Published online Jul 14, 2014. doi: 10.3748/wjg.v20.i26.8681
Revised: February 19, 2014
Accepted: April 8, 2014
Published online: July 14, 2014
Processing time: 179 Days and 17.1 Hours
AIM: To investigate the expression of P450 enzyme genes by using end-stage liver disease samples and trimmed normal Chinese donor livers.
METHODS: The end-stage liver disease samples [n = 93, including hepatocellular carcinoma (HCC), peri-HCC tissue, hepatitis B virus cirrhosis, alcoholic cirrhosis, and severe cirrhosis] and trimmed normal Chinese donor livers (n = 35) from The Institute of Organ Transplantation in Beijing, China. Total RNA was extracted, purified, and subjected to real-time RT-PCR analysis.
RESULTS: For cytochrome P450 enzymes 1 (CYP1) family, the expression of CYP1A2 was decreased 90% in HCC, 80% in alcoholic cirrhosis, and 65% in severe cirrhosis. For CYP2 family, the expression of CAR was decreased 50% in HCC, but increased 50% in peri-HCC tissues. Similar decreases (about 50%) of CYP2B6, CYP2C9, CYP2C19, CYP2D6 and CYP2E1 were observed in HCC, as compared to peri-HCC tissues and normal livers. CYP2C19 were decreased in all end-stage liver diseases and CYP2E1 also decreased in alcoholic cirrhosis and severe cirrhosis. For CYP3 family, the expression of PXR was decreased 60% in HCC, together with decreases in CYP3A4, CYP3A5, and CYP3A7. In contrast, the expression of CYP3A7 was slightly increased in HBV cirrhosis. The expression of CYP4A11 was decreased 85% in HCC, 7% in alcoholic cirrhosis and severe liver cirrhosis, along with decreases in PPARα. The 93 end-stage livers had much higher inter-individual variations in gene expression than 35 normal livers.
CONCLUSION: The expression of CYP enzyme genes and corresponding nuclear receptors was generally decreased in end-stage liver diseases, and significant differences in gene expression were evident between peri-HCC and HCC.
Core tip: This study examined the expression of P450 enzyme (CYP) genes in end-stage liver diseases, including hepatocellular carcinoma (HCC), peri-HCC tissue, hepatitis B virus cirrhosis, alcoholic cirrhosis, and severe cirrhosis, and compared to normal donor livers by real-time RT-PCR analysis. The 93 end-stage livers had much higher inter-individual variations in gene expression than 35 normal livers. In general, the end-stage livers had decreased expression of CYP1A2, CYP2B6, CYP2C9, CYP2C19, CYP2D6, CYP2E1, CYP3A4, CYP3A5, CYP3A7 and CYP4A11, as well as their corresponding nuclear receptor AhR, CAR, PXR, and PPARα. HCC had dramatic decreases in CYP gene expression and nuclear receptors compared with peri-HCC tissues.
- Citation: Chen H, Shen ZY, Xu W, Fan TY, Li J, Lu YF, Cheng ML, Liu J. Expression of P450 and nuclear receptors in normal and end-stage Chinese livers. World J Gastroenterol 2014; 20(26): 8681-8690
- URL: https://www.wjgnet.com/1007-9327/full/v20/i26/8681.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i26.8681
Cytochrome P450 enzymes are important in the metabolism of endogenous substances and xenobiotics[1]. Functional human P450 isozyme variations are implicated with drug toxicities, such as S-acenocoumarol, phenytoin and warfarin (CYP2C9), metoclopramide, codeine, mirtazapin and tramadol (CYP2D6), acetaminophen and alcohol (CYP2E1), methadone and efavirenz (CYP2B6), clopidogrel and desipramine (CYP2C19), and tacrolimus (CYP3A5)[2-4]. The expression of CYP enzymes is influenced by endogenous factors, such as genetic polymorphisms[5,6], sex[7], age[8], and hormone levels[7]. The expression of CYP enzymes is also influenced by exogenous factors such as drugs and environmental chemicals[9] as well as the physiopathological conditions[10].
Hepatic cytochrome P450 (CYP) expression is subjected to changes during various disease conditions, such as steatosis[11], chronic hepatitis C[12], alcoholic liver diseases[13], nonalcoholic fatty liver disease[14], liver fibrosis[15], cirrhosis[16], hepatoblastoma[17], and hepatocellular carcinoma[18,19]. Hepatic CYP expression is also under control of nuclear receptors in that AhR regulates CYP1 gene family, CAR regulates CYP2 gene family, PXR regulates CYP3 family, and PPARα regulates CYP4 family[20-22].
The end-stage livers removed for liver transplantation are precious resources to investigate gene expression changes, including P450 enzyme genes[10,23-25]. However, little is known about end-stage liver P450 profiling from the Chinese population. This study was initiated to fill the gap. A total of 93 end-stage livers and 35 trimmed normal donor livers were obtained from the Oriental Liver Transplant Center in Beijing, China, and total RNA was isolated and purified to profile the expression of CYP1-4 family genes and corresponding nuclear receptors. The results demonstrated the generalized reduction of CYP gene expression in end-stage liver diseases.
The end-stage human liver samples (n = 93) were collected from Oriental Liver Translation Center (Beijing, China), and the sample demographics are shown in Table 1. Samples included hepatocellular carcinoma (HCC, n = 24), and the corresponding HCC surrounding tissues (peri-HCC, n = 24), hepatitis B virus (HBV) cirrhosis (n = 27), severe cirrhosis (n = 13), and alcoholic cirrhosis (n = 5). The diagnosis of end-stage liver diseases was made by Pathology Department of the Institute of Organ Transplantation. This study received Institutional Review Board exemption status by the Oriental Liver Transplant Center and the Guiyang Medical College Human Subject Committee because the specimens were obtained without identification to the authors of the study.
| Liver disease | Gender | Age (yr) | ||||
| Men | Women | 21-40 | 40-60 | 61-80 | ||
| Normal donors | Trimmed normal livers | 32 | 3 | 34 | 1 | 0 |
| End-stage livers | HCC | 22 | 2 | 4 | 16 | 4 |
| Peri-HCC | 22 | 2 | 4 | 16 | 4 | |
| HBV cirrhosis | 21 | 6 | 2 | 18 | 7 | |
| Severe cirrhosis | 11 | 2 | 4 | 7 | 2 | |
| Alcoholic cirrhosis | 5 | 0 | 0 | 5 | 0 | |
All 128 human liver samples were used for real-time RT-PCR analysis. Total RNA was extracted with TRIzol (Invitrogen, Carlsbad, CA) and purified with RNeasy columns (Qiagen, Valencia, CA). The quality and integrity of purified RNA was determined by spectrophotometry with 260/280 ratio > 1.8 and 260/230 ratio > 1.5, as well as by agarose gel electrophoresis showing clear 18S and 28S bands without degradation.
