INTRODUCTION
Gastric cancer (GC) is the second highest cause of global cancer mortality. GC is a heterogeneous disease with multiple environmental etiologies and alternative pathways of carcinogenesis[1,2]. One of the major etiologic risk factors for GC is Helicobacter pylori (H. pylori) infection, but only a small proportion of individuals infected with H. pylori develop GC[3,4]. There is an increasing understanding of the roles that genetic and epigenetic alterations play in GCs (Figure 1). Consequently, the development of appropriate biomarkers that reflect an individual’s cancer risk is essential to reduce the mortality from GC[5,6]. Recent advances in molecular research of GC have brought new diagnostic and therapeutic strategies into clinical settings.
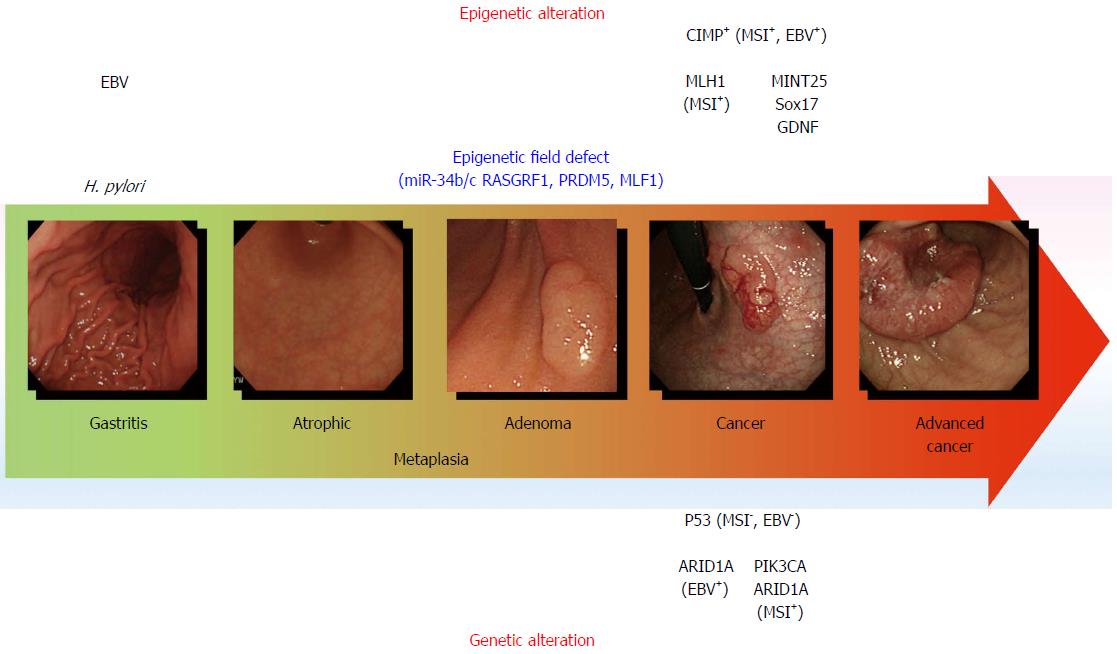
Figure 1 Genetic and epigenetic alterations in gastric carcinogenesis.
The model for gastric carcinogenesis is presented based on genetic and epigenetic alterations. Methylation of the genes in blue appears to be involved in an epigenetic field defect. H. pylori: Helicobacter pylori; MSI: Microsatellite instability; EBV: Epstein-Barr virus; CIMP: CpG island methylator phenotype.
Next-generation sequencing (NGS) is a technology that involves the parallel sequencing of enormous amounts of short DNA strands from randomly fragmented copies of a genome[7,8]. NGS methods used for genome[9], exome[10], epigenome[11] and transcriptome[12] sequencing have the potential to provide novel avenues towards achieving a comprehensive understanding of diseases, including cancer[13,14]. Such advances have also shown puzzling tumor heterogeneity with limited somatic alterations shared between tumors of the same histopathologic subtype[15-17]. Although NGS techniques are just beginning to expand our abilities to detect genome-wide alterations in GC, several NGS studies in GC have recently been published[18].
