Published online May 21, 2013. doi: 10.3748/wjg.v19.i19.2913
Revised: April 2, 2013
Accepted: April 9, 2013
Published online: May 21, 2013
AIM: To investigate the association of p42.3 expression with clinicopathological characteristics and the biological function of p42.3 in human hepatocellular carcinoma (HCC).
METHODS: We used reverse transcription-polymerase chain reaction (RT-PCR), quantitative real-time RT-PCR and western blotting to detect p42.3 mRNA and protein expression in hepatic cell lines. We examined primary HCC samples and matched adjacent normal tissue by immunohistochemistry to investigate the correlation between p42.3 expression and clinicopathological features. HepG2 cells were transfected with a pIRES2-EGFP-p42.3 expression vector to examine the function of the p42.3 gene. Transfected cells were analyzed for their viability and malignant transformation abilities by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay, colony formation assay, and tumorigenicity assay in nude mice.
RESULTS: p42.3 is differentially expressed in primary HCC tumors and cell lines. Approximately 69.6% (96/138) of cells were p42.3-positive in hepatic tumor tissues, while 30.7% (35/114) were p42.3-positive in tumor-adjacent normal tissues. Clinicopathological characteristics of the HCC specimens revealed a significant correlation between p42.3 expression and tumor differentiation (P = 0.031). However, p42.3 positivity was not related to tumor tumor-node-metastasis classification, hepatitis B virus status, or hepatoma type. Regarding p42.3 overexpression in stably transfected HepG2 cells, we discovered significant enhancement of cancer cell growth and colony formation in vitro, and significantly enhanced tumorigenicity in nude mice. Western blot analysis of cell cycle proteins revealed that enhanced p42.3 levels promote upregulation of proliferating cell nuclear antigen, cyclin B1 and mitotic arrest deficient 2.
CONCLUSION: p42.3 promotes tumorigenicity and tumor growth in HCC and may be a potential target for future clinical cancer therapeutics.
Core tip: p42.3 is a novel tumor-specific and mitosis phase-dependent expression gene. It is believed to be involved in tumorigenesis in gastric and colorectal cancer. To the best of our knowledge, this is the first study to investigate the expression and function of p42.3 in hepatocellular carcinoma (HCC). We found that p42.3 promotes tumorigenicity and tumor growth in HepG2 cells and is overexpressed in HCC. These results suggest that p42.3 may act as a novel tumor biomarker and aid in the development of improved therapeutic strategies.
- Citation: Sun W, Dong WW, Mao LL, Li WM, Cui JT, Xing R, Lu YY. Overexpression of p42.3 promotes cell growth and tumorigenicity in hepatocellular carcinoma. World J Gastroenterol 2013; 19(19): 2913-2920
- URL: https://www.wjgnet.com/1007-9327/full/v19/i19/2913.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i19.2913
Hepatocellular carcinoma (HCC) is a major world health problem due to its high incidence and fatality rate. The annual number of new HCC cases worldwide is over one million, making it the 5th most common cancer and the 3rd leading cause of cancer-related deaths[1], accounting for more than 1 million deaths annually[2]. Despite improvements in monitoring and clinical treatment strategies, HCC prognosis remains poor[3,4]. Discovering novel biomarkers that correlate with HCC development or progression may present opportunities to reduce the severity of this disease through early and novel therapeutic interventions.
In our previous research, we cloned the full-length cDNA of the p42.3 gene by using mRNA differential display in a synchronized gastric cancer (GC) cell lines. We found that p42.3 expression is frequently upregulated in primary tumors and embryonic tissues but not in normal tissues from adult organs. Moreover, stable silencing of p42.3 in BGC823 cells suppresses tumorigenicity and cell proliferation with accumulation of cells at G2/M stage of the cell cycle[5]. In addition, Jung et al[6] reported that the expression of p42.3 mRNA was significantly elevated in colorectal cancer (CRC) tissues compared to normal tissues. All these data indicate that p42.3 plays an important role in tumorigenesis, suggesting that it may be a potential tumor biomarker. In order to elucidate the role of p42.3 in tumorigenesis, we characterized p42.3 expression and validated its biologic significance in HCC.
HCC specimens (n = 138) were collected from 98 men and 40 women (age, 31-74 years; mean ± SD, 52.6 ± 8.7 years) who were inpatients at Beijing Cancer Hospital, Beijing, China, from January 2006 to September 2009. Patient data are shown in Table 1. All patients underwent a radical resection with curative intent and had sufficient clinical information available. No patients had received chemotherapy or radiation therapy. Moreover, 114 adjacent normal hepatic tissues (at least 5 cm distant from the tumor edge) were also collected from HCC patients. Tumor stage was classified according to the American Joint Committee on Cancer tumor-node-metastasis (TNM) classification. The investigation project and its informed consent have been examined and certified by the Ethics Committee of Beijing Cancer Hospital.