Total RNA was reverse-transcribed into cDNA with Multiscript reverse transcriptase using High Capacity RT kits from Applied Biosystems (Foster City, CA), and amplified with Power SYBR® Green PCR Master Mix in a 7900HT PCR System (Applied Biosystems, Foster City, CA). Oligonucleotide primers were designed with Primer3 software (version 4), and are listed in Table 2. Relative expression of genes was calculated by the 2-ΔΔCt method and normalizing to the house-keeping gene GAPDH.
| Gene | GenBank | Forward | Reverse |
| number | |||
| AhR | NM_001621 | CTGCCTTTCCCACAAGATGT | AGTTATCCTGGCCTCCGTTT |
| CAR | NM_005122 | CTTCTCTCCTGACCGACCTG | AGGCCTAGCAACTTCGCATA |
| CYP1A2 | NM_000761 | GGACAGCACTTCCCTGAGAG | GAGGCAGTCTCCACGAACTC |
| CYP2B6 | NM_000767 | TCCAGTCTCAGCTCCCAAGT | CTGGCCAACATGTCCCTACT |
| CYP2C9 | NM_000771 | CCACATGCCCTACACAGATG | TGCCCTTGGGAATGAGATAG |
| CYP2C19 | NM_000769 | CAACAACCCTCGGGACTTTA | GTCTCTGTCCCAGCTCCAAG |
| CYP2D6 | NM_000106 | CAGAGATGGAGAAGGCCAAG | AGAACAGGTCAGCCACCACT |
| CYP2E1 | NM_000773 | CCCAAAGGATATCGACCTCA | AGGGTGTCCTCCACACACTC |
| CYP3A4 | NM_017460 | ACCGTGACCCAAAGTACTGG | GTTTCTGGGTCCACTTCCAA |
| CYP3A5 | NM_000777 | GGAGATGTTCCCCATCATTG | GCCCCAAAGATGTCTTTCAA |
| CYP3A7 | NM_000765 | GAAACACAGATCCCCCTGAA | AGGGAAATCAGGCTCCACTT |
| CYP4A11 | NM_000778 | CACCACAACCCAAAAGTGTG | ATTGTTTCCCGATGCAGTTC |
| FXR | NM_005123 | ATTTTGACGGAAATGGCAAC | AGACCCTTTCAGCAAAGCAA |
| GADPH | NM_002046 | ACAGTCAGCCGCATCTTCTT | ACGACCAAATCCGTTGACTC |
| PPARα | Y07619 | ACGATTCGACTCAAGCTGGT | GTTGTGTGACATCCCGACAG |
| PXR | NM_005123 | GGAATGTTGGCTGAATGCTT | CTGCATGCTGCTTCACATTT |
Data was calculated as mean ± SE. The SPSS17 software was used for statistical analysis. For comparisons among three or more groups, data were analyzed using a one-way analysis of variance (ANOVA), followed by Duncan’s multiple range test. For the comparisons between HCC and peri-HCC, Student t-test was used. The significance level was set at P < 0.05 in all cases.
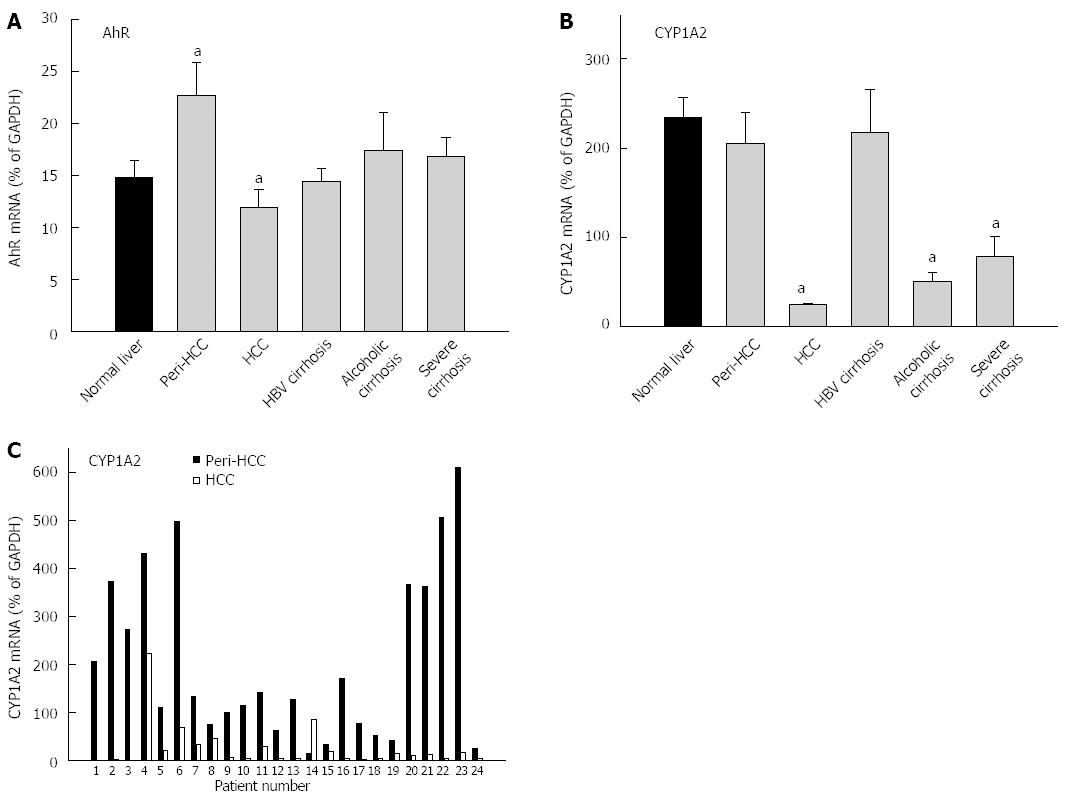
The expression of AhR and AhR-regulated CYP1A2 is shown in Figure 1 and Table 3. The average expression of AhR in 35 normal livers (14.7% ± 0.7% of GADPH) was not significantly different from 93 diseased livers (17.4% ± 1.5% of GADPH), but diseased livers had huge inter-individual variations compared with normal livers (47-fold vs 5-fold) (Table 3). peri-HCC had 50% higher expression of AhR compared with normal livers, but this was not statistically significant. Compared with peri-HCC, AhR mRNA levels in HCC were significantly lower (Figure 1). There were no major changes in AhR expression in HBV cirrhosis, alcoholic cirrhosis, and severe cirrhosis.
