). Ann Forum Collab HIV Res. 2012;14:1-8. [Cited in This Article: ] |
| 195. | Wyles DL. Beyond telaprevir and boceprevir: resistance and new agents for hepatitis C virus infection. Top Antivir Med. 2012;20:139-145. [PubMed] [Cited in This Article: ] |
| 196. | McHutchison JG, Everson GT, Gordon SC, Jacobson IM, Sulkowski M, Kauffman R, McNair L, Alam J, Muir AJ. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N Engl J Med. 2009;360:1827-1838. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 851] [Cited by in F6Publishing: 916] [Article Influence: 61.1] [Reference Citation Analysis (0)] |
| 197. | Sullivan JC, De Meyer S, Bartels DJ, Dierynck I, Zhang EZ, Spanks J, Tigges AM, Ghys A, Dorrian J, Adda N. Evolution of treatment-emergent resistant variants in telaprevir phase 3 clinical trials. Clin Infect Dis. 2013;57:221-229. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 134] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 198. | Asselah T, Bièche I, Paradis V, Bedossa P, Vidaud M, Marcellin P. Genetics, genomics, and proteomics: implications for the diagnosis and the treatment of chronic hepatitis C. Semin Liver Dis. 2007;27:13-27. [PubMed] [Cited in This Article: ] |
| 199. | Massard J, Ratziu V, Thabut D, Moussalli J, Lebray P, Benhamou Y, Poynard T. Natural history and predictors of disease severity in chronic hepatitis C. J Hepatol. 2006;44:S19-S24. [PubMed] [Cited in This Article: ] |
| 200. | Thio CL. Host genetic factors and antiviral immune responses to hepatitis C virus. Clin Liver Dis. 2008;12:713-726, xi. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 60] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 201. | Yee LJ. Host genetic determinants in hepatitis C virus infection. Genes Immun. 2004;5:237-245. [PubMed] [Cited in This Article: ] |
| 202. | Ahlenstiel G, Booth DR, George J. IL28B in hepatitis C virus infection: translating pharmacogenomics into clinical practice. J Gastroenterol. 2010;45:903-910. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 40] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 203. | Doyle JS, Hellard ME, Thompson AJ. The role of viral and host genetics in natural history and treatment of chronic HCV infection. Best Pract Res Clin Gastroenterol. 2012;26:413-427. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 204. | Balagopal A, Thomas DL, Thio CL. IL28B and the control of hepatitis C virus infection. Gastroenterology. 2010;139:1865-1876. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 185] [Cited by in F6Publishing: 201] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 205. | Zhang L, Jilg N, Shao RX, Lin W, Fusco DN, Zhao H, Goto K, Peng LF, Chen WC, Chung RT. IL28B inhibits hepatitis C virus replication through the JAK-STAT pathway. J Hepatol. 2011;55:289-298. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 108] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 206. | Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798-801. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1667] [Cited by in F6Publishing: 1635] [Article Influence: 109.0] [Reference Citation Analysis (0)] |
| 207. | Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399-401. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2776] [Cited by in F6Publishing: 2666] [Article Influence: 177.7] [Reference Citation Analysis (0)] |
| 208. | Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, Bochud M, Battegay M, Bernasconi E, Borovicka J. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338-145, 1338-145. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 850] [Cited by in F6Publishing: 854] [Article Influence: 61.0] [Reference Citation Analysis (0)] |
| 209. | Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100-1104. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1505] [Cited by in F6Publishing: 1482] [Article Influence: 98.8] [Reference Citation Analysis (0)] |
| 210. | Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105-1109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1747] [Cited by in F6Publishing: 1746] [Article Influence: 116.4] [Reference Citation Analysis (0)] |