| Tissues parameters | No. of cases | Positive | Negative | P value |
| Gender | NS | |||
| Male | 98 (71.0) | 42 (42.9) | 56 (57.1) | |
| Female | 40 (29.0) | 23 (58.0) | 17 (42.0) | |
| Age at diagnosis (yr) | NS | |||
| < 60 | 117 (84.8) | 52 (44.4) | 63 (55.6) | |
| ≥ 60 | 21 (15.2) | 12 (57.1) | 9 (42.9) | |
| Carcinoma and adjacent tissue | 0.0008 | |||
| Carcinoma tissue | 138 (54.8) | 96 (69.6) | 42 (30.4) | |
| Adjacent tissue | 114 (45.2) | 35 (30.7) | 79 (69.3) | |
| Degree of differentiation | 0.031 | |||
| Well | 42 (30.4) | 11 (26.2) | 31 (73.8) | |
| Moderate | 87 (63.0) | 39 (44.8) | 48 (55.2) | |
| Poor | 9 (6.5) | 6 (66.7) | 3 (33.3) | |
| TNM classification | NS | |||
| Stage I/II | 101 (73.2) | 43 (42.6) | 58 (57.4) | |
| Stage III/IV | 37 (26.8) | 19 (51.4) | 18 (48.6) | |
| HBV | NS | |||
| Negative | 41 (29.7) | 15 (36.6) | 26 (63.4) | |
| Positive | 97 (70.3) | 47 (48.5) | 50 (51.5) | |
| Type of hepatoma | NS | |||
| Nodular | 94 (68.1) | 44 (46.8) | 50 (53.2) | |
| Massive | 35 (25.4) | 13 (37.1) | 22 (62.9) | |
| Diffuse | 9 (6.5) | 5 (55.6) | 4 (44.4) | |
The hepatic tissue microarray was constructed using a tissue array instrument as previously described[7]. For immunohistochemistry studies, sections were deparaffinized and rehydrated. Endogenous peroxidase activity was blocked by incubation in 3% H2O2 solution for 10 min at room temperature. After blocking with 5% skim milk, sections were incubated with specific murine p42.3 mAb (1:1000, our lab) at 4 °C overnight, followed by the incubation with the peroxidase-based EnVision TM kit (Dako Cytomation, Cambridgeshire, United Kingdom) for 30 min at room temperature. The reaction product was visualized with diaminobenzidine (DAB, Dako, Glostrup, Denmark) for 5 min at room temperature. Sections were counterstained with hematoxylin.
Purified IgG from normal mouse sera was used as a negative control. The number of tumor cells or normal hepatic cells was evaluated by two independent pathologists. A specimen with more than 20% immunostained cells was classified as a positive case.
The 6 human HCC cell lines MHCC97L, MHCC97M3, BEL7402, Huh7, HepG2, and SMMC7721 and the immortal human hepatocyte line HL7702 were routinely maintained as previously described[8]. HL7702 was cultured in Roswell Park Memorial Institute medium (RPMI 1640; Gibco, Grand Island, NY, United States), supplemented with 20% fetal bovine serum (FBS; Gibco). BEL7402 and SMMC7721 cell lines were cultured in RPMI 1640 medium supplemented with 10% FBS. The remaining cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% FBS. All media contained 100 units/mL penicillin and 100 μg/mL streptomycin. All cell lines were maintained at 37 °C in 5% CO2.
Total RNA was extracted from cell lines using TRIzol (Qiagen, United States). The prepared RNA (5 μg) was mixed with oligo-dT primers and reverse-transcribed with moloney murine leukemia virus reverse transcriptase (Promega, United States) for 60 min at 37 °C, followed by polymerase chain reaction (PCR) amplification with specific primers for p42.3 (forward: 5′-TGGACTGCGGCCTGCTGAA-3′; reverse: 5′-ACTCCATCGCTGTGTTTCAAT-3′). PCR amplification was performed in 20 μL using a thermocycler (Biometra, Germany) with the following PCR program: pre-denaturation for 5 min at 94 °C, denaturation for 45 s at 94 °C, annealing for 45 s at 61 °C, extension for 45 s at 72 °C, and a final elongation at 72 °C for 10 min. β-Actin served as an internal positive control (forward: 5′-TCACCCACACTGTGCCCATCTACGA-3′; reverse: 5′-CAGCGGAACCGCTCATTGCCAATGG-3′). PCR was performed for 24 or 32 cycles (β-actin 24 cycles; p42.3 32 cycles). PCR products were separated by electrophoresis on a 1.5% agarose gel. Quantitative real-time reverse transcription-PCR (Q-RT-PCR) using SYBR-Green Master PCR mix (Applied Biosystems, Carls-bad, CA) was performed in triplicate (p42.3 forward: 5′-CCTGGCATCTTTACTGGACTGGA-3′; p42.3 reverse: 5′-GTGCCAGCCTGTCTCACATTTC-3′). Quantification was normalized to the endogenous control β-actin (forward: 5′-TTAGTTGCGTTACACCCTTTC-3′; reverse: 5′-ACCTTCACCGTTCCAGTTT-3′).