| Normal liver (n = 35) | End-stage livers (n = 93) | |||||
| Gene | Expression% | -fold | mean ± SE | Expression% | -fold | mean ± SE |
| AhR | 5.52-27.1 | 4.91 | 14.7 ± 0.74 | 2.71-128 | 47.2 | 17.4 ± 1.52 |
| CYP1A2 | 20.5-537 | 26.2 | 234 ± 94.6 | 0.78-175 | 224 | 136 ± 23.9a |
| CAR | 0.55-14.1 | 25.6 | 7.87 ± 0.50 | 0.21-293 | 1395 | 7.73 ± 0.86 |
| CYP2B6 | 1.31-102 | 77.9 | 17.1 ± 2.74 | 0.81-128 | 158 | 27.2 ± 2.77a |
| CYP2C9 | 14.4-1544 | 107 | 786 ± 57.9 | 3.74-1486 | 387 | 418 ± 34.8a |
| CYP2C19 | 49.6-1254 | 25.2 | 594 ± 40.2 | 1.75-1379 | 788 | 256 ± 23.9a |
| CYP2D6 | 10.7-316 | 29.5 | 149 ± 12.2 | 4.31-930 | 216 | 156 ± 15.6 |
| CYP2E1 | 210-3577 | 17.2 | 1708 ± 115 | 17.7-4373 | 247 | 990 ± 75.4a |
| PXR | 14.2-189 | 13.3 | 41.4 ± 4.95 | 3.01-87.1 | 29.0 | 33.0 ± 1.81 |
| CYP3A4 | 171-1357 | 7.94 | 798 ± 49.2 | 8.21-1966 | 240 | 498 ± 48.6a |
| CYP3A5 | 22.2-659 | 30.0 | 278 ± 26.9 | 9.92-979 | 98.7 | 203 ± 19.1a |
| CYP3A7 | 86.3-1054 | 12.2 | 450 ± 36.7 | 1.51-2137 | 1415 | 479 ± 47.7 |
| PPARα | 13.7-57.7 | 4.21 | 29.4 ± 1.89 | 2.61-169 | 64.8 | 27.6 ± 2.43 |
| CYP4A11 | 15.1-478 | 31.6 | 198 ± 17.2 | 1.41-740 | 524 | 89.2 ± 11.1a |
The average expression of CYP1A2 in normal livers (234% ± 94% of GADPH) was significantly higher than diseased livers (136% ± 24% of GADPH), and diseased livers had 8.5-fold higher inter-individual variations than normal livers (224-fold vs 26-fold) (Table 3). CYP1A2 in HCC was decreased by 90%, significantly lower than normal livers and peri-HCC (Figure 1). CYP1A2 expression was also decreased in alcoholic cirrhosis by 79% and severe cirrhosis by 66%, as compared to normal livers, but was unchanged in HBV cirrhosis and peri-HCC tissues.
The paired individual values from peri-HCC and HCC from 24 patients are shown in the bottom panel of Figure 1. CYP1A2 expression was low in HCC, except for patient No. 14, as compared to peri-HCC. Some patients showed huge differences (i.e., patient No. 22), while others were moderate (i.e., patient No. 4).
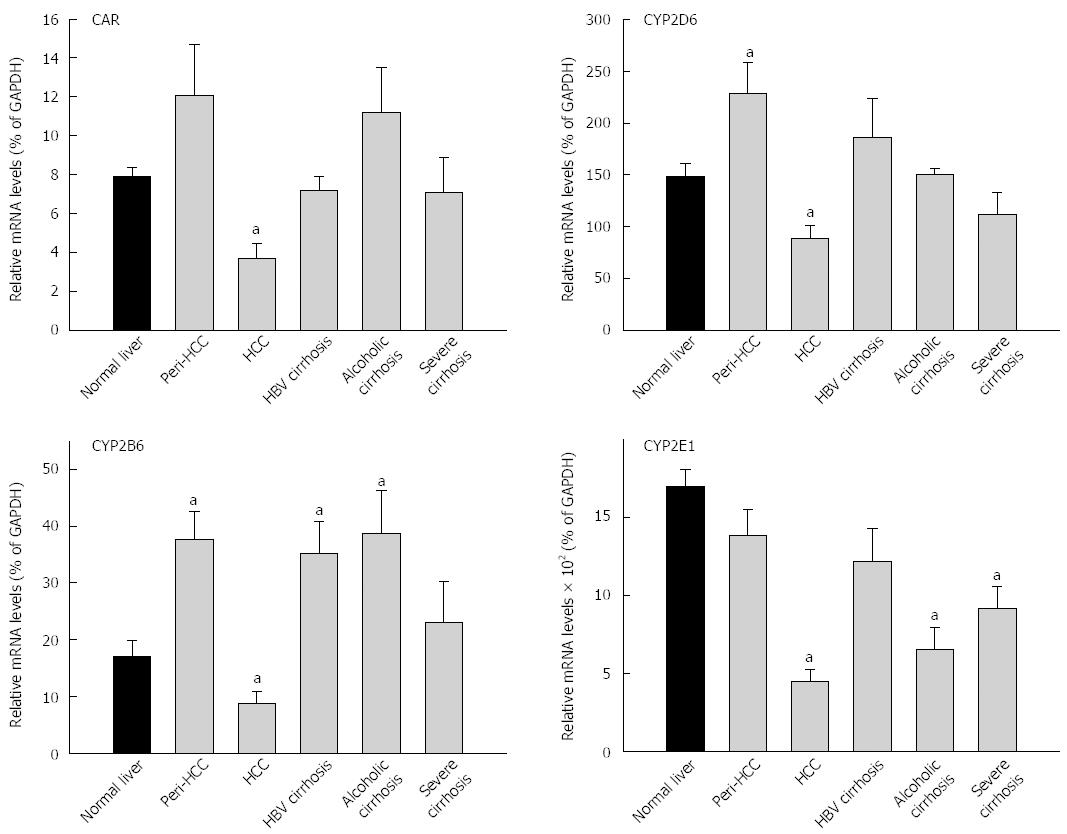
The expression of CAR and CAR-regulated CYPs are shown in Figure 2 and Table 3. The average expression of CAR in 35 normal livers (7.87% ± 0.5% of GADPH) was not significantly different from 93 diseased livers (7.73% ± 0.8% of GADPH), but diseased livers had huge inter-individual variations (1390-fold vs 26-fold) (Table 3). The basal expression of CAR was about 8% of GADPH. CAR mRNA levels in HCC was decreased to 3.7% of GADPH, 50% lower than normal livers and 70% lower than peri-HCC (Figure 2). There were no major changes in CAR expression in HBV cirrhosis and severe cirrhosis. Alcoholic cirrhosis tended to be 45% higher than normal livers, but was not significant. The average expression of CYP2B6 in normal livers (17.1% ± 2.7% of GADPH) was lower than 93 diseased livers (27.2% ± 2.8% of GADPH), and diseased livers had higher inter-individual variations (158-fold vs 78-fold) (Table 3). CYP2B6 mRNA levels in peri-HCC were increased 120%, and were decreased 50% in HCC as compared to normal livers (Figure 2). Expression of CYP2B6 was also increased in HBV cirrhosis and alcoholic cirrhosis, but was not altered in severe cirrhosis.