In this review, we summarize the key findings of past reports pertaining to the genetics and epigenetics of GC and their relationship to and future application in NGS. We also describe the recurrently mutated genes and alterations in GC identified by NGS technology and discuss the basic framework for future investigations, including the challenges of using NGS as a tool for biomarker and therapeutic target discovery.
MICROSATELLITE INSTABILITY
A type of genetic instability characterized by alterations in length within simple repeat microsatellite sequences, termed microsatellite instability (MSI), occurs in approximately 15% of sporadic GCs, mainly as a result of epigenetic changes[19-22]. Genetic and epigenetic inactivation of DNA mismatch repair (MMR) genes leads to the mutator phenotype, mutations in cancer-related genes and cancer development (Figure 2). MSI underlies a distinctive carcinogenic pathway because MSI-positive (MSI+) GCs exhibit many differences in clinical, pathological and molecular characteristics compared with MSI-negative (MSI-) GCs[19-22]. The differences in genotype occur because defective MMR results in a strong mutator phenotype with a very specific mutation spectrum. MSI mainly accumulates frameshift mutations in the repeated sequences located in the coding regions of a target tumor suppressor or other tumor-related genes[23-26]. The atypical genotype of MSI+ GCs also includes specific patterns of gene dysregulation. MSI+ GCs often show epigenetic alterations, such as hypermethylation of various genes, including the key MMR gene MLH1. The differences in genotype and phenotype between MSI+ and MSI- GCs are likely linked to their differences in biological and clinical features. Recent findings from NGS analysis, such as the frequent mutation of the AT-rich interactive domain 1A (ARID1A) in MSI+ GCs, support this notion[27,28].
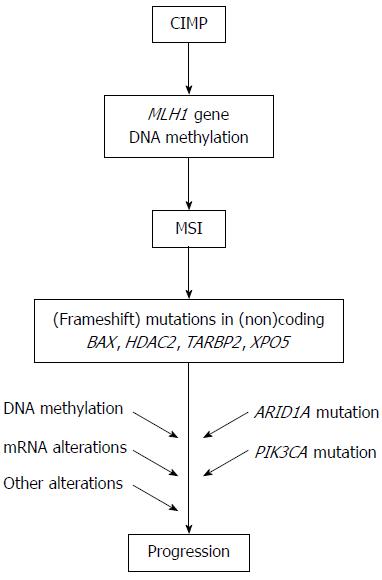
Figure 2 Molecular pathway for microsatellite instability+ gastric cancer.
The model for the carcinogenesis of microsatellite instability (MSI)+ gastric cancer is presented. CIMP: CpG island methylator phenotype.
The clinicopathological, genetic, epigenetic, prognostic and therapeutic characteristics of MSI+ GCs are becoming clearer, but further research is still required. Because molecular targeting therapeutics are being used in clinical settings and trials, the differential regulation of molecular target genes in MSI+ and MSI- GCs[29,30] needs to be clarified. Diagnostic characterization of the MSI status of GCs thus has important implications for basic and clinical oncology.
Frequent inactivating mutations of ARID1A in molecular subtypes of GC identified by exome sequencing
Holbrook et al[31] analyzed 50 GC samples with targeted deep sequencing of the DNA of 384 genes. In addition to the previously reported mutations in genes belonging to various pathways, the authors found tractable target genes, such as the genes for the thyrotropin receptor and the Rho-associated coiled-coil containing protein kinases ROCK1 and ROCK2. Wang et al[27] performed exome sequencing of 22 GC samples and found novel mutated genes and pathway alterations involved in chromatin modification. A validation study confirmed frequent inactivating mutations or protein loss of the ARID1A gene, which encodes one of the subunits in the Switch/Sucrose Nonfermentable (SWI-SNF) chromatin remodeling complex. The mutation spectrum for ARID1A differed among molecular subtypes of GC; mutations were detected in 83% of GCs with MSI, 73% of GCs with EBV infection and 11% of GCs without EBV and MSI. Moreover, ARID1A mutations were negatively associated with TP53 mutations. ARID1A alterations were associated with better prognosis in a stage-independent manner. These results suggest the importance of altered chromatin remodeling in the pathogenesis of GC.
