Published online May 28, 2013. doi: 10.3748/wjg.v19.i20.3143
Revised: April 8, 2013
Accepted: April 18, 2013
Published online: May 28, 2013
Processing time: 166 Days and 13.1 Hours
AIM: To investigate the value of combined detection of circulating cell-free DNA (cfDNA), α-fetal protein (AFP) and α L-fucosidase (AFU) for diagnosis of hepatocellular carcinoma (HCC).
METHODS: Serum samples from 39 HCC patients and 45 normal controls were collected. Branched DNA (bDNA) was used to detect the level of cfDNA, and a receiver operating characteristic curve was employed to evaluate the diagnostic sensitivity, specificity, accuracy, positive predictive value, negative predictive value, positive likelihood ratio, negative likelihood ratio and Youden index, and to assess the diagnostic efficiency and their correlations with the clinicopathological features. AFP and AFU were detected by chemiluminescence and colorimetry, respectively. The significance of combined detection of the three biomarkers was discussed.
RESULTS: cfDNA level was increased in 22 of the 39 HCC samples and in 2 of the 45 normal controls. cfDNA level in HCC samples was significantly higher than that in normal controls (P < 0.05). There were significant differences in sex and extra- and intrahepatic metastasis (P < 0.05). There was no significant correlation between cfDNA, AFP and AFU in the detection of HCC. The sensitivity of combined detection of cfDNA with one marker (AFP or AFU) and cfDNA with two markers (AFP and AFU) was 71.8%, 87.2% and 89.7% vs 56.4%, 53.8% and 66.7% for cfDNA, AFP and AFU used alone, respectively, the difference being statistically significant (P < 0.05).
CONCLUSION: Quantitative analysis of cfDNA is sensitive and feasible, and the combined detection of cfDNA with AFP or AFU or both could improve the diagnostic sensitivity for HCC.
Core tip: Various techniques are available to quantify serum circulating free cell DNA (cfDNA), some of these techniques require abstraction and purification as there is only a very small amount of cfDNA in the serum. This study investigates the value of combined detection of cfDNA, α-fetal protein (AFP) and α L-fucosidase (AFU) for diagnosis of hepatocellular carcinoma. Quantitative analysis of cfDNA is sensitive and feasible, and the combined detection of cfDNA with AFP or AFU or both could improve the diagnostic sensitivity for hepatocellular carcinoma.
- Citation: Chen K, Zhang H, Zhang LN, Ju SQ, Qi J, Huang DF, Li F, Wei Q, Zhang J. Value of circulating cell-free DNA in diagnosis of hepatocelluar carcinoma. World J Gastroenterol 2013; 19(20): 3143-3149
- URL: https://www.wjgnet.com/1007-9327/full/v19/i20/3143.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i20.3143
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors worldwide, with an incidence which ranks third and mortality which ranks second in all malignant tumors. The one-year survival in patients with HCC without surgical ablation is less than 30%, and the 5-year recurrence rate is about 80%, even in patients who undergo radical resection. Therefore, the key to HCC treatment is early detection and diagnosis[1,2]. Existing biomarkers for the diagnosis of HCC are alpha-fetal protein (AFP), γ-glutamyltranspeptidase isoenzymesII (GGT-II), α L-fucosidase (AFU) and acidic isoferritin[3], of which AFP remains the most specific marker due to its recognized effect in the diagnosis of HCC and assessment of the curative effect, prognosis and recurrence. However, not all HCC patients are diagnosed by AFP alone as the false negative rate of AFP is about 30%. Markers like AFU and GGT-II have assisted in diagnosing patients with HCC whose AFP levels are negative, but they can not replace AFP in the diagnosis of HCC. Therefore, there is an urgent need to find a new biomarker which is superior or at least equivalent to AFP for the diagnosis of HCC.