The average expression of CYP2D6 in normal livers (149% ± 12%) was not different from diseased livers (156% ± 16% of GADPH), and diseased livers had greater inter-individual variations (220-fold vs 30-fold) (Table 3). Similarly, CYP2D6 mRNA levels in peri-HCC were increased 53%, while in HCC CYP2D6 mRNA was decreased 40% as compared to normal livers (Figure 2). There were no major changes in CYP2D6 expression in HBV cirrhosis, alcoholic cirrhosis and severe cirrhosis. The average expression of CYP2E1 in normal livers (1710% ± 115%) was significantly higher than diseased livers (990% ± 75% of GADPH), and diseased livers had huge inter-individual variations than normal livers (250-fold vs 17-fold) (Table 3). CYP2E1 mRNA levels in HCC were decreased by 75%, significantly lower than normal livers and peri-HCC. CYP2E1 expression was also decreased in alcoholic cirrhosis and in severe cirrhosis.
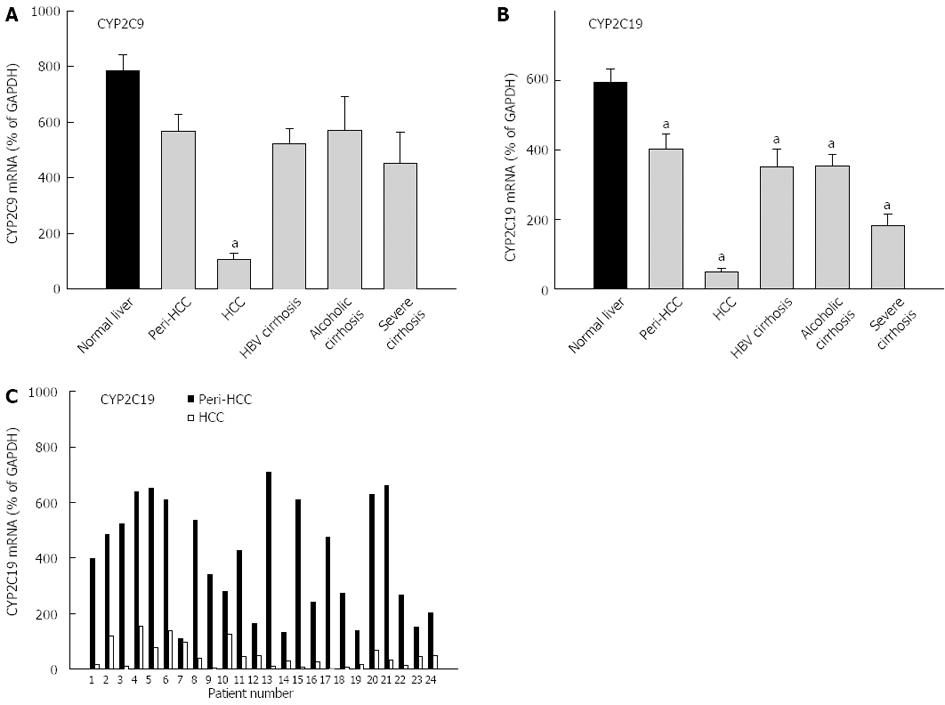
Both CYP2C9 and CYP2C19 are highly expressed in liver and are shown in Figure 3. The average expression of CYP2C9 in normal livers (786% ± 58% of GAPDH) was higher than in diseased livers (418% ± 35% of GADPH), and diseased livers had greater inter-individual variations (390-fold vs 110-fold) (Table 3). HCC had dramatic decreases in mRNA levels of CYP2C9 (13% of normal livers), which is also significantly lower than peri-HCC. The expression of CYP2C9 tended to be lower in HBV cirrhosis, alcoholic cirrhosis and severe cirrhosis, but was not significant. The average expression of CYP2C19 in normal livers (594% ± 40% of GAPDH) was higher than diseased livers (256% ± 39% of GADPH), and diseased livers had huge inter-individual variations compared with normal livers (788-fold vs 25-fold) (Table 3). CYP2C19 expression was decreased in all end-stage livers, with dramatic decreases in HCC by over 90% (Figure 3). peri-HCC had decreased CYP2C19, but the levels were significantly higher than HCC. The individual pair values of CYP2C19 in peri-HCC and HCC are shown in the bottom panel of Figure 3. In all 24 patients, CYP2C9 mRNA levels were consistently lower in HCC, some showed huge differences (i.e., patient No. 13), while others moderate (i.e., patient No. 4) or mild (i.e., patient No. 7).