Recurrent somatic mutations in cell adhesion and chromatin remodeling genes identified by exome sequencing
Zang et al[28] also analyzed a spectrum of somatic alterations in GC by sequencing the exomes of 15 GC specimens, including 11 intestinal-type, 1-mixed-type, and 3 diffuse-type adenocarcinomas and their matched normal DNAs. TP53 (11/15 tumors), PIK3CA (3/15) and ARID1A (3/15) were frequently mutated. Among the frequently mutated genes, cell adhesion was the most significant biological pathway affected. A prevalence screening confirmed mutations in FAT4, a member of the cadherin gene family, in 5% of GCs (6/110) and FAT4 genomic deletions in 4% (3/83) of GCs. Mutations in chromatin remodeling genes (ARID1A, MLL3 and MLL) were also found in 47% of GCs. ARID1A mutations were detected in 8% of GCs (9/110) and were associated with concurrent PIK3CA mutations and MSI. Both FAT4 and ARID1A showed tumor-suppressor activity in functional assays. Somatic inactivation of FAT4 and ARID1A may thus be key tumorigenic events in a subset of GCs. Because PI3K inhibitors are currently in clinical testing as treatment for GC[32], it will be interesting to evaluate whether the tumor responses to these compounds are affected by the genomic status of ARID1A.
Frequent loss of ARID1A expression in GC with EBV infection or MSI
Mutations of ARID1A lead to a loss of protein expression in GC and are particularly associated with EBV infection or MSI. Abe et al[33] investigated the significance of the loss of ARID1A in 857 GC cases, including 67 EBV+ and 136 MLH1-lost MSI+ GCs. Loss of ARID1A expression was significantly more frequent in cases of EBV+ (23/67; 34%) and MSI+ (40/136; 29%) GCs than in cases of EBV-/MSI- (32/657; 5%) GCs. Loss of ARID1A was correlated with larger tumor size, deeper depth of invasion, lymph node metastasis and poorer prognosis in cases of EBV-/MSI- GC. A correlation with tumor size and diffuse-type histology was found only in the MSI+ GC; no correlation was observed in EBV+ GC. Loss of ARID1A expression in EBV+ GC was frequent in the early stage of GC, but EBV infection did not cause loss of ARID1A in GC cell lines. Thus, loss of ARID1A may be an early event in EBV+ GC and may precede EBV infection in gastric epithelial cells. On the other hand, loss of ARID1A may be involved in the progression of EBV-/MSI- GCs. Thus, loss of ARID1A appears to have different, pathway-dependent roles in GC.
WHOLE-GENOME SEQUENCING ANALYSIS OF GC
To explore the complete list of somatic alterations in GC, Nagarajan et al[34] combined massively parallel short read and DNA paired-end tag sequencing for the first whole-genome analysis of two GCs, one with CIN and the other with MSI. Integrative analysis and de novo assemblies revealed the architecture of a wild-type KRAS amplification, a common driver event in GC[35]. Three distinct mutational signatures were discovered against a genome-wide backdrop of oxidative and MSI-associated mutational signatures. Combining sequencing data from 40 complete GC exomes and targeted screening of an additional 94 independent GCs led to the discovery of ACVR2A, RPL22 and LMAN1 as recurrently mutated genes in MSI+ GC and the identification of PAPPA as a recurrently mutated gene in TP53 wild-type GC. These results highlight how whole-genome sequencing analysis can provide relevant information about tissue-specific carcinogenesis that would otherwise be missed in exome-sequencing data. WGS of more GCs will uncover more recurrently altered genes.
miRNA alterations
A microRNA (miRNA) is a small noncoding RNA that regulates gene expression at the posttranscriptional level and is critical in many biological processes and cellular pathways[36-40]. The causes of aberrant miRNA expression patterns in cancer include DNA copy number amplification or deletion, inappropriate transactivation, transcriptional repression by oncogenic and other factors, failure of miRNA post-transcriptional regulation and genetic mutation or transcriptional silencing associated with hypermethylation of the CpG island promoters.
There is accumulating evidence to support the notion that miRNA alterations play a key role in the pathogenesis of GC[41-44]. A large number of miRNAs with different biological functions have been found to be altered and correlated with clinicopathological characteristics and/or prognosis in GC. Moreover, the clinical potential of miRNA alterations as minimally invasive diagnostic biomarkers and therapeutic targets has been extensively reported[37,40,42,44]. Recent studies have shown that tumor-derived miRNAs are present and stable in circulation, and the levels of circulating miRNAs are detectable and quantifiable. Both tissue and soluble miRNAs are candidates for diagnostic biomarkers and therapeutic targets in GCs[44]. The basic strategy of current miRNA-based treatment studies is to either antagonize the expression of target oncogenic miRNAs with antisense therapy and other technology or to restore the function of impaired tumor suppressor miRNAs[42].