Cell-free fetal DNA (cfDNA) has been extensively studied over the past few decades. Supported by theory and observation, two major sources of cfDNA have been postulated: first, fragmented DNA released as a consequence of cell death (apoptotic/necrotic) and, second, active metabolic secretion of DNA from cells. Considerable research effort has been made on the use of cfDNA as a biomarker in cancer diagnosis. cfDNA from malignancies exhibits characteristic changes such as mutations, deletions, methylations and microsatellite aberrations which are distinct from those in benign conditions, and thus may be useful in the diagnosis of cancer[4].
This study was designed to validate a bDNA-based Alu assay for quantifying human cfDNA in blood, and explore the feasibility of Alu quantification for HCC screening.
Included in this study were 39 patients randomly selected from those who had been diagnosed with HCC by clinical and radiographic evaluations at the Affiliated Hospital of Nantong University (Nantong, China) between May and October 2011. They included 32 males and 7 females ranging in age from 40 to 85 years. Forty-five healthy volunteers (36 males and 9 females; age range 28-83 years) who were negative for physical indices following health examination and documented to have normal liver biochemistry served as controls. Fresh blood was collected in serum separator tubes (Vacuette, Austria) coated with a clot activator and barrier gel, and EDTA tubes (Kangshi, China) for plasma, were centrifuged at 1600 ×g for 10 min, and then microcentrifuged at 16000 ×g. The separated serum was immediately stored at -80 °C. The samples tested were diluted using the standard human genomic DNA qualification kit according to the manufacturer’s instructions, with an initial concentration of 2.73× 105.
Quantitation of cfDNA: Ten μL diluted serum was added to lysis solution to a final volume of 100 μL containing 50%Lysis Mixture (Panomics) and 1 g/L proteinase K. Target gene probe sets containing 250, 250 and 250 fmol of capture extender probe (CE), blocking probe and label extender probe (LE) were added to the blood lysate. The lysate was then transferred to an assay well in a 96-well plate (Panomics, Redwood City, CA, United States) covalently coated with capture probe (CP) oligo, incubated at 55 °C for 1 h, washed 3 times with 200 μL wash buffer (0.1 × standard 4 saline citrate containing 0.3 g/L lithium lauryl sulfate), sequentially hybridized with 100 μL 1:1000 dilution of bDNA preamplifier (amplifier) (Panomics) at 55 °C for 30 min and then with 100 μL (50 fmol) 3’-alkaline phosphatase-conjugated Label Probe oligo (Panomics) at 50 °C for 30 min, and then washed three times. After the final wash, the alkaline phosphatase substrate dioxetane (Panomics) was added to the wells and incubated at room temperature for 5-10 min to detect luminescent signals using an Lmax microtiter plate luminometer (a molecular device). The reagent for the QuantiGene 2.0 DNA Assay was provided by Professor Zhang Lurong from Florida State University, United States.
In the same 96-well-plate, samples with a known amount of Human Genomic DNA were added as follows: 0, 50, 100, 200 and 400 ng/mL, respectively. A standard curve was mapped out by the computer. The standard Human Genomic DNA solution was obtained from Promega, Madison, United States.
Quantitation of AFP: The concentration of AFP in the serum samples from HCC patients was measured manually with the ARCHITECT AFP Reagent Kit (Abbott, Chicago, IL, United States) according to the calibrator dose response curve by the chemiluminescence method using an Abbott 2000 immunoassay instrument.
Quantitation of AFU: The activity of AFU in the serum samples from HCC patients was measured manually with a Shanghai Institute of Naval Medicine Biotechnology Center Kit (Shanghai, China) by a colorimetric method using a BA-88 semi-automatic biochemical analyzer.
Reference value of cfDNA: The content of cfDNA in each sample was calculated according to the standard curve. A receiver operating characteristic (ROC) curve was drawn to select the Youden’s index[5], and the highest sensitivity and specificity (Youden’s index = sensitivity + specificity-1) were selected as the cut-off values. A value greater than or equal to the cut-off value was regarded as positive, and smaller values were regarded as negative.