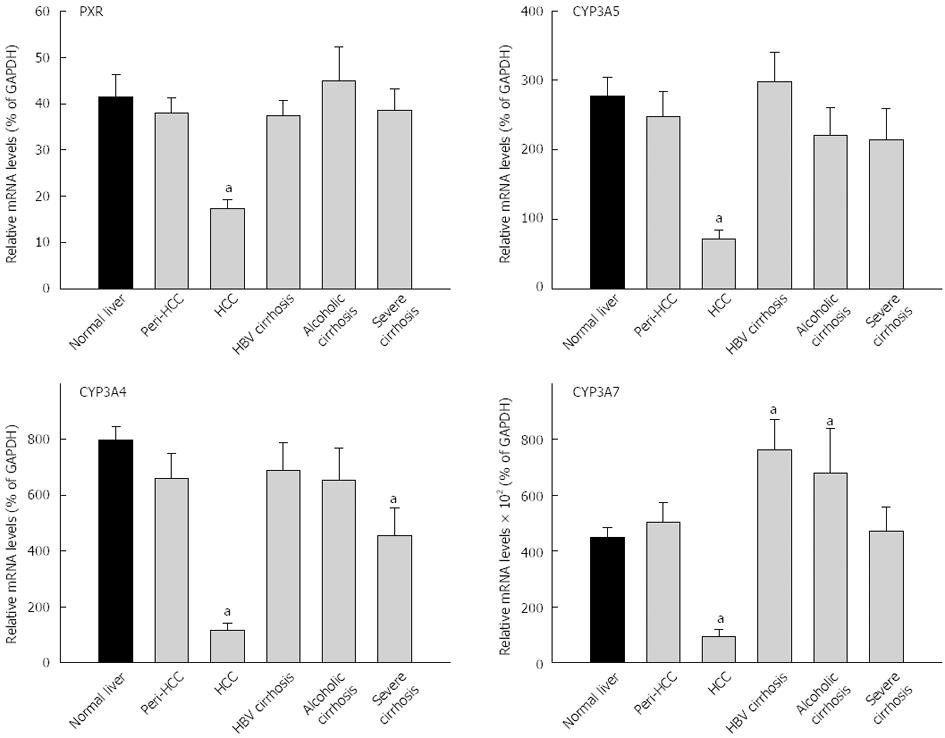
The expression of PXR and PCR-regulated CYPs are shown in Figure 4 and Table 3. The average expression of PXR in normal livers (41.4% ± 5.0% of GAPDH) was not significantly different from diseased livers (33.0% ± 1.8% of GADPH), and diseased livers had higher inter-individual variations (29-fold vs 13-fold) (Table 3). PXR mRNA levels in HCC were decreased by 60%, significantly lower than peri-HCC and normal livers (Figure 4). There were no major changes in PXR expression in HBV cirrhosis, alcoholic cirrhosis, and severe cirrhosis. The average expression of CYP3A4 in normal livers (798% ± 49% of GAPDH) was significantly higher than diseased livers (498% ± 49% of GADPH), and diseased livers had much higher inter-individual variations than normal livers (240-fold vs 8-fold) (Table 3). The mRNA expression of CYP3A4 in HCC was decreased 85%, significantly lower than normal livers and peri-HCC. The mRNA levels of CYP3A4 were also lower in severe cirrhosis.
The average expression of CYP3A5 in normal livers (278% ± 27% of GAPDH) was significantly different from diseased livers (203% ± 19% of GADPH), and diseased livers had higher inter-individual variations (99-fold vs 30-fold) (Table 3). The expression of CYP3A5 was decreased 75% in HCC, significantly lower than normal livers and peri-HCC (Figure 4). The average expression of CYP3A7 in normal livers (450% ± 37% of GAPDH) was not significantly different from diseased livers (479% ± 48% of GADPH), and diseased livers had huge inter-individual variations compared with normal livers (1415-fold vs 12-fold) (Table 3). The expression of CYP3A7 was decreased 80% in HCC, significantly lower than normal livers and peri-HCC (Figure 4). The mRNAs of CYP3A7 in HBV cirrhosis were 70% significantly higher than normal livers and 50% higher than in alcoholic cirrhosis (not significant). The mRNAs of CYP3A7 in peri-HCC and severe cirrhosis were not different from normal livers.
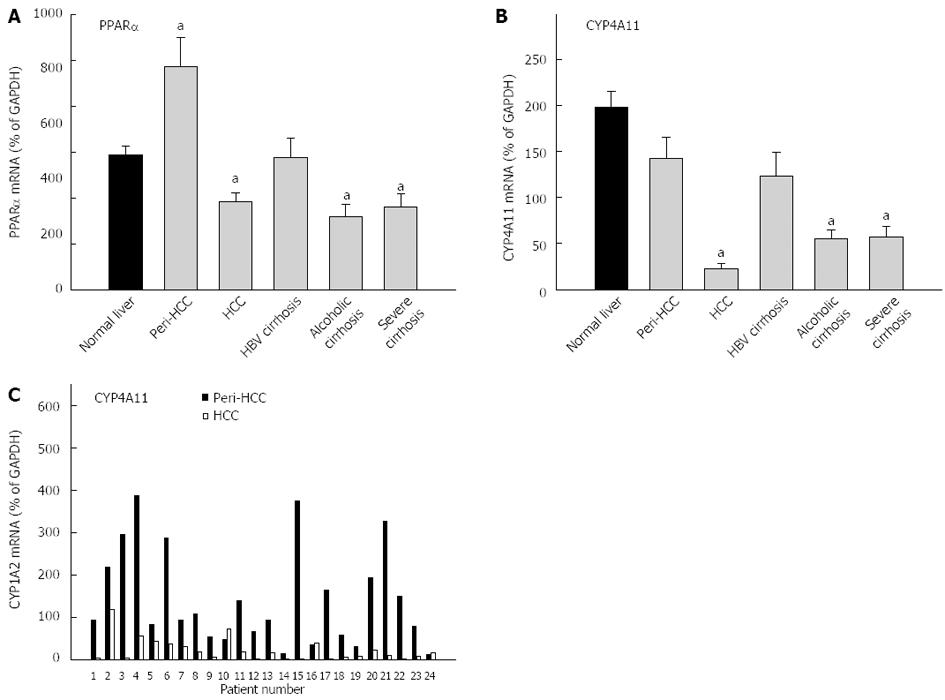
The average expression of PPARα in normal livers (29.4% ± 1.9% of GAPDH) was not different from diseased livers (27.6% ± 2.4% of GADPH), and diseased livers had much higher inter-individual variations than normal livers (65-fold vs 4-fold) (Table 3). PPARα mRNAs were 65% higher in peri-HCC, but were 35% lower in HCC, as compared to normal livers. The mRNAs of PPARα were also lower in alcoholic cirrhosis and severe cirrhosis (Figure 5). The average expression of CYP4A11 in normal livers (198% ± 17% of GAPDH) was significantly higher than in diseased livers (89.2% ± 11% of GADPH), but diseased livers had much higher inter-individual variations than normal livers (524-fold vs 32-fold) (Table 3). The expression of CYP4A11 was decreased nearly 90% in HCC, followed by severe cirrhosis (78%), and alcoholic cirrhosis (72%), but was not significantly altered in peri-HCC and HBV cirrhosis. The individual paired mRNAs of CYP4A11 in peri-HCC and HCC are shown in the bottom panel of Figure 5C. CYP4A11 mRNAs were decreased in 21 of 24 patients, except for patient No. 10, No. 16 and No. 24, as compared to peri-HCC. Some patients showed huge differences (patient No. 15, No. 17, and No. 21), while others had mild differences (patient No. 2 and No. 5).