The inclusion of different isoforms of miRNA (isomiRs) that are natural variants of mature miRNAs will form a detailed miRnome. Because expression of isomiRs can be estimated by NGS, NGS platforms provide the most effective method of miRNA profiling, leading to the identification of the miRNA alterations with clinical applications. Li et al[45] sequenced small RNAs from one pair of GC and noncancerous tissue and found that isomiR patterns are significantly different between these tissues. Moreover, these authors found that the 5p arm and 3p arm miRNAs derived from the same pre-miRNAs have different tissue preferences in GC and noncancerous tissue, suggesting a novel mechanism regulating mature miRNA selection.
WHOLE-TRANSCRIPTOME SEQUENCING OF GC
The first comprehensive RNA-seq study in GC has been recently published. Kim et al[46] applied a whole-transcriptome sequencing approach to 24 GC samples and six noncancerous tissue specimens. Importantly, these authors developed a multilayered integrative analysis to identify various types of transcriptional aberrations, such as differentially expressed mRNAs and miRNAs, as well as recurrently mutated genes. A central metabolic regulator gene, AMPKa2 (PRKAA2), was identified as a potential functional target in GC. Six key miRNAs (miR-548d-3p, miR-20b, miR-135b, miR-140-3p, miR-93 and miR-19a) in GC were also identified.
Epigenetic alterations
Epigenetic regulation is essential for the normal development and maintenance of tissue-specific gene expression patterns in mammals. Disruption of epigenetic regulation can lead to altered gene function and malignant cellular transformation[47]. Recent cancer epigenetic studies have revealed various alterations in the epigenetic machinery in GC, including DNA methylation, histone modifications, nucleosome positioning, noncoding RNAs and miRNAs[48-52]. Aberrant DNA methylation in the promoter CpG islands of genes results in inactivation of tumor suppressor and other tumor-related genes in cancer cells and is the most well-defined epigenetic hallmark in GC. Methylation of a large number of genes with different biological functions has been found to be correlated with the clinicopathological characteristics and prognosis in GC[48-52]. DNA methylation with its advantages as a biomarker for the detection of cancer in biopsy specimens and body fluids that can be obtained non-invasively, such as serum and gastric washes, may have a clinical application in GC. Detection of aberrant DNA methylation of genes, such as CDH1, DAPK, GSTP1, p15, p16, RARβ, RASSF1A, RUNX3 and TFPI2, in the serum may be a useful biomarker for the detection of GC[50]. Studies of DNA methylation and histone modification using NGS technologies, such as whole-genome bisulfite sequencing and targeted bisulfite sequencing, will lead to new discoveries and improve our knowledge of the epigenomics of GC[11].
Association of the aberrant methylation of RASGRF1 with an epigenetic field defect and an increased risk of GC
Aberrant DNA methylation is implicated in the epigenetic field defect seen in GC. Thus, it is important to identify predictive biomarkers by screening for DNA methylation in the noncancerous background gastric mucosa of patients with GC. Using methylated-CpG island amplification coupled with CpG island microarray (MCAM) analysis, Takamaru et al[53] found 224 genes that were methylated in the noncancerous gastric mucosa of patients with GC. Among them, RASGRF1 methylation was significantly elevated in the gastric mucosa from patients with either intestinal- or diffuse-type GC, compared with the mucosa from healthy individuals. RASGRF1 methylation was independent of mucosal atrophy and could be used to distinguish both serum pepsinogen test-positive and -negative patients with GC from healthy individuals. Ectopic expression of RASGRF1 suppressed colony formation and Matrigel invasion by GC cells. RASGRF1 methylation appears to be significantly involved in the epigenetic field defect of the stomach and to be a useful biomarker to identify individuals at high risk for GC.