Cell lysates were prepared by incubating cells at 4 °C for 1 h in a buffer containing 50 mmol/L Tris-HCl, pH 8.0, 0.5% Nonidet P-40, 2 mmol/L dithiothreitol, 5 mmol/L ethylene diamine tetraacetic acid, 100 mmol/L NaCl, and 2 mmol/L phenylmethylsulfonyl fluoride. Equal amounts of protein were electrophoresed on a 12% sodium dodecylsulfate polyacrylamide gel and transferred to a polyvinylidene difluoride membrane using standard techniques. We used four specific antibodies obtained from Santa Cruz Biotechnology: proliferating cell nuclear antigen (PCNA) (diluted 1:300; F-2), cyclin B1 (diluted 1:500; H-433), cell division cycle 25 A (Cdc25A) (diluted 1:500; DCS-122), and cell division cycle 25 homolog C (Cdc25C) (diluted 1:500; C-20). The following specific antibodies were also used: mitotic arrest deficient 2 (MAD2) (diluted 1:1000; Ab70383; Abcam, United Kingdom), actin (diluted 1:10000, AC-15; Sigma, United States), and p42.3 (diluted 1:1000; our lab). Nonspecific binding was blocked using a 5% fat-free milk solution. Signals were detected using an enhanced chemiluminescence system (Amersham Pharmacia Biotech).
The whole coding region of p42.3 was cloned into the pIRES2-EGFP vector at the BamHI and HindIII sites. Nucleotide sequences of the subcloned cDNAs were verified by sequencing. HepG2 were selected and cultured at 60%-70% confluence in 35 mm plates. Cells were transfected with recombinant p42.3 plasmids or an empty vector using Lipofectamine 2000 (Invitrogen, Carlsbad, United States). At 48 h post-transfection, cells were seeded for 21 d in selection medium containing 400 μg/mL G418 to screen for stable clones. To confirm the transfection efficiency, RT-PCR and Western blot analysis were performed.
Stably transfected cells in were seeded (2 × 103) in duplicates into each well of a 96-well culture plate and grown in 200 μL DMEM with 5% FBS; 10 μL 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Genview, Florida, United States) (5 mg/mL) was added at 0, 24, 48, 72, 96 and 120 h. The MTT was removed after 4 h incubation; 100 μL of dimethylsulfoxide (Amresco, Cochran, United States) was added to each well, then incubated for 30 min. Absorbency was measured at 570 nm using an iMark Microplate Reader (Bio-Rad, CA, United States).
For the soft agar assay, cells (2 × 103) were trypsinized and resuspended in 4 mL of 0.3% agar in DMEM containing 10% FBS, and overlaid with 0.6% agar in 60-mm culture dishes. The dishes were incubated routinely for 21 d. Colonies were stained with 0.2% p-iodonitrotetrazolium violet, then photographed and counted.