The present study examined the expression of major drug metabolizing P450 genes and their corresponding nuclear receptors in 93 end-stage Chinese livers and compared with 35 normal Chinese donor liver trimmings from the Oriental Liver Transplant Center (Beijing, China), including HCC and peri-HCC tissues, HBV cirrhosis, sever cirrhosis and alcoholic cirrhosis. The results clearly demonstrate that the CYP1-4 family genes were generally down-regulated in end-stage liver diseases, with much greater inter-individual variations in gene expression than normal livers. Significant differences in gene expression were also evident between peri-HCC and HCC.
CYP1 family genes are regulated by AhR. CYP1A2 is an abundant CYP1A isoform in the liver. In chronic hepatitis C in association with fibrosis, both AhR and CYP1A2 were decreased with the development of fibrosis[12,14]; their down-regulation appeared to be associated with oxidative stress in the liver diseases[19]. CYP1A2 was reported to be decreased in end-stage liver diseases[16,23,25], and the same phenomenon occurs for Chinese end-stage livers in the present study. CYP1A1 and CYP1B1 are also AhR target genes; although CYP1A1 was down-regulated in end-stage liver diseases[10,15], CYP1B1 was increased together with ABCB4[15], indicating they are differentially regulated. The down-regulation of CYP1A2 in end-stage liver diseases is consistent with liver dysfunction with the progress of the diseases.
CYP2 family had large members of enzyme genes and 5 of them (CYP2B6, CYP2C9, CYP2C19, CYP2E1, and CYP2D6) were investigated in the present study. CYP2E1 is a major P450 isoform for ethanol and is responsible for the metabolism of a variety of xenobiotics[2], CYP2C19 is important in the metabolism of clopidogrel and lacosamide[26,27], while CYP2B6 is an “overlooked” P450 isozyme, which is now recognized to be important for xenobiotic metabolism[3]. In general, CYP2E enzyme genes were decreased depending on the end-stage liver diseases, with most decreases seen in HCC. CAR is a nuclear receptor regulating CYP2 family gene expression[19]. In end-stage livers, the expression of CAR was generally decreased[10,12], consistent with present observations. Accordingly, CYP2 family enzyme genes were decreased with the liver diseases progressed to the end-stage. In nonalcoholic fatty liver disease[28] and early stages of HCC[18], increased expression of CYP2E1 was reported, but at the end-stage of liver diseases, CYP2E1 was decreased[12-13,25]. In the present study, CYP2E1 decreased not only in HCC, but also in alcoholic cirrhosis and severe cirrhosis (Figure 4). The function of cytochrome P450 2C19 (CYP2C19) is decreased in patients with end-stage liver disease that require liver transplantation[25,29]. In the present study, CYP2C19 was decreased in all end-stage liver diseases, including decreases in peri-HCC. Among CYP2 family genes, CYP2C19 was affected the most, and decreased up to 90% in HCC. All CYP2 family genes are dramatically down-regulated in HCC as compared to peri-HCC and normal tissues, and decreases in CYP2C (2C9 and 2C19) were also evident in end-stage liver diseases.
CYP3 family metabolizes the majority of drugs[1]. CYP3 family genes are mainly regulated by PXR and three major CYP3A family members (CYP3A4, CYP3A5 and CYP3A7) were studied in the present study. In end-stage liver diseases, the expression of these genes was decreased only in HCC. The nuclear receptor PXR is expressed in liver and intestine and is activated by a wide variety of endobiotics and xenobiotics, including the majority of drugs. PXR serves as a master transcriptional regulator of CYP3A isozymes[22]. PXR is decreased in end-stage liver diseases[10,12]. CYP3A4 is the most important drug metabolizing enzyme in adult humans because of its prominent expression in liver and its broad substrate specificity[30]. Expression of CYP3A4 decreased in the end-stage liver diseases[13,25]. CYP3A5 is important for metabolizing the immunosuppressive drugs used for solid organ transplantation, such as tacrolimus, sirolimus, and ciclosporin[31], and in the present study, it was decreased in end-stage HCC, but it tended to be slightly higher in HBV cirrhosis. Little is known about the expression of CYP3A7 in end-stage liver diseases, and this research would add to our understanding of this CYP3A isoforms in human liver diseases. In general, CYP3A family gene expressions were down-regulated in HCC, but were variably affected in other end-stage liver diseases.
CYP4 family genes are regulated by PPARα. Both PPARα and CYP4A11 were investigated in the present study. PPAR is important in lipid metabolism and PPAR-regulated fatty acid oxidation gene CYP4A11 plays important roles in lipid metabolism[32]. In the present study, the expression of PPARα was slightly decreased in HCC only, but the expression of CYP4A11 was decreased over 85% in HCC, and 70% in severe cirrhosis and alcoholic cirrhosis. The down-regulation of PPARα and its target genes would affect the function of liver in lipid metabolism.