Association of aberrant methylation of miR-34b/c with an epigenetic field defect and an increased risk of GC
The silencing of miRNAs is often associated with CpG island hypermethylation. Thus, to identify epigenetically silenced miRNAs in GC, Suzuki et al[54] screened for miRNAs that were induced by treatment of GC cells with 5-aza-2’-deoxycytidine and 4-phenylbutyrate. Hypermethylation of the neighboring CpG island epigenetically silenced miR-34b and miR-34c. Methylation of the miR-34b/c CpG island was frequently observed in GC cell lines (13/13, 100%) but not in normal gastric mucosa from healthy H. pylori-negative individuals. Transfection of the precursors of miR-34b and miR-34c into GC cells suppressed growth and changed the gene expression profile. Methylation of miR-34b/c was found in a majority of primary GCs (83/118, 70%). Notably, analysis of the non-cancerous gastric mucosae from GC patients (n = 109) and healthy individuals (n = 85) revealed that methylation levels were higher in the gastric mucosae of patients with multiple GC lesions than in the mucosae from those patients with single GC and the mucosae from healthy H. pylori-positive individuals. These results suggest that miR-34b and miR-34c are novel tumor suppressors frequently silenced by DNA methylation in GC. Methylation of miR-34b/c appears to be significantly involved in an epigenetic field defect in the stomach and to be a useful biomarker to identify individuals at high risk for multiple GC.
Methylation of miR-34b/c in the mucosa of the noncancerous gastric body may be a useful biomarker for predicting the risk of metachronous GC
Metachronous GC can develop after endoscopic resection of GC and is not predictable based on the clinical characteristics alone. Aberrant DNA methylation in noncancerous gastric mucosa has been implicated in gastric carcinogenesis and may be a useful biomarker of GC risk. Suzuki et al[55] evaluated the clinical utility of DNA methylation as a biomarker of metachronous GC risk. Scheduled follow-up endoscopy was performed in 129 patients after curative endoscopic resection of early GC. Biopsy specimens were collected from noncancerous mucosa in the gastric antrum and body. A quantitative methylation analysis of miR-34b/c, SFRP1, SFRP2, SFRP5, DKK2 and DKK3 using bisulfite pyrosequencing was performed on the collected biopsy specimens. The utility of the methylation status for predicting the risk of developing metachronous GC was analyzed using Kaplan-Meier and Cox proportional hazards models. During the follow-up period, 17 patients (13%) developed metachronous GCs. The cumulative incidence of metachronous GC was significantly higher among patients with elevated miR-34b/c, SFRP2 and DKK2 methylation in the gastric body. Elevated methylation of miR-34b/c showed the most significant association with the risk of metachronous GC; the cumulative incidence of metachronous GC was much higher in the high miR-34b/c-methylation group than in the low methylation group. Multivariate analysis adjusted for age, sex, H. pylori status and pathological findings showed that miR-34b/c methylation in the gastric body was an independent predictor of metachronous GC risk. Methylation of miR-34b/c in the mucosa of the noncancerous gastric body may be a useful biomarker for predicting the risk of metachronous GC. Finally, NGS technologies may characterize an epigenetic field defect more clearly and highlight more useful biomarkers.
Sensitive and specific detection of early GC by DNA methylation analysis of gastric washes
Because many mucosal cells can be found in the gastric juice, the detection of molecular markers in the gastric juice was a possible noninvasive approach to detect GC. However, the use of gastric juice as a molecular diagnostic or predictive tool has been previously reported to be impractical because the DNA is easily degraded by gastric acidity. In this regard, Watanabe et al[56] have developed a new method for GC detection by DNA methylation in gastric washes but not in gastric juice. These authors analyzed 51 candidate genes in 7 GC cell lines and 24 GC samples (training set). They then selected 6 genes (MINT25, RORA, GDNF, ADAM23, PRDM5 and MLF1) for further analyses. The methylation status of these genes was analyzed in a test set consisting of 131 GCs at various stages. The 6 candidate genes were validated in a different population of 40 primary GC samples and 113 noncancerous gastric mucosa samples. The 6 genes showed differential methylation in GC and normal mucosa in the training, test and validation sets. GDNF and MINT25 were the most sensitive molecular markers of early-stage GC, whereas PRDM5 and MLF1 were markers of a field defect. A close correlation between methylation levels in tumor biopsy samples and gastric washes was noted. MINT25 methylation showed the best sensitivity (90%) and specificity (96%), and it had the greatest area under the receiver operating characteristic curve (0.961) in terms of tumor detection in gastric washes. MINT25 methylation in gastric washes may be a sensitive and specific marker for the screening of GC.