| 211. | Honda M, Sakai A, Yamashita T, Nakamoto Y, Mizukoshi E, Sakai Y, Yamashita T, Nakamura M, Shirasaki T, Horimoto K. Hepatic ISG expression is associated with genetic variation in interleukin 28B and the outcome of IFN therapy for chronic hepatitis C. Gastroenterology. 2010;139:499-509. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 330] [Cited by in F6Publishing: 339] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 212. | Feld JJ, Hoofnagle JH. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature. 2005;436:967-972. [PubMed] [Cited in This Article: ] |
| 213. | Tillmann H, Thompson AJ, Patel K, Wiese M, Tenckhoff H, Nischalke HD, Kullig U, Goebel U, Capka E, Wiegand J, Schiefke I, Spengler U, McCarthy J, Shianna K, Goldstein DB, McHutchison JG, Timm J, Nattermann J, Anti-D Study Group. IL28B polymorphism is associated with jaundice during acute HCV infection and is a strong predictor for spontaneous clearance in the prospective german anti-D cohort. J Hepatol. 2010;52 Suppl 1:S56. [DOI] [Cited in This Article: ] |
| 214. | Grebely J, Petoumenos K, Hellard M, Matthews GV, Suppiah V, Applegate T, Yeung B, Marks P, Rawlinson W, Lloyd AR. Potential role for interleukin-28B genotype in treatment decision-making in recent hepatitis C virus infection. Hepatology. 2010;52:1216-1224. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 134] [Cited by in F6Publishing: 146] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 215. | Luo Y, Jin C, Ling Z, Mou X, Zhang Q, Xiang C. Association study of IL28B: rs12979860 and rs8099917 polymorphisms with SVR in patients infected with chronic HCV genotype 1 to PEG-INF/RBV therapy using systematic meta-analysis. Gene. 2013;513:292-296. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 216. | Fischer J, Böhm S, Scholz M, Müller T, Witt H, George J, Sarrazin C, Susser S, Schott E, Suppiah V. Combined effects of different interleukin-28B gene variants on the outcome of dual combination therapy in chronic hepatitis C virus type 1 infection. Hepatology. 2012;55:1700-1710. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 66] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 217. | Afdhal NH, McHutchison JG, Zeuzem S, Mangia A, Pawlotsky JM, Murray JS, Shianna KV, Tanaka Y, Thomas DL, Booth DR. Hepatitis C pharmacogenetics: state of the art in 2010. Hepatology. 2011;53:336-345. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 119] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 218. | Soriano V, Poveda E, Vispo E, Labarga P, Rallón N, Barreiro P. Pharmacogenetics of hepatitis C. J Antimicrob Chemother. 2012;67:523-529. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 219. | Jacobson IM, Catlett I, Marcellin P, Bzowej NH, Muir AJ, Adda N, Bengtsson L, George S, Seepersaud S, Ramachandran R. Telaprevir substantially improved SVR rates across all IL28B genotypes in the ADVANCE trial. J Hepatol. 2011;54 Suppl 1:S542-S543. [DOI] [Cited in This Article: ] |
| 220. | Fellay J, Thompson AJ, Ge D, Gumbs CE, Urban TJ, Shianna KV, Little LD, Qiu P, Bertelsen AH, Watson M. ITPA gene variants protect against anaemia in patients treated for chronic hepatitis C. Nature. 2010;464:405-408. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 366] [Cited by in F6Publishing: 359] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 221. | Sakamoto N, Tanaka Y, Nakagawa M, Yatsuhashi H, Nishiguchi S, Enomoto N, Azuma S, Nishimura-Sakurai Y, Kakinuma S, Nishida N. ITPA gene variant protects against anemia induced by pegylated interferon-α and ribavirin therapy for Japanese patients with chronic hepatitis C. Hepatol Res. 2010;40:1063-1071. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 55] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 222. | Naggie S, Rallon NI, Benito JM, Morello J, Rodriguez-Novoa S, Clark PJ, Thompson AJ, Shianna KV, Vispo E, McHutchison JG. Variants in the ITPA gene protect against ribavirin-induced hemolytic anemia in HIV/HCV-coinfected patients with all HCV genotypes. J Infect Dis. 2012;205:376-383. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 223. | Tanaka Y, Kurosaki M, Nishida N, Sugiyama M, Matsuura K, Sakamoto N, Enomoto N, Yatsuhashi H, Nishiguchi S, Hino K. Genome-wide association study identified ITPA/DDRGK1 variants reflecting thrombocytopenia in pegylated interferon and ribavirin therapy for chronic hepatitis C. Hum Mol Genet. 2011;20:3507-3516. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 75] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 224. | Rau M, Baur K, Geier A. Host genetic variants in the pathogenesis of hepatitis C. Viruses. 2012;4:3281-3302. [PubMed] [Cited in This Article: ] |