Reference value of AFP: According to the clinical diagnosis and staging criteria of HCC of the Chinese Anti-Cancer Association, a content of AFP greater than or equal to 400 ng/mL is considered positive, and a content smaller than this value is negative, excluding cases of pregnancy, embryonic tumors of the reproductive system, active liver disease and metastatic liver cancer.
Reference value of AFU: According to the reagent kit manual, the activity of serum AFU = (A determination tube-A control tube)/average A standard tube × 400, a value greater than 650 μmol/L per hour is considered positive, 550-650 μmol/L per hour is suspicious, and less than 550 μmol/L per hour is negative.
Statistical analysis of the data was performed using the SPSS 13.0 software package. As the results of the concentrations of cfDNA, AFP and AFU in each group were normally distributed, they were presented as the median (range of detection). Measurement data were based on mean ± SD. Correlations between the three indices were evaluated with Pearson’s correlation coefficient. The rates of interclass were determined using the χ2 test. A P value less than 0.05 was considered statistically significant.
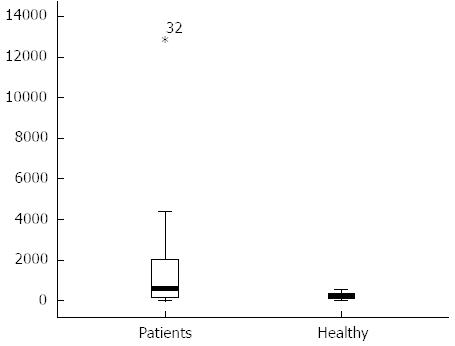
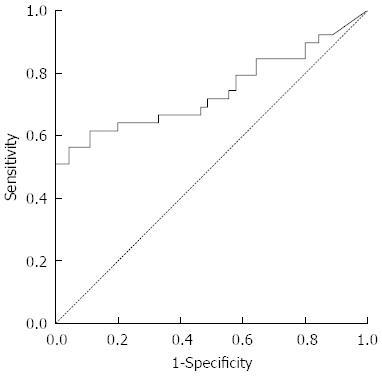
The content of serum cfDNA in the 39 HCC samples ranged from 12866.8 ng/mL to 0 ng/mL, with a median of 557.59 ng/mL compared with 569.2963 ng/mL, 0 ng/mL and 240.07 ng/mL in the 45 normal control samples, and a significant difference was observed between them (P < 0.05) (Figure 1). According to the ROC curve (Figure 2), the Youden index was maximal when the concentration of cfDNA was 509.9774 ng/mL, therefore 509.9774 ng/mL was defined as the positive value.
The ROC area under the curve (AUC) was 0.742 ± 0.058, therefore a sum score of the classifier at 509.9774 ng/mL was chosen as the optimal cut-off, as it had the highest Youden’s index of 0.520. At this cut-off value, the sensitivity was 56.4%, the specificity was 95.6%, the accuracy was 77.4%, the positive predictive value was 91.7%, the negative predictive value was 71.7%, the positive likelihood ratio was 12.692 and the negative likelihood ratio was 0.048.
Correlations between clinicopathological features and serum cfDNA level were observed, suggesting that serum cfDNA level in HCC patients was correlated with intrahepatic and extrahepatic metastasis and gender (Table 1).
| Observation index | No. | No. of positive cfDNA (%) | P value |
| Age (yr) | |||
| ≤ 60 | 26 | 16 (61.5) | 0.453255 |
| > 60 | 13 | 6 (46.2) | |
| Sex | |||
| Male | 32 | 19 (59.4) | 0.024449 |
| Female | 7 | 3 (42.9) | |
| HBsAg | |||
| Positive | 31 | 16 (51.6) | 0.080857 |
| Negative | 8 | 6 (75) | |
| Intrahepatic and extrahepatic metastasis | |||
| Positive | 5 | 4 (80) | 0.000242 |
| Negative | 34 | 18 (52.9) | |
| Cirrhosis of liver | |||
| Positive | 20 | 10 (50) | 0.831179 |
| Negative | 19 | 12 (63.2) | |
The content of serum AFP in HCC samples ranged from > 10000 ng/mL to 3.92 ng/mL, with a median of 392.07 ng/mL, compared with 526.10 ng/mL, 1.31 ng/mL and 6.02 ng/mL in the normal control samples, and a significant difference was observed between them (P < 0.05).