The end-stage liver diseases are characterized by a decrease in functional hepatocytes, a decrease in P450 enzyme activities, and reduced blood flow to the liver, which could affect drug metabolism and systemic clearance[33,34]. The decreased CYP1-4 family gene expressions could have a significant impact on drug metabolism and xenobiotic elimination. For example, ropivacaine is metabolized by CYP1A2 and CYP3A4, and its clearance is decreased in chronic end-stage liver diseases[24]; Fluvoxamine-induced inhibition of theophylline clearance decreased from 62% in healthy subjects to 52% and 12% in patients with mild cirrhosis and those with severe cirrhosis, respectively, due to CYP1A2 dysfunction[35]. Thus, during end-stage liver diseases, liver dysfunction would dramatically reduce drug efficacy and may increase drug toxicity.
It should also be noted that end-stage livers had huge individual variations in CYP gene expression, as compared to normal livers. Such a huge individual variation in gene expression among end-stage livers was reported before[24]. CYP polymorphisms and mutations[29,31,32] could also contribute to this huge inter-individual variation. Epigenetic mechanisms such as aberrant microRNA expression would also affect CYP expression[36]. Compared to effects of steatosis on CYP expression[11,37], cirrhosis is the end-stage liver disease and the effects are much more pronounced[16,38].
In summary, this study demonstrates that end-stage livers had compromised liver P450 enzyme gene expression, together with their corresponding nuclear receptors. All the tested P450s were decreased in HCC, as compared to peri-HCC and normal livers. Other end-stage liver diseases, namely severe cirrhosis, alcoholic cirrhosis and HBV cirrhosis also had decreased hepatic P450 enzyme gene expressions, albeit to varying degrees. The decreased P450 enzymes would result in liver dysfunction in reducing drug efficacy and increasing drug toxicities.
The end-stage livers removed for liver transplantation are precious resources to investigate gene expression changes, including P450 enzyme genes. However, little is known about end-stage liver P450 expression from the Chinese population.
Hepatic cytochrome P450 (CYP) expression is subjected to changes in liver diseases, and to profile their expression is of basic and clinical significance.
The end-stage livers had compromised liver P450 enzyme gene expression, together with varying expression of their corresponding nuclear receptors. All the tested P450s were decreased in hepatocellular carcinoma, as compared to peri-HCC and normal livers.
The decreased P450 enzymes would result in liver dysfunction, in reducing drug efficacy and increasing drug toxicities.
Real-time RT-PCR is a sensitive technology to study the expression of CYP genes in the end-stage liver diseases.
This study is among the first to study CYP and nuclear receptors in the Chinese population.
P- Reviewers: Buchler C, Dietrich CG, Laguna JC S- Editor: Ma YJ L- Editor: O’Neill M E- Editor: Zhang DN
| 1. | Parkinson A, Ogilvie BW. Biotransformation of xenobiotics. Casarett and Doull’s Toxicology: The Basic Science of Poisons. Sydney: McGraw Hill 2013; 185-366. |
| 2. | Gonzalez FJ. The 2006 Bernard B. Brodie Award Lecture. Cyp2e1. Drug Metab Dispos. 2007;35:1-8. [PubMed] |
| 3. | Wang H, Tompkins LM. CYP2B6: new insights into a historically overlooked cytochrome P450 isozyme. Curr Drug Metab. 2008;9:598-610. [PubMed] |
| 4. | Johansson I, Ingelman-Sundberg M. Genetic polymorphism and toxicology--with emphasis on cytochrome p450. Toxicol Sci. 2011;120:1-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 152] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 5. | Xu BY, Guo LP, Lee SS, Dong QM, Tan Y, Yao H, Li LH, Lin CK, Kung HF, He ML. Genetic variability of CYP2B6 polymorphisms in four southern Chinese populations. World J Gastroenterol. 2007;13:2100-2103. [PubMed] |
| 6. | Neafsey P, Ginsberg G, Hattis D, Sonawane B. Genetic polymorphism in cytochrome P450 2D6 (CYP2D6): Population distribution of CYP2D6 activity. J Toxicol Environ Health B Crit Rev. 2009;12:334-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 80] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 7. | Waxman DJ, Holloway MG. Sex differences in the expression of hepatic drug metabolizing enzymes. Mol Pharmacol. 2009;76:215-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 568] [Cited by in RCA: 546] [Article Influence: 34.1] [Reference Citation Analysis (0)] |
| 8. | Hines RN. Ontogeny of human hepatic cytochromes P450. J Biochem Mol Toxicol. 2007;21:169-175. [PubMed] |
| 9. | Guengerich FP. Cytochrome p450 and chemical toxicology. Chem Res Toxicol. 2008;21:70-83. [PubMed] |
| 10. | Kurzawski M, Dziedziejko V, Post M, Wójcicki M, Urasińska E, Miętkiewski J, Droździk M. Expression of genes involved in xenobiotic metabolism and transport in end-stage liver disease: up-regulation of ABCC4 and CYP1B1. Pharmacol Rep. 2012;64:927-939. [PubMed] |
| 11. | Gómez-Lechón MJ, Jover R, Donato MT. Cytochrome p450 and steatosis. Curr Drug Metab. 2009;10:692-699. [PubMed] |
| 12. | Hanada K, Nakai K, Tanaka H, Suzuki F, Kumada H, Ohno Y, Ozawa S, Ogata H. Effect of nuclear receptor downregulation on hepatic expression of cytochrome P450 and transporters in chronic hepatitis C in association with fibrosis development. Drug Metab Pharmacokinet. 2012;27:301-306. [PubMed] |
| 13. | Theile D, Haefeli WE, Seitz HK, Millonig G, Weiss J, Mueller S. Association of liver stiffness with hepatic expression of pharmacokinetically important genes in alcoholic liver disease. Alcohol Clin Exp Res. 2013;37 Suppl 1:E17-E22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 14. | Fisher CD, Lickteig AJ, Augustine LM, Ranger-Moore J, Jackson JP, Ferguson SS, Cherrington NJ. Hepatic cytochrome P450 enzyme alterations in humans with progressive stages of nonalcoholic fatty liver disease. Drug Metab Dispos. 2009;37:2087-2094. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 273] [Cited by in RCA: 265] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 15. | Haas S, Merkelbach-Bruse S, Justenhoven C, Brauch H, Fischer HP. Expression of xenobiotic and steroid hormone metabolizing enzymes in hepatocellular tumors of the non-cirrhotic liver. Pathol Res Pract. 2009;205:716-725. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 16. | Elbekai RH, Korashy HM, El-Kadi AO. The effect of liver cirrhosis on the regulation and expression of drug metabolizing enzymes. Curr Drug Metab. 2004;5:157-167. [PubMed] |
| 17. | Schmidt A, Braeuning A, Ruck P, Seitz G, Armeanu-Ebinger S, Fuchs J, Warmann SW, Schwarz M. Differential expression of glutamine synthetase and cytochrome P450 isoforms in human hepatoblastoma. Toxicology. 2011;281:7-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 18. | Wang YX, Meng XW, Wang YD, Liu Y. MRNA levels of two different enzymes in hepatocellular carcinoma. Hepatogastroenterology. 2010;57:1187-1190. [PubMed] |
| 19. | Tanaka S, Mogushi K, Yasen M, Ban D, Noguchi N, Irie T, Kudo A, Nakamura N, Tanaka H, Yamamoto M. Oxidative stress pathways in noncancerous human liver tissue to predict hepatocellular carcinoma recurrence: a prospective, multicenter study. Hepatology. 2011;54:1273-1281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 20. | Waxman DJ. P450 gene induction by structurally diverse xenochemicals: central role of nuclear receptors CAR, PXR, and PPAR. Arch Biochem Biophys. 1999;369:11-23. [PubMed] |
| 21. | Aleksunes LM, Klaassen CD. Coordinated regulation of hepatic phase I and II drug-metabolizing genes and transporters using AhR-, CAR-, PXR-, PPARα-, and Nrf2-null mice. Drug Metab Dispos. 2012;40:1366-1379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 213] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 22. | Koutsounas I, Theocharis S, Patsouris E, Giaginis C. Pregnane X receptor (PXR) at the crossroads of human metabolism and disease. Curr Drug Metab. 2013;14:341-350. [PubMed] |
| 23. | George J, Murray M, Byth K, Farrell GC. Differential alterations of cytochrome P450 proteins in livers from patients with severe chronic liver disease. Hepatology. 1995;21:120-128. [PubMed] |
| 24. | Jokinen MJ, Neuvonen PJ, Lindgren L, Höckerstedt K, Sjövall J, Breuer O, Askemark Y, Ahonen J, Olkkola KT. Pharmacokinetics of ropivacaine in patients with chronic end-stage liver disease. Anesthesiology. 2007;106:43-55. [PubMed] |
| 25. | De Bock L, Boussery K, Van Winckel M, De Paepe P, Rogiers X, Stephenne X, Sokal E, Van Bocxlaer J. In vitro cytochrome p450 activity decreases in children with high pediatric end-stage liver disease scores. Drug Metab Dispos. 2013;41:390-397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 26. | Bentué-Ferrer D, Tribut O, Verdier MC. [Therapeutic drug monitoring of lacosamide]. Therapie. 2012;67:151-155. [PubMed] |
| 27. | Lee JB, Lee KA, Lee KY. Cytochrome P450 2C19 polymorphism is associated with reduced clopidogrel response in cerebrovascular disease. Yonsei Med J. 2011;52:734-738. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 28. | Aubert J, Begriche K, Knockaert L, Robin MA, Fromenty B. Increased expression of cytochrome P450 2E1 in nonalcoholic fatty liver disease: mechanisms and pathophysiological role. Clin Res Hepatol Gastroenterol. 2011;35:630-637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 219] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 29. | Chiu KW, Nakano T, Hu TH, Tseng HP, Cheng YF, Jawan B, Eng HL, Goto S, Chen CL. Influence of CYP2C19 genotypes on graft pathological findings and postoperative liver function in recipients after living-donor liver transplantation. Ann Transplant. 2010;15:38-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 30. | Klein K, Zanger UM. Pharmacogenomics of Cytochrome P450 3A4: Recent Progress Toward the “Missing Heritability” Problem. Front Genet. 2013;4:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 143] [Cited by in RCA: 170] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 31. | MacPhee IA. Pharmacogenetic biomarkers: cytochrome P450 3A5. Clin Chim Acta. 2012;413:1312-1317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 32. | Goldwasser J, Cohen PY, Yang E, Balaguer P, Yarmush ML, Nahmias Y. Transcriptional regulation of human and rat hepatic lipid metabolism by the grapefruit flavonoid naringenin: role of PPARalpha, PPARgamma and LXRalpha. PLoS One. 2010;5:e12399. [PubMed] |
| 33. | Parkinson A, Mudra DR, Johnson C, Dwyer A, Carroll KM. The effects of gender, age, ethnicity, and liver cirrhosis on cytochrome P450 enzyme activity in human liver microsomes and inducibility in cultured human hepatocytes. Toxicol Appl Pharmacol. 2004;199:193-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 184] [Cited by in RCA: 159] [Article Influence: 10.6] [Reference Citation Analysis (1)] |
| 34. | Johnson TN, Boussery K, Rowland-Yeo K, Tucker GT, Rostami-Hodjegan A. A semi-mechanistic model to predict the effects of liver cirrhosis on drug clearance. Clin Pharmacokinet. 2010;49:189-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 169] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 35. | Orlando R, Padrini R, Perazzi M, De Martin S, Piccoli P, Palatini P. Liver dysfunction markedly decreases the inhibition of cytochrome P450 1A2-mediated theophylline metabolism by fluvoxamine. Clin Pharmacol Ther. 2006;79:489-499. [PubMed] |
| 36. | Rieger JK, Klein K, Winter S, Zanger UM. Expression variability of absorption, distribution, metabolism, excretion-related microRNAs in human liver: influence of nongenetic factors and association with gene expression. Drug Metab Dispos. 2013;41:1752-1762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 93] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 37. | Buechler C, Weiss TS. Does hepatic steatosis affect drug metabolizing enzymes in the liver? Curr Drug Metab. 2011;12:24-34. [PubMed] |
| 38. | Albarmawi A, Czock D, Gauss A, Ehehalt R, Lorenzo Bermejo J, Burhenne J, Ganten TM, Sauer P, Haefeli WE. CYP3A activity in severe liver cirrhosis correlates with Child-Pugh and model for end-stage liver disease (MELD) scores. Br J Clin Pharmacol. 2014;77:160-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 49] [Article Influence: 4.5] [Reference Citation Analysis (0)] |