Detection of early GC by DNA methylation analysis of Sox17 in gastric washes
Although minimally invasive treatment is widely accepted for early-stage GC, appropriate risk markers to detect residual cancer after endoscopic resection and the potential for recurrence are not available. To find candidate genes that might be markers for the detection of early GC, Oishi et al[57] performed methylated CpG island amplification microarray analysis on 12 gastric washes (from the pre- and post-endoscopic treatment of six patients). Among the candidate genes, the Sox17 gene was selected for further analysis. The DNA methylation status of Sox17 was examined in a validation set consisting of 128 gastric wash samples (64 pre-treatment and 64 post-treatment) from cases of early GC. Sox17 showed significant differential methylation in the pre- and post-treatment gastric washes of early GC patients (Figure 3). Moreover, the treatment of GC cells that lacked Sox17 expression with the methyltransferase inhibitor 5-aza-2′-deoxycytidine restored the gene’s expression. Additionally, the introduction of exogenous Sox17 into silenced GC cells suppressed colony formation. The data suggest that the silencing of Sox17 occurs frequently in early GC and plays a key role in the disease. Gastric wash-based DNA methylation analysis could be useful for the early detection of recurrence following endoscopic resection in early GC patients. Interestingly, the usefulness of gastric wash-based molecular testing for antibiotic resistance in H. pylori has also been reported[58]. It will be interesting to analyze gastric washes using NGS.
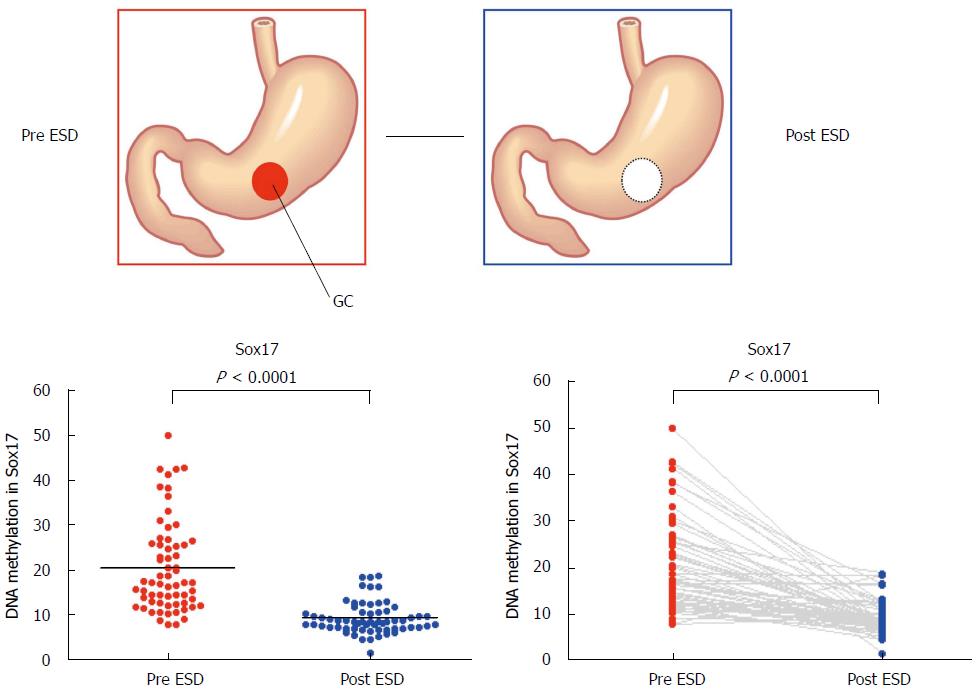
Figure 3 Methylation levels of Sox17 before and after endoscopic submucosal dissection.
Methylation levels of Sox17 were analyzed by pyrosequencing using the DNA recovered from gastric washes before and after endoscopic submucosal dissection[57].
Anti-HER2 antibody trastuzumab has led to an era of personalized therapy in GC
Trastuzumab is an antibody that targets the HER2 extracellular domain and induces antibody-dependent cellular cytotoxicity and inhibition of the HER2 downstream signals (Figure 4). In the ToGA study, standard chemotherapy regimens (capecitabine plus cisplatin or fluorouracil plus cisplatin) combined with trastuzumab resulted in a longer survival time than standard regimens without trastuzumab in patients with HER2-positive GC[59]. Thus, HER2 expression has become a major concern in GC[60]. HER2 overexpression is observed in 7%-34% of GC cases. Mechanisms of resistance to trastuzumab have been reported in breast cancer. There are various mechanisms underlying trastuzumab resistance, such as alterations of the HER2 structure or surroundings, dysregulation of HER2 downstream signal effectors and interaction of HER2 with other membrane receptors (Figure 4). The PI3K-Akt pathway is one of the main downstream signaling pathways of HER2. It is well known that PIK3CA mutations and PTEN inactivation cause over-activation of a downstream signal without activation of an upstream signal. The frequencies of PIK3CA mutations and PTEN inactivation in GC have been reported to be 4%-25% and 16%-77%, respectively. However, little is known about the association between HER2 expression and PI3K-Akt pathway alterations in GC. Sukawa et al[29] have found that HER2 overexpression was significantly correlated with pAkt expression in GC tissues. Furthermore, pAkt expression was correlated with poor prognosis. These results suggest that the PI3K-Akt pathway plays an important role in HER2-positive GC. Moreover, PIK3CA mutations and PTEN inactivation could affect the effectiveness of HER2-targeting therapy. Thus, it is necessary to clarify not only HER2 alterations but also PI3K-Akt pathway alterations to optimize HER2-targeting therapy in patients with GC. In this regard, NGS will be useful for the identification of complicated mechanisms of trastuzumab resistance in GC. The only approved targeted therapy for patients with advanced GC is trastuzumab. It is hoped that NGS will reveal a driver gene alteration that will make other targeted therapies possible[13,61].
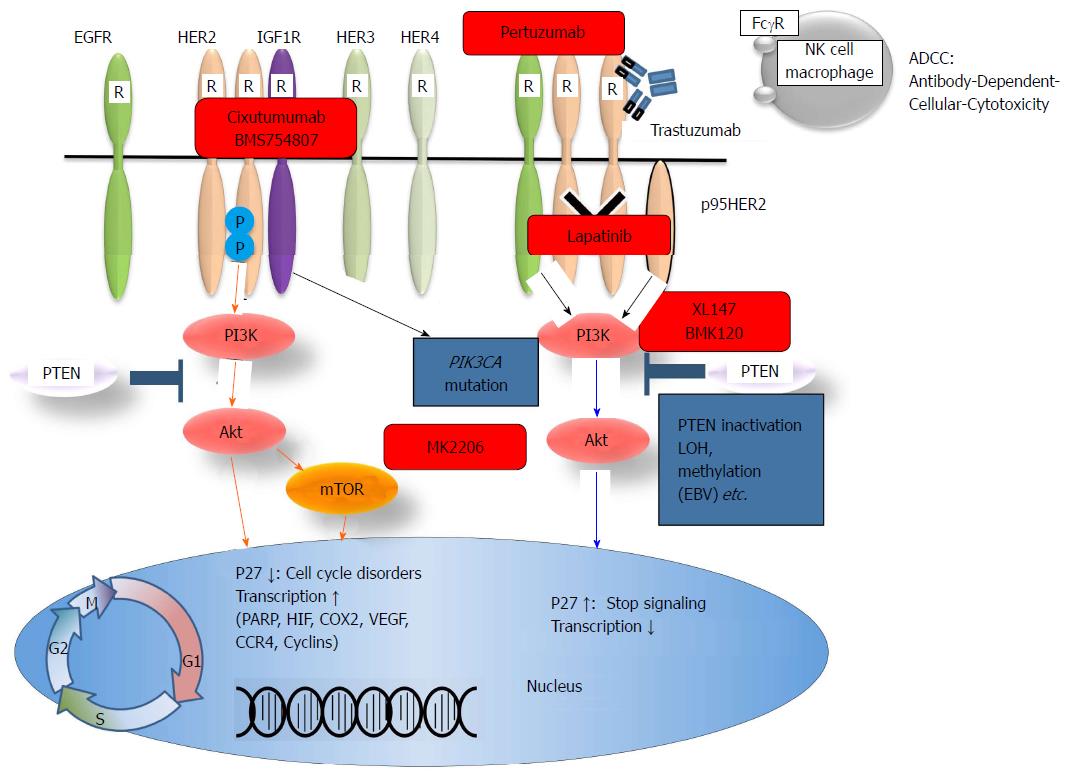
Figure 4 Human epidermal growth receptor family members, the PI3K/Akt pathway, and targeted drugs.
HER: Human epidermal growth receptor; NK: Natural killer; IGF1R: α-insulin-like growth factor 1-receptor; EGFR: Epidermal growth factor receptor; PI3K: Phosphatidylinositol-3-kinase; PTEN: Phosphatase and tensin homologue.
Monoclonal antibodies targeting VEGF (AVAGAST trial) and VEGFR-2 (REGARD trial) in advanced GC
Several vascular endothelial growth factor (VEGF)-targeted agents have been developed, including neutralizing monoclonal antibodies (MoAbs) to VEGF/VEGFRs, soluble VEGF receptors and tyrosine kinase inhibitors (TKIs). The anti-VEGF MoAb bevacizumab has been approved for colorectal cancers. VEGF and VEGF receptor-2 (VEGFR-2)-mediated signaling and angiogenesis contribute to the pathogenesis and progression of GC. The Avastin in Gastric Cancer (AVAGAST) trial was a multinational, randomized, placebo-controlled trial designed to evaluate the efficacy of adding bevacizumab to capecitabine-cisplatin in the first-line treatment of advanced GC[62]. The study showed that adding bevacizumab to the chemotherapy regimen in patients with advanced GC improved the progression-free survival and tumor response rate but not the overall survival. A following biomarker evaluation analysis revealed that plasma VEGF-A and tumor neuropilin-1 are strong biomarker candidates for predicting the clinical outcome in patients with advanced GC treated with bevacizumab[63]. In this regard, NGS will be a powerful method for the identification of predictive biomarkers.
To analyze whether ramucirumab, a monoclonal antibody targeting VEGFR-2, prolongs survival in patients with advanced GC, an international, randomized, double-blind, placebo-controlled, phase 3 trial was conducted in 29 countries[64]. In total, 355 patients with advanced gastric or gastro-esophageal junction adenocarcinoma and disease progression after first-line chemotherapy were randomly assigned (2:1) to receive best supportive care plus either ramucirumab 8 mg/kg (n = 238) or placebo (n = 117), intravenously once every 2 wk. The primary endpoint was overall survival. The median overall survival was 5.2 mo in the ramucirumab group and 3.8 mo in the placebo group (HR = 0.776, 95%CI: 0.603-0.998, P = 0.047). The survival benefit with ramucirumab remained unchanged after multivariate adjustment for other prognostic factors (multivariate HR = 0.774, 95%CI: 0.605-0.991, P = 0.042). Thus, ramucirumab is the first biological treatment given as a single drug that showed survival benefits in patients with advanced gastric or gastro-esophageal junction adenocarcinoma who progressed after first-line chemotherapy. The findings also validate VEGFR-2 signaling as an important therapeutic target in advanced GC.
Potential targeted drugs for GC
Using NGS to target a subset of druggable genes becomes a more effective way to discover therapeutic targets[13,14,61]. There are several potential targeted drugs, either MoAb or small-molecule TKIs, that are being investigated either in synergy with, or in place of, established treatments. These drugs include inhibitors of growth factors and their receptors [i.e., VEGF, epidermal growth factor receptor, HER2, insulin-like growth factor 1 (IGF1) receptor, c-MET], MEK inhibitors and drugs targeting the Hedgehog pathway[65].
Dysregulation of the IGF1 and IGF2/IGF1R system has been implicated in the pathogenesis of GC[66-69]. The expression levels of both IGFs and IGF1R are increased in GC. IGF1R is also involved in angiogenesis and lymphangiogenesis through the modulation of VEGF expression in a GC cell line[70]. IGF1R blockade reduced tumor angiogenesis and enhanced the effects of bevacizumab in a GC cell line. Thus, targeting IGF1R in combination with agents that block the VEGF pathway may have therapeutic utility in GC. Moreover, targeting the novel miR-7/IGF1R/Snail axis has been reported to be useful as a therapeutic approach to block GC metastasis[71].