The content of serum AFU in HCC samples ranged from 2366 ng/mL to 362 ng/mL, with a median of 744 ng/mL compared with 792 ng/mL, 121 ng/mL and 431 ng/mL in the normal control samples, and a significant difference was observed between them (P < 0.05).
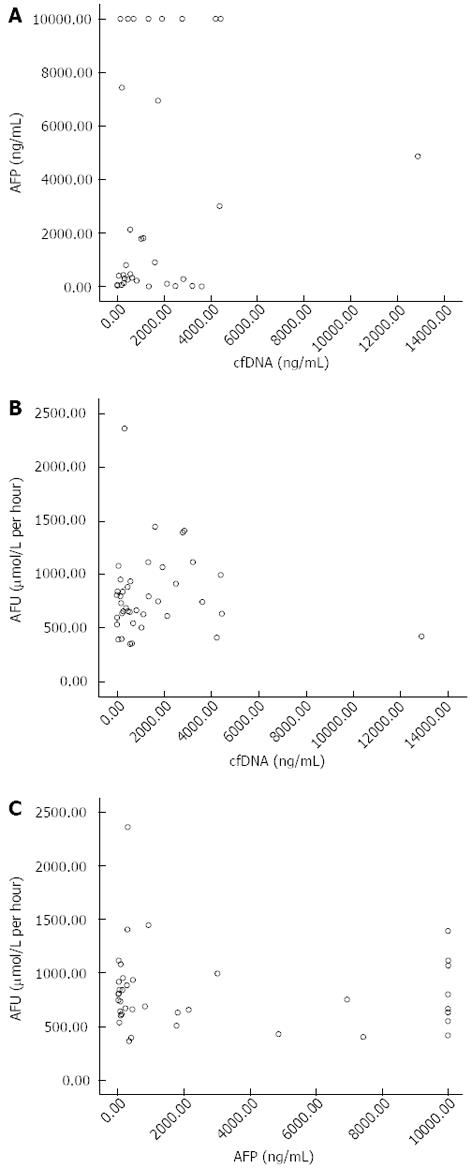
Scatter point diagrams were mapped out to analyze correlations between the three indices using SPSS13.0 (Figure 3).
Compared with the diagnostic efficiency of cfDNA, AFP or AFU alone, combined detection improved the diagnostic efficiency of HCC to some extent (Table 2).
| Groups | Sensitivity | Specificity | Accuracy | Positive predictive value | Negative predictive value |
| cfDNA | 56.4ce | 95.6ce | 77.4 | 91.7ce | 71.7 |
| (22/39) | (43/45) | (65/84) | (22/24) | (43/60) | |
| AFP | 53.8ce | 91.1ce | 73.8 | 84.0e | 69.5ce |
| (21/39) | (41/45) | (62/84) | (21/25) | (41/59) | |
| AFU | 66.7e | 75.6 | 71.4 | 70.3 | 72.3 |
| (26/39) | (34/45) | (60/84) | (26/37) | (34/47) | |
| cfDNA + AFP | 71.8e | 86.7e | 79.8 | 82.4 | 78 |
| (28/39) | (39/45) | (67/84) | (28/34) | (39/50) | |
| cfDNA + AFU | 87.2 | 71.1 | 78.6 | 72.3 | 86.5 |
| (34/39) | (32/45) | (66/84) | (34/47) | (32/37) | |
| cfDNA + AFP + AFU | 89.7a | 64.4a | 76.2 | 68.6 | 87.9 |
| (35/39) | (29/45) | (64/84) | (35/51) | (29/33) |
Circulating free cell DNA is the fragmentation of nucleic acids in body fluids, and has been described in the serum of healthy persons and patients with a variety of diseases, including systemic lupus erythematosus (SLE), diabetes, cerebral stroke, rheumatoid arthritis, myocardial infarction, pulmonary embolism, preeclampsia, Whipple’s disease and malignant tumors[6-10]. Investigations have detected increased cfDNA in the plasma of patients with severe injuries, organ failure and multiple organ dysfunction syndromes. Elevated levels of cfDNA were also found in patients who had undergone organ transplantation[11]. Fetal cfDNA has been detected in maternal plasma and has been used for the prenatal diagnosis of fetal abnormalities[12]. Blunt traumatic injury and burn injury can also cause cfDNA release into the blood circulation[13]. Many studies have reported the detection of cfDNA in the serum of patients with various malignant and benign tumors. It is hypothesized that cfDNA may originate from lymphocytes and other nucleated cells in healthy individuals, however, the origin of cfDNA in malignant tumors is not fully understood[14].
Previous studies have suggested the use of serum cfDNA in the diagnosis of HCC. One study showed that cfDNA level was significantly higher in HBV carriers than in normal controls[15]. Ren et al[16] suggested that cfDNA level correlated inversely with HCC prognosis, and was attributable to HBV infection in most cases, suggesting that cfDNA may be a predictive marker for the prognosis of HBV-related HCC. In HCV-related HCC patients, serum cfDNA levels were positively correlated with the increased expression of several inflammatory cytokine genes, suggesting that serum cfDNA is associated with local inflammation. cfDNA is a potential non-invasive cancer biomarker as it is easily isolated from the circulation[17]. Positive correlations were observed between cfDNA level, aspartate aminotransferase (AST) level and the number of leukocytes and neutrophils in HCV-related HCC patients, but not in HCV carriers[18]. The level of cfDNA in serum can function as a predictor of overall survival in distant organs after curative hepatectomy in patients with HCV-related HCC[19]. In our study, serum cfDNA level was significantly higher in HCC patients than in normal controls; patients with intrahepatic and extrahepatic metastasis had a significantly higher cfDNA level; and male patients had higher cfDNA levels as compared with female patients. Therefore, it is clear that serum cfDNA level is associated with intrahepatic and extrahepatic metastasis and gender in HCC patients, but is not closely associated with age, HbsAg or liver cirrhosis. The significance of serum cfDNA in the clinical diagnosis of HCC is as follows: First, serum cfDNA level is positively correlated with the degree of differentiation and metastasis and negatively correlated with the prognosis of HCC patients, suggesting that cfDNA may be useful in monitoring the pathogenic condition of HCC. Second, serum cfDNA level is relevant to the survival of HCC patients after hepatectomy, suggesting that cfDNA may be a useful marker for surgical or non-surgical treatment decision making.
Various techniques are available to quantify serum cfDNA, some of these techniques require abstraction and purification as there is only a very small amount of cfDNA in the serum, and these technologies have failed to reach expectations. The bDNA amplification-based Alu assay is a new technology to quantify serum cfDNA directly and does not require extraction of cfDNA from the serum. The Alu family is the largest of the short interspersed elements (SINEs). Each Alu sequence is approximately 300 bp with a high homology of 70%-98%[20,21]. The sequence AGCT at the 170 degree position can be recognized and cut by the restriction enzyme Alu. Recent studies[22,23] on the Alu sequence showed that it had a unique structure and physiological function, and was associated with a variety of malignant tumors and other human diseases such as gene deletion or duplication of homologous recombination, gene rearrangement of chromosome translocation, and change in transcriptional level. The results of our study suggest that the bDNA technique may be a useful approach in the diagnosis of HCC, as amplification of the hybridization signal is needed instead of amplification of the target sequence. In addition, it is simple, economical, time saving and reproducible.
AFP is the most commonly used biomarker for HCC screening, and a continuous elevation in AFP is considered a risk factor for HCC. AFP has been widely used in HCC survey, diagnosis, evaluation of treatment effects and prediction of recurrence[24,25]. AFP can also be used to monitor treatment and prognosis. Vigilance is required when AFP is higher than 400 μg/L or rises continuously. The survival period is very short in patients with an AFP value > 500 μg/L and bilirubin > 2 mg/L. A rapid increase in AFP often suggests the occurrence of HCC metastasis. Postoperative elevation of AFP > 200 μg/L often indicates incomplete resection of HCC tissue, or the occurrence of metastasis. AFP can also be used in other cancers, such as testicular cancer, teratoma, gastric carcinoma, and pancreatic carcinoma. However, not all HCC cases are diagnosed by AFP alone, as the false negative rate is approximately 30%. False positive AFP results are also seen in some patients with non malignant conditions such as viral hepatitis and cirrhosis. The level of AFP can also increase continuously in the blood or urine of pregnant women, but usually does not exceed 300 μg/L.
AFU is a lysosomal acid hydrolase found extensively in human tissues, with higher activity in the liver and kidney. Some researchers[26] believe that increased serum AFU in HCC patients is associated with an increase in its synthesis, while others reported that hepatoma cells can produce a type of inhibitor to reduce substrate hydrolysis, resulting in substrate accumulation and a compensatory increase in AFU, which is an effective marker of HCC[27,28]. In the present study, the sensitivity of AFU, cfDNA and AFP was 66.7% 56.4% and 53.8%, respectively; the specificity of AFU was only 75.6% compared with 95.6% for cfDNA and AFP, and 91.1% for AFP; the accuracy of cfDNA, AFP and AFU was 77.4%, 73.8% and 71.4%, respectively, with no significant differences between them. Although the sensitivity of AFU was the highest of the three, its specificity was very low. In contrast, the sensitivity of cfDNA and AFP was lower than that of AFU, and their specificity was relatively high. There were no significant correlations between cfDNA, AFP and AFU. The specificity of detection of cfDNA combined with AFP or AFU was lower. However, the sensitivity improved significantly, and the accuracy was similar. The sensitivity of combined detection with the three items was slightly improved, while the specificity was decreased. Taking into account the economic burden of HCC treatment, we believe that combined detection of any two of the three biomarkers is better than combined detection of all three biomarkers in the diagnosis of HCC. In our opinion, cfDNA in combination with AFP or AFU may effectively improve the clinical detection of HCC.
In conclusion, cfDNA for HCC detection has a high sensitivity and specificity, and the clinical significance is similar to AFP, and cfDNA is not correlated with AFP and AFU. The combined detection of cfDNA with one or two markers can significantly improve the diagnostic rate of HCC.
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors worldwide. Existing biomarkers for the diagnosis of HCC are α-fetal protein (AFP), γ-glutamyltranspeptidase isoenzymesII (GGT-II), α L-fucosidase (AFU) and acidic isoferritin, of which AFP remains the most specific marker for the diagnosis of HCC. However, not all HCC patients are diagnosed using AFP alone. Circulating free cell DNA (cfDNA) has been extensively studied over the past few decades, and has a high sensitivity and specificity in the detection of HCC.
cfDNA results from the fragmentation of nucleic acids in body fluids, and has been described in the serum of healthy persons and patients with a variety of diseases. This research was designed to investigate the value of cfDNA in the diagnosis of HCC, determine correlations between cfDNA and clinicopathological features, and explore the value of combined detection of cfDNA, AFP and AFU in the clinical diagnosis of HCC.
Various techniques are available to quantify serum cfDNA, some of these techniques require abstraction and purification as there is only a very small amount of cfDNA in the serum, and these technologies have failed to reach expectations. The bDNA amplification-based Alu assay is a new technology to quantify serum cfDNA directly and does not require the extraction of cfDNA from serum.
The results of this study suggest that the quantitative analysis of cfDNA is sensitive and feasible, and the combined detection of cfDNA with AFP or AFU or both could improve the diagnostic sensitivity for HCC.
cfDNA: cfDNA results from the fragmentation of nucleic acids in body fluids, and has been described in the serum of healthy persons and patients with a variety of diseases. Alu: The Alu family of short (-300 bp) interspersed elements is one of the most successful mobile genetic elements, having increased to a copy number in excess of one million in primate genomes in the last 65 million years.
The study provides some new information. However, a brief mention regarding the importance of studying circulating DNA and its pathophysiological relevance in cancer needs to be highlighted. The authors should be encouraged to present the observed levels of DNA in healthy controls and patients. This will facilitate the readers to understand the significance of the observed differences in both groups studied. The clinical translational potential of cfDNA as a diagnostic/prognostic biomarker for HCC is not addressed in detail, and needs to be elaborated.
P- Reviewer Mishra PK S- Editor Huang XZ L- Editor A E- Editor Lu YJ
| 1. | Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907-1917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3241] [Cited by in RCA: 3282] [Article Influence: 149.2] [Reference Citation Analysis (0)] |
| 2. | Yuen MF, Cheng CC, Lauder IJ, Lam SK, Ooi CG, Lai CL. Early detection of hepatocellular carcinoma increases the chance of treatment: Hong Kong experience. Hepatology. 2000;31:330-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 308] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 3. | Yao DF, Dong ZZ, Yao M. Specific molecular markers in hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2007;6:241-247. [PubMed] |
| 4. | Mittra I, Nair NK, Mishra PK. Nucleic acids in circulation: are they harmful to the host? J Biosci. 2012;37:301-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 57] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 5. | Fluss R, Faraggi D, Reiser B. Estimation of the Youden Index and its associated cutoff point. Biom J. 2005;47:458-472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1260] [Cited by in RCA: 1749] [Article Influence: 87.5] [Reference Citation Analysis (0)] |
| 6. | Vlassov VV, Laktionov PP, Rykova EY. Circulating nucleic acids as a potential source for cancer biomarkers. Curr Mol Med. 2010;10:142-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 73] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 7. | Galeazzi M, Morozzi G, Piccini M, Chen J, Bellisai F, Fineschi S, Marcolongo R. Dosage and characterization of circulating DNA: present usage and possible applications in systemic autoimmune disorders. Autoimmun Rev. 2003;2:50-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 61] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 8. | Antonatos D, Patsilinakos S, Spanodimos S, Korkonikitas P, Tsigas D. Cell-free DNA levels as a prognostic marker in acute myocardial infarction. Ann N Y Acad Sci. 2006;1075:278-281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 85] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 9. | Sai S, Ichikawa D, Tomita H, Ikoma D, Tani N, Ikoma H, Kikuchi S, Fujiwara H, Ueda Y, Otsuji E. Quantification of plasma cell-free DNA in patients with gastric cancer. Anticancer Res. 2007;27:2747-2751. [PubMed] |
| 10. | Pathak AK, Bhutani M, Kumar S, Mohan A, Guleria R. Circulating cell-free DNA in plasma/serum of lung cancer patients as a potential screening and prognostic tool. Clin Chem. 2006;52:1833-1842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 59] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 11. | Lui YY, Woo KS, Wang AY, Yeung CK, Li PK, Chau E, Ruygrok P, Lo YM. Origin of plasma cell-free DNA after solid organ transplantation. Clin Chem. 2003;49:495-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 63] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 12. | Chiu RW, Lo YM. Recent developments in fetal DNA in maternal plasma. Ann N Y Acad Sci. 2004;1022:100-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 13. | Breitbach S, Tug S, Simon P. Circulating cell-free DNA: an up-coming molecular marker in exercise physiology. Sports Med. 2012;42:565-586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 160] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 14. | Ziegler A, Zangemeister-Wittke U, Stahel RA. Circulating DNA: a new diagnostic gold mine? Cancer Treat Rev. 2002;28:255-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 170] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 15. | Chen H, Sun LY, Zheng HQ, Zhang QF, Jin XM. Total serum DNA and DNA integrity: diagnostic value in patients with hepatitis B virus-related hepatocellular carcinoma. Pathology. 2012;44:318-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 16. | Ren N, Qin LX, Tu H, Liu YK, Zhang BH, Tang ZY. The prognostic value of circulating plasma DNA level and its allelic imbalance on chromosome 8p in patients with hepatocellular carcinoma. J Cancer Res Clin Oncol. 2006;132:399-407. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 58] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 17. | Zhou J, Shi YH, Fan J. Circulating cell-free nucleic acids: promising biomarkers of hepatocellular carcinoma. Semin Oncol. 2012;39:440-448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 18. | Iida M, Iizuka N, Sakaida I, Moribe T, Fujita N, Miura T, Tamatsukuri S, Ishitsuka H, Uchida K, Terai S. Relation between serum levels of cell-free DNA and inflammation status in hepatitis C virus-related hepatocellular carcinoma. Oncol Rep. 2008;20:761-765. [PubMed] |
| 19. | Tokuhisa Y, Iizuka N, Sakaida I, Moribe T, Fujita N, Miura T, Tamatsukuri S, Ishitsuka H, Uchida K, Terai S. Circulating cell-free DNA as a predictive marker for distant metastasis of hepatitis C virus-related hepatocellular carcinoma. Br J Cancer. 2007;97:1399-1403. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 83] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 20. | Gu Z, Wang H, Nekrutenko A, Li WH. Densities, length proportions, and other distributional features of repetitive sequences in the human genome estimated from 430 megabases of genomic sequence. Gene. 2000;259:81-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 61] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 21. | Deininger P. Alu elements: know the SINEs. Genome Biol. 2011;12:236. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 363] [Cited by in RCA: 454] [Article Influence: 32.4] [Reference Citation Analysis (0)] |
| 22. | Andreassen R. [Alu elements in the human genome]. Tidsskr Nor Laegeforen. 2004;124:2345-2349. [PubMed] |
| 23. | Qi J, Qian C, Shi W, Wu X, Jing R, Zhang L, Wang Z, Ju S. Alu-based cell-free DNA: a potential complementary biomarker for diagnosis of colorectal cancer. Clin Biochem. 2013;46:64-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 24. | Marubashi S, Nagano H, Wada H, Kobayashi S, Eguchi H, Takeda Y, Tanemura M, Umeshita K, Doki Y, Mori M. Clinical significance of alpha-fetoprotein mRNA in peripheral blood in liver resection for hepatocellular carcinoma. Ann Surg Oncol. 2011;18:2200-2209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 25. | Ogawa C, Kudo M, Minami Y, Chung H, Kawasaki T. Tumor markers after radiofrequency ablation therapy for hepatocellular carcinoma. Hepatogastroenterology. 2008;55:1454-1457. [PubMed] |
| 26. | Deugnier Y, David V, Brissot P, Mabo P, Delamaire D, Messner M, Bourel M, Legall JY. Serum alpha-L-fucosidase: a new marker for the diagnosis of primary hepatic carcinoma? Hepatology. 1984;4:889-892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 65] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 27. | Fawzy Montaser M, Amin Sakr M, Omar Khalifa M. Alpha-L-fucosidase as a tumour marker of hepatocellular carcinoma. Arab J Gastroenterol. 2012;13:9-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 28. | Malaguarnera G, Giordano M, Paladina I, Berretta M, Cappellani A, Malaguarnera M. Serum markers of hepatocellular carcinoma. Dig Dis Sci. 2010;55:2744-2755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 141] [Article Influence: 9.4] [Reference Citation Analysis (0)] |