Stably transfected cells were washed twice and resuspended in 1 × Hank’s buffer at a concentration of 1 × 106 cells/mL. A 100-μL cell suspension of HepG2-p42.3 was then injected subcutaneously into the left dorsal flank of 10, 4-wk-old female nude mice. As a control, the right side was inoculated with HepG2-vector. Tumor diameters were checked every 3 d, and tumor volume was calculated according to ab2/2 (a > b). Tumor specimens were collected at 15 d after injections and split. Immunohistochemistry (IHC) analysis was used to detect p42.3 protein expression. Three independent experiments were performed and yielded similar results.
To evaluate the possible differences of p42.3 expression in different hepatic specimens, we performed Pearson’s χ2 test. The Student’s two-sided t-test was used to compare test and control sample values in MTT assay, soft agar colony formation assay and tumorigenicity assay. All statistical analyses were carried out using the SPSS statistical software package 16.0 (SPSS Inc., United States). P values < 0.05 were considered statistically significant.
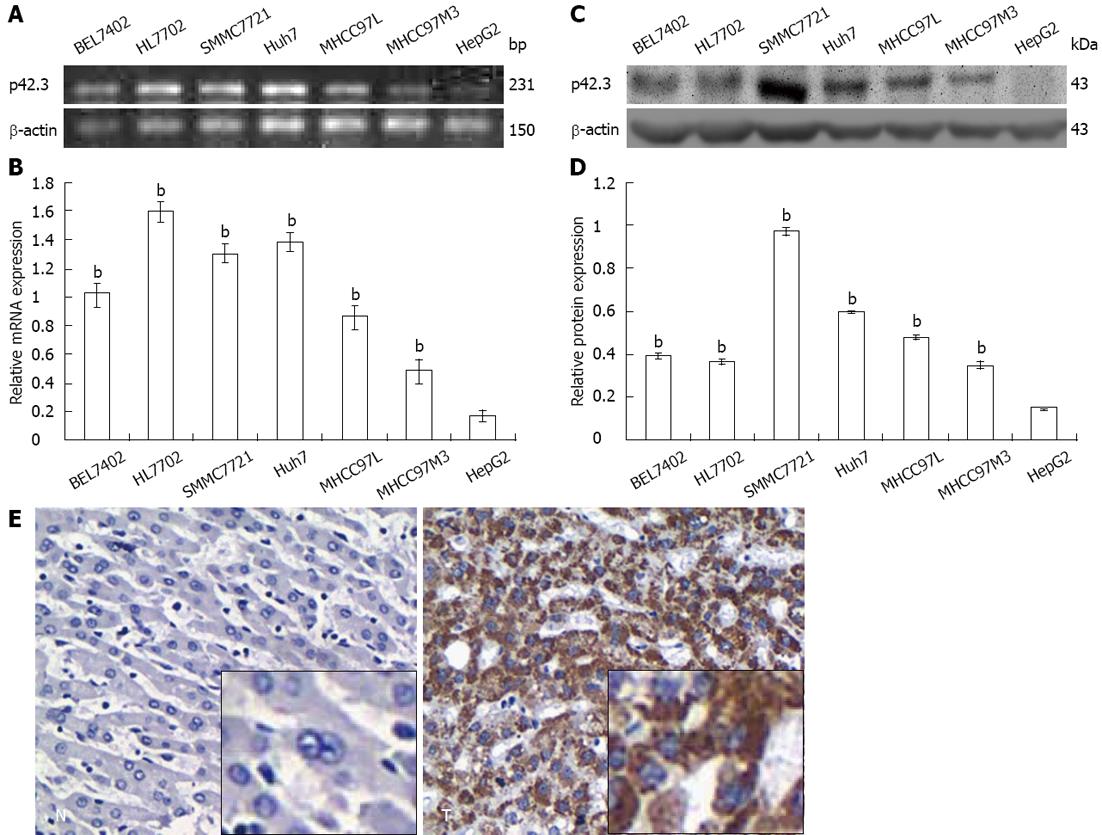
p42.3 mRNA and protein expression were examined in 6 human HCC cell lines and the immortal human hepatocyte HL7702. RT-PCR and Q-RT-PCR showed that p42.3 mRNA was expressed in all of 7 cell lines (7/7, 100%), and the lowest expression was found in HepG2 cells (Figure 1A and B). Consistent with mRNA expression levels, p42.3 protein was expressed at high levels in all cell lines except HepG2 cells (6/7, 85.7%) (Figure 1C and D). Thus, we confirmed that the HepG2 cell line is a p42.3-deficient line and could therefore be used as a model to investigate p42.3 protein function.
To characterize p42.3 expression in HCC specimens, IHC was performed on tumor tissues and tumor-adjacent normal tissues. We found p42.3 protein was detected in 69.6% (96/138) of hepatic tumor tissues. However, p42.3 expression was less apparent, with significantly less positive cells (30.7%, 35/114) in tumor-adjacent normal tissues (P < 0.001, Table 1 and Figure 1E). The results indicate that p42.3 protein is highly expressed in primary HCC tissues rather than tumor-adjacent normal tissues. Analysis of the clinicopathological characteristics of the 138 HCC specimens revealed a significant correlation between p42.3 expression and tumor differentiation (P = 0.031, Table 1). However, we found no relationship between p42.3 positivity and tumor TNM classification, hepatitis B virus status, or type of hepatoma.
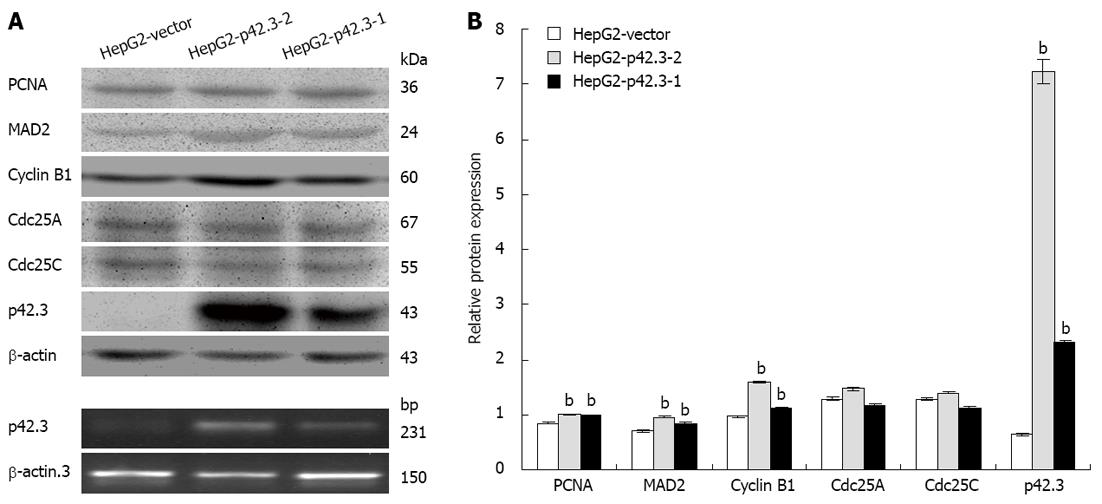
To examine the gene function of p42.3 overexpression on HCC cells, we stably transfected the pIRES2-EGFP-p42.3 expression vector into HepG2 cells. A cell line that stably expresses p42.3 (HepG2-p42.3) was generated and analyzed by western blotting. As shown in Figure 2, p42.3 protein was not detected in cells stably transfected with the empty vector. However, p42.3 protein was significantly increased in the p42.3 overexpressing cells, HepG2-p42.3-1 and HepG2-p42.3-2. These results indicated that the eukaryotic vector for p42.3 used in this study sufficiently upregulates p42.3 expression in HepG2 cells.
Since p42.3 is a novel cell cycle-dependent protein, we investigated cyclin B1 and other M phase-related proteins in p42.3-expressing HepG2 cells and control cells (HepG2-vector). We found that p42.3 expression resulted in a significant upregulation in PCNA, cyclin B1 and MAD2 protein levels. However, Cdc25A and Cdc25C protein levels only slightly changed with p42.3 expression (Figure 2).
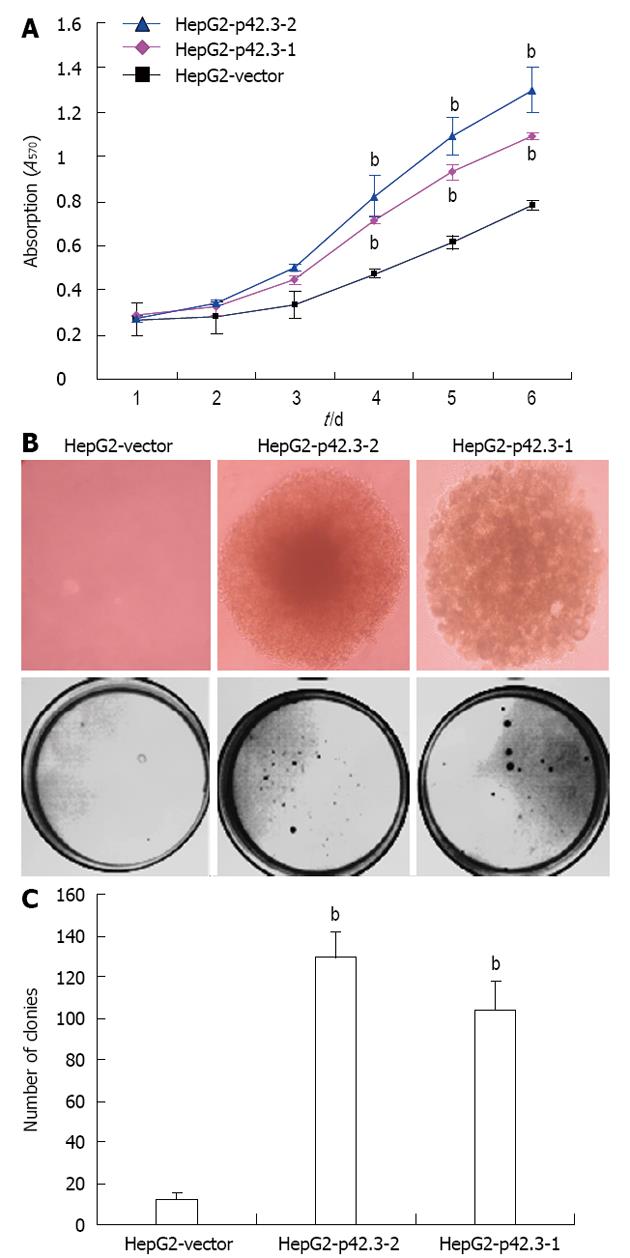
The effects of p42.3 overexpression on the viability of HepG2 cells were measured using an MTT colorimetric assay. We found that transfection with pIRES2-EGFP-p42.3 promotes HepG2 cell growth. A stable single clone of HepG2-p42.3-1 and HepG2-p42.3-2 cells grew much faster over a 6-day period when compared to parental HepG2-vector cells, indicating that p42.3 may confer a strong growth capability in HepG2 cells (P < 0.01, Figure 3A).
The colony formation assay was used to evaluate the ability for anchorage-independent growth of cells in soft agar medium. Our data showed a significant increase in HepG2-p42.3-1 and HepG2-p42.3-2 colony formation in both number and size; however, the HepG2-vector cells only formed a few small colonies (P < 0.01, Figure 3B and C). This suggests that p42.3 confers anchorage-independent growth to cells.
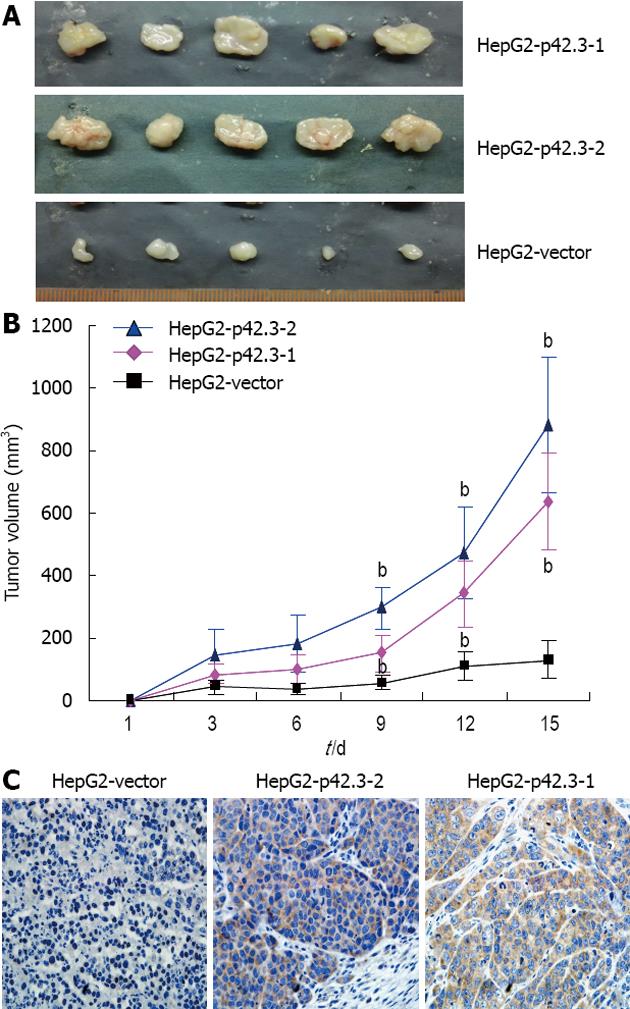
We tested p42.3 tumorigenicity in nude mice. HepG2-p42.3 cells were injected subcutaneously into the left dorsal flank of female nude mice (BALB/c), the right side was injected with HepG2-vector cells as a control. Of the 5 animals injected with HepG2-p42.3-1 or HepG2-p42.3-2 cells, tumors appeared faster and were larger than in the controls (HepG2-vector) (P < 0.01, Figure 4A and B). After the animals were sacrificed, the xenografts were removed and collected for immunohistochemistry analysis. The results showed that p42.3 protein was found in all HepG2-p42.3 xenografts, but that p42.3 protein was not found in HepG2-vector xenografts (Figure 4C). These results further confirmed that the p42.3 overexpression promotes tumorigenicity of HepG2 cells.
p42.3 is a highly conserved mammalian gene and strong G2 induction[5,9-11]. Moreover, p42.3 is involved in Chromosome segregation[12], it may play a key role in tumorigenicity. Our previous studies have shown that p42.3 was overexpressed in GC tissue and its expression is cell cycle dependent in the BGC823 cell line, expression peaked at early G1 phase[5,13]. Additionally, reduced proliferation and tumorigenic properties were detected in the BGC823 cell line that lacked p42.3[5], suggesting that p42.3 is involved in gastric carcinogenesis. While most studies have focused on the roles of p42.3 in GC development[5,11,14], the roles of p42.3 in other cancer remain to be elucidated. Therefore, we characterized p42.3 expression in HCC tissues from patients. Moreover, we investigated p42.3 functions and potential mechanisms of action in HepG2 cells.
In 7 human HCC cell lines, consistent with the mRNA expression, p42.3 protein was highly expressed with the exception of the HepG2 line. In concert with our previous study in GC cells, we found that the p42.3 gene was highly expressed in the majority of the tested tumor cell lines. This suggests that the p42.3 gene is overexpressed in tumor cells. In previous research, data showed that the p42.3 gene is closely related to GC and CRC[5,6]. Similarly, our current data revealed that the p42.3 protein was expressed in 64.7% of hepatic tumor tissues compared to only 35.3% in tumor-adjacent normal tissues. The clinical p42.3 data in GC, CRC and HCC tissues suggest that upregulation of p42.3 may be a common feature in a variety of tumors.
Our results further support the hypothesis that p42.3 might stimulate cellular viability and malignant transformation since overexpression of p42.3, by stable transfection of the pIRES2-EGFP-p42.3 into HepG2 cells, significantly promotes cancer cell growth by MTT colorimetry and colony formation in vitro, and significantly induced tumorigenicity in nude mice. Thus, these findings provide evidence that p42.3 plays an important role in tumorigenesis. Therefore, we investigated the molecular mechanism responsible for stimulating cell growth and malignant transformation. Since the expression of p42.3 is cell cycle dependent and G2/M checkpoint abrogated[5,11,13,15], we analyzed the effects of elevated p42.3 levels on a series of cell cycle proteins. Our results indicate that p42.3 expression significantly upregulates PCNA, cyclin B1 and MAD2 protein levels. However, Cdc25A and Cdc25C protein levels hardly change with p42.3 expression. Cyclin B1 is one of the key genes involved in M-phase regulation[16-20]. Together with Cdc2, cyclin B1 forms a complex that controls M-phase entry and exit[17,21]. Cyclin B1 can promote the G2-M transition, and even leads to a loss cell proliferation control, thus leading to malignant transformation[22,23]. The alteration of cyclin B1 protein levels shown here is consistent with our previous study[5], it shows p42.3 may a regulator of cyclin B1. Furthermore, MAD2 is a component of the mitotic spindle assembly checkpoint that prevents the onset of anaphase until all chromosomes are properly aligned at the metaphase plate[24-29], and MAD2 is involved in mediating the upregulation of cyclin B1 proteins[30,31]. Our results showed that the significant increase in expression of cyclin B1 and Mad2 may correlation with G2/M checkpoint abrogation. On the other hand, though Cdc25 phosphatases involved in cell cycle checkpoints as key regulators of normal cell division and the cell’s response to DNA damage[32-35], our data did not reveal any obvious change in total Cdc25A and Cdc25C levels with p42.3 overexpression, Cdc25 phosphatases may have no role in the cell’s response to induced p42.3 expression.
In summary, the data obtained in this study demonstrate that p42.3 protein is upregulated in primary HCC tissues but not tumor-adjacent normal tissues, and that cyclin B1 might be responsible for the cellular proliferation and malignant transformation induced by p42.3. These insights may help to identify p42.3 as a potential target for improved cancer therapies or as a diagnostic marker in clinical cancer treatment.
Hepatocellular carcinoma (HCC) is a major world health problem due to its high incidence and fatality rate. Discovering novel biomarkers that correlate with HCC may present opportunities to reduce the severity of this disease. As a novel tumor-specific and mitosis phase-dependent expression gene, p42.3 is involved in cell proliferation and tumorigenesis in gastric cancer (GC). Previous data also indicate that p42.3 expression is significantly elevated in GC and colorectal cancer (CRC), thus raising the possibility that it may act as a potential tumor biomarker.
p42.3 is a novel tumor-specific and mitosis phase-dependent expression gene. It is involved in tumorigenesis in GC and CRC. However, the research concerning p42.3 in HCC is lacking. In this study, the authors investigate p42.3 expression and function in primary HCC. These results suggest that p42.3 may act as a novel disease biomarker and aid in the development of improved therapy strategies.
Recent reports have highlighted that p42.3 is involved in GC and CRC. This is the first study to investigate the expression and function of p42.3 in HCC. The authors found that p42.3 promotes tumorigenicity and tumor growth in HepG2 cells and is overexpressed in HCC.
In understanding the expression and function of p42.3 in HCC, this study may represent a future strategy as a therapeutic target and/or improve clinical cancer HCC treatment.
The authors examined the expression of p42.3 and its function in HCC. These data revealed that p42.3 was increased in HCC and in all HCC cells with the exception of HepG2 cells. Moreover, p42.3 expression was correlated with tumor differentiation. p42.3 promotes tumorigenicity and tumor growth in HCC; therefore, it may be used as a potential target to improve the clinical treatment of HCC.
P- Reviewers Butterworth J, Grassi G, Yu DY S- Editor Wen LL L- Editor A E- Editor Xiong L
| 1. | Yang JD, Roberts LR. Epidemiology and management of hepatocellular carcinoma. Infect Dis Clin North Am. 2010;24:899-919, viii. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 152] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 2. | El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557-2576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3846] [Cited by in RCA: 4262] [Article Influence: 236.8] [Reference Citation Analysis (2)] |
| 3. | Yang Y, Nagano H, Ota H, Morimoto O, Nakamura M, Wada H, Noda T, Damdinsuren B, Marubashi S, Miyamoto A. Patterns and clinicopathologic features of extrahepatic recurrence of hepatocellular carcinoma after curative resection. Surgery. 2007;141:196-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 176] [Article Influence: 9.8] [Reference Citation Analysis (1)] |
| 4. | Tralhão JG, Dagher I, Lino T, Roudié J, Franco D. Treatment of tumour recurrence after resection of hepatocellular carcinoma. Analysis of 97 consecutive patients. Eur J Surg Oncol. 2007;33:746-751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 60] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 5. | Xu X, Li W, Fan X, Liang Y, Zhao M, Zhang J, Liang Y, Tong W, Wang J, Yang W. Identification and characterization of a novel p42.3 gene as tumor-specific and mitosis phase-dependent expression in gastric cancer. Oncogene. 2007;26:7371-7379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 6. | Jung Y, Lee S, Choi HS, Kim SN, Lee E, Shin Y, Seo J, Kim B, Jung Y, Kim WK. Clinical validation of colorectal cancer biomarkers identified from bioinformatics analysis of public expression data. Clin Cancer Res. 2011;17:700-709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 74] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 7. | Kang B, Guo RF, Tan XH, Zhao M, Tang ZB, Lu YY. Expression status of ataxia-telangiectasia-mutated gene correlated with prognosis in advanced gastric cancer. Mutat Res. 2008;638:17-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 48] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 8. | Dong WW, Mou Q, Chen J, Cui JT, Li WM, Xiao WH. Differential expression of Rab27A/B correlates with clinical outcome in hepatocellular carcinoma. World J Gastroenterol. 2012;18:1806-1813. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 67] [Cited by in RCA: 70] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 9. | Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA. 2002;99:16899-16903. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1288] [Cited by in RCA: 1366] [Article Influence: 59.4] [Reference Citation Analysis (0)] |
| 10. | Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977-2000. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 801] [Cited by in RCA: 1133] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 11. | Segal E, Friedman N, Koller D, Regev A. A module map showing conditional activity of expression modules in cancer. Nat Genet. 2004;36:1090-1098. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 517] [Cited by in RCA: 497] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 12. | Hutchins JR, Toyoda Y, Hegemann B, Poser I, Hériché JK, Sykora MM, Augsburg M, Hudecz O, Buschhorn BA, Bulkescher J. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593-599. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 444] [Cited by in RCA: 421] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 13. | Mao L, Sun W, Li W, Cui J, Zhang J, Xing R, Lu Y. Cell cycle-dependent expression of p42.3 promotes mitotic progression in malignant transformed cells. Mol Carcinog. 2012;[Epub ahead of print]. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 14. | Cui Y, Su WY, Xing J, Wang YC, Wang P, Chen XY, Shen ZY, Cao H, Lu YY, Fang JY. MiR-29a inhibits cell proliferation and induces cell cycle arrest through the downregulation of p42.3 in human gastric cancer. PLoS One. 2011;6:e25872. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 84] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 15. | Zhang J, Lu C, Shang Z, Xing R, Shi L, Lv Y. p42.3 gene expression in gastric cancer cell and its protein regulatory network analysis. Theor Biol Med Model. 2012;9:53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 16. | Pines J. Mitosis: a matter of getting rid of the right protein at the right time. Trends Cell Biol. 2006;16:55-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 206] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 17. | Gavet O, Pines J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev Cell. 2010;18:533-543. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 703] [Cited by in RCA: 659] [Article Influence: 43.9] [Reference Citation Analysis (0)] |
| 18. | Sakai K, Barnitz RA, Chaigne-Delalande B, Bidère N, Lenardo MJ. Human immunodeficiency virus type 1 Vif causes dysfunction of Cdk1 and CyclinB1: implications for cell cycle arrest. Virol J. 2011;8:219. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 19. | Chen H, Huang Q, Dong J, Zhai DZ, Wang AD, Lan Q. Overexpression of CDC2/CyclinB1 in gliomas, and CDC2 depletion inhibits proliferation of human glioma cells in vitro and in vivo. BMC Cancer. 2008;8:29. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 45] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 20. | Beauman SR, Campos B, Kaetzel MA, Dedman JR. CyclinB1 expression is elevated and mitosis is delayed in HeLa cells expressing autonomous CaMKII. Cell Signal. 2003;15:1049-1057. [PubMed] |
| 21. | Ren Y, Wang Q, Shi L, Yue W, Zhang C, Lei F. Effects of maternal and dietary selenium (Se-enriched yeast) on the expression of p34(cdc2) and CyclinB1 of germ cells of their offspring in goats. Anim Reprod Sci. 2011;123:187-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 23. | Li YZ, Zhao P. [Expressions of cyclinB1, FHIT and Ki-67 in 336 gastric carcinoma patients and their clinicopathologic significance]. Zhonghua Yixue Zazhi. 2009;89:2337-2341. [PubMed] |
| 24. | Ho CY, Wong CH, Li HY. Perturbation of the chromosomal binding of RCC1, Mad2 and survivin causes spindle assembly defects and mitotic catastrophe. J Cell Biochem. 2008;105:835-846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 25. | Wu CW, Chi CW, Huang TS. Elevated level of spindle checkprotein MAD2 correlates with cellular mitotic arrest, but not with aneuploidy and clinicopathological characteristics in gastric cancer. World J Gastroenterol. 2004;10:3240-3244. [PubMed] |
| 26. | Saitoh S, Ishii K, Kobayashi Y, Takahashi K. Spindle checkpoint signaling requires the mis6 kinetochore subcomplex, which interacts with mad2 and mitotic spindles. Mol Biol Cell. 2005;16:3666-3677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 27. | Kim HS, Park KH, Kim SA, Wen J, Park SW, Park B, Gham CW, Hyung WJ, Noh SH, Kim HK. Frequent mutations of human Mad2, but not Bub1, in gastric cancers cause defective mitotic spindle checkpoint. Mutat Res. 2005;578:187-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 28. | Yu H. Structural activation of Mad2 in the mitotic spindle checkpoint: the two-state Mad2 model versus the Mad2 template model. J Cell Biol. 2006;173:153-157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 85] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 29. | Lee SH, Sterling H, Burlingame A, McCormick F. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 2008;22:2926-2931. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 119] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 30. | Choi HJ, Fukui M, Zhu BT. Role of cyclin B1/Cdc2 up-regulation in the development of mitotic prometaphase arrest in human breast cancer cells treated with nocodazole. PLoS One. 2011;6:e24312. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 68] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 31. | Mukherjee S, Manna S, Pal D, Mukherjee P, Panda CK. Sequential loss of cell cycle checkpoint control contributes to malignant transformation of murine embryonic fibroblasts induced by 20-methylcholanthrene. J Cell Physiol. 2010;224:49-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 32. | Boutros R, Dozier C, Ducommun B. The when and wheres of CDC25 phosphatases. Curr Opin Cell Biol. 2006;18:185-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 299] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 33. | Arantes GM. Flexibility and inhibitor binding in cdc25 phosphatases. Proteins. 2010;78:3017-3032. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 34. | Albert H, Santos S, Battaglia E, Brito M, Monteiro C, Bagrel D. Differential expression of CDC25 phosphatases splice variants in human breast cancer cells. Clin Chem Lab Med. 2011;49:1707-1714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 24] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 35. | Aressy B, Ducommun B. Cell cycle control by the CDC25 phosphatases. Anticancer Agents Med Chem. 2008;8:818-824. [PubMed] |