| 225. | Macías J, Vispo E, Pineda JA, Soriano V. Host genetics. Curr Opin HIV AIDS. 2011;6:491-500. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 226. | Venegas M, Brahm J, Villanueva RA. Genomic determinants of hepatitis C virus antiviral therapy outcomes: toward individualized treatment. Ann Hepatol. 2012;11:827-837. [PubMed] [Cited in This Article: ] |
| 227. | Hennig BJ, Hellier S, Frodsham AJ, Zhang L, Klenerman P, Knapp S, Wright M, Thomas HC, Thursz M, Hill AV. Association of low-density lipoprotein receptor polymorphisms and outcome of hepatitis C infection. Genes Immun. 2002;3:359-367. [PubMed] [Cited in This Article: ] |
| 228. | Pineda JA, Caruz A, Di Lello FA, Camacho A, Mesa P, Neukam K, Rivero-juárez A, Macías J, Gómez-Mateos J, Rivero A. Low-density lipoprotein receptor genotyping enhances the predictive value of IL28B genotype in HIV/hepatitis C virus-coinfected patients. AIDS. 2011;25:1415-1420. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 229. | Fischer DG, Tal N, Novick D, Barak S, Rubinstein M. An antiviral soluble form of the LDL receptor induced by interferon. Science. 1993;262:250-253. [PubMed] [Cited in This Article: ] |
| 230. | Petta S, Cammà C, Scazzone C, Tripodo C, Di Marco V, Bono A, Cabibi D, Licata G, Porcasi R, Marchesini G. Low vitamin D serum level is related to severe fibrosis and low responsiveness to interferon-based therapy in genotype 1 chronic hepatitis C. Hepatology. 2010;51:1158-1167. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 305] [Cited by in F6Publishing: 313] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 231. | Abu-Mouch S, Fireman Z, Jarchovsky J, Zeina AR, Assy N. Vitamin D supplementation improves sustained virologic response in chronic hepatitis C (genotype 1)-naïve patients. World J Gastroenterol. 2011;17:5184-5190. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 150] [Cited by in F6Publishing: 149] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 232. | Lange CM, Bibert S, Kutalik Z, Burgisser P, Cerny A, Dufour JF, Geier A, Gerlach TJ, Heim MH, Malinverni R. A genetic validation study reveals a role of vitamin D metabolism in the response to interferon-alfa-based therapy of chronic hepatitis C. PLoS One. 2012;7:e40159. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 233. | Nimer A, Mouch A. Vitamin D improves viral response in hepatitis C genotype 2-3 naïve patients. World J Gastroenterol. 2012;18:800-805. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 103] [Cited by in F6Publishing: 104] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 234. | Matsumura T, Kato T, Sugiyama N, Tasaka-Fujita M, Murayama A, Masaki T, Wakita T, Imawari M. 25-Hydroxyvitamin D3 suppresses hepatitis C virus production. Hepatology. 2012;56:1231-1239. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 64] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 235. | Gal-Tanamy M, Bachmetov L, Ravid A, Koren R, Erman A, Tur-Kaspa R, Zemel R. Vitamin D: an innate antiviral agent suppressing hepatitis C virus in human hepatocytes. Hepatology. 2011;54:1570-1579. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 137] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 236. | Lange CM, Miki D, Ochi H, Nischalke HD, Bojunga J, Bibert S, Morikawa K, Gouttenoire J, Cerny A, Dufour JF. Genetic analyses reveal a role for vitamin D insufficiency in HCV-associated hepatocellular carcinoma development. PLoS One. 2013;8:e64053. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 237. | Jorquera F, Monte MJ, Guerra J, Sanchez-Campos S, Merayo JA, Olcóz JL, González-Gallego J, Marin JJ. Usefulness of combined measurement of serum bile acids and ferritin as additional prognostic markers to predict failure to reach sustained response to antiviral treatment in chronic hepatitis C. J Gastroenterol Hepatol. 2005;20:547-554. [PubMed] [Cited in This Article: ] |
| 238. | Iwata R, Stieger B, Mertens JC, Müller T, Baur K, Frei P, Braun J, Vergopoulos A, Martin IV, Schmitt J. The role of bile acid retention and a common polymorphism in the ABCB11 gene as host factors affecting antiviral treatment response in chronic hepatitis C. J Viral Hepat. 2011;18:768-778. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |