Published online Aug 7, 2012. doi: 10.3748/wjg.v18.i29.3849
Revised: May 2, 2012
Accepted: May 5, 2012
Published online: August 7, 2012
AIM: To investigate the reciprocal modulation between microRNA (miRNA) and DNA methylation via exploring the correlation between miR-373 and methyl-CpG-binding domain protein (MBD)2.
METHODS: MiR-373 expression was examined using the TaqMan miRNA assay. Methylation of miR-373 was investigated using methylation-specific polymerase chain reaction, and recruitment of methyl binding proteins was studied using the chromatin immunoprecipitation assay. Mutation analysis was conducted using the QuikChange™ Site-Directed Mutagenesis kit. The activity of miR-373 gene promoter constructs and targeting at MBD2-three prime untranslated region (3’UTR) by miR-373 were evaluated by a dual-luciferase reporter gene assay.
RESULTS: In hilar cholangiocarcinoma, miR-373 decreased and was closely associated with poor cell differentiation, advanced clinical stage, and shorter survival. The promoter-associated CpG island of miR-373 gene was hypermethylated and inhibited expression of miR-373. MBD2 was up-regulated and enriched at the promoter-associated CpG island of miR-373. Methylation-mediated suppression of miR-373 required MBD2 enrichment at the promoter-associated CpG island, and miR-373 negatively regulated MBD2 expression through targeting the 3’UTR.
CONCLUSION: MiR-373 behaves as a direct transcriptional target and negative regulator of MBD2 activity through a feedback loop of CpG island methylation.
- Citation: Chen YJ, Luo J, Yang GY, Yang K, Wen SQ, Zou SQ. Mutual regulation between microRNA-373 and methyl-CpG-binding domain protein 2 in hilar cholangiocarcinoma. World J Gastroenterol 2012; 18(29): 3849-3861
- URL: https://www.wjgnet.com/1007-9327/full/v18/i29/3849.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i29.3849
Hilar cholangiocarcinoma, known as Klatskin tumor[1], is an uncommon cancer with an incidence of 0.01% to 0.2% per year[2]. Although it is relatively rare, hilar cholangiocarcinoma displays a highly aggressive malignancy and is considered to be an incurable and rapidly lethal disease despite recent progress in diagnostic and therapeutic techniques. The 5-year overall survival after curative resection ranges from 15% to 35%, but the 10-year survival is almost zero[3]. Patients with inoperable, recurrent, or metastatic disease can only be treated with palliative therapy, such as endoscopic, percutaneous biliary drainage in combination with radiotherapy and chemotherapy. However, hilar cholangiocarcinoma is not sensitive to chemotherapy and radiotherapy, and the median survival rate of those cases is only about 5.8 mo[4].
Studies have established a direct link between aberrant DNA methylation and regulation of gene expression in human cancer[5]. Once a given sequence becomes methylated, it can directly repress transcription by either impeding recognition of transcriptional activators to DNA sequences[6], or by recruiting methyl-CpG binding domain proteins (MBPs) to modify chromatin compaction and control gene silencing[7]. The MBP family consists of five isoforms, including Mecp2, methyl-CpG-binding domain protein (MBD)1, MBD2, MBD3, and MBD4. With the exception of MBD4, which is primarily a thymine glycosylase involved in DNA repair[8], all MBPs are implicated in transcriptional repression mediated by DNA methylation. Mecp2, MBD1, and MBD2 have been demonstrated to be involved in methylation-based gene repression and also affect chromatin structure[9-11]. MBD3 lacks a functional MBD, but is an integral subunit of the histone deacetylase Mi2-NuRD complex that is recruited through MBD2[12].
MicroRNAs (miRNAs) are non-coding, single-stranded RNAs of 18 to 24 nucleotides in length that constitute a novel class of gene regulators[13]. In general, miRNAs negatively regulate gene expression by targeting the three prime untranslated region (3’UTR), which consequently triggers mRNA degradation or translational suppression[14,15] on the basis of complementary value. miRNAs have recently been shown to play important roles in cancers, as more than 50% of miRNA genes reside in cancer-associated genomic regions, and their expression has been found to be dysregulated in various cancers[16]. Depending on the target genes, miRNAs can function as tumor suppressor genes or oncogenes[17].
Mature miRNAs are transcribed from miRNA genes by RNA polymerase II. Hence, the expression of miRNAs share the same genetic and epigenetic regulation of gene function including methylation[18]. Although only subsets of miRNA genes harbor CpG islands in their promoter regions or are embedded in CpG islands, DNA methylation-mediated down-regulation of miRNAs have been reported by a number of groups[19]. Moreover, miRNA interference with DNA methylation through DNA methyltransferases (DNMTs) 3a, 3b, and DNMT1, have been observed[20-22]. These results suggest that miRNA and DNA methylation regulate one another. However, the literature has only revealed a one-way effect of miRNA on DNA methylation or miRNA modification by promoter methylation[23]. We speculate that a particular miRNA may act as a bidirectional regulator by not only impacting DNA methylation, but also via regulation by methylation itself.
In this study, we demonstrate that miR-373 functions as a negative regulator of MBD2 by targeting the 3’UTR. Inversely, miR-373 is restrained by MBD2 enrichment at the methylated promoter-associated CpG island. We functionally demonstrate the fact that miR-373 serves as a one directional transcriptional target and negative regulator of MBD2 through a feedback loop of CpG methylation in hilar cholangiocarcinoma.
A total of 48 patients with both tumor and normal bile duct tissues, which were successfully obtained from operations conducted from January 2005 to December 2008 at Tongji Hospital in the Tongji Medical College of the Huazhong University of Science and Technology (China), were used in this study. The fresh tissues were harvested immediately after surgery, washed twice with chilled phosphate buffered saline, and immediately stored in liquid nitrogen and at -80 °C in our tissue bank until further use. The detailed clinical data of these patients is provided in Table 1. Written informed consent was obtained from each patient before sample collection. Ethical approval was obtained from the Cancer Center Research Ethics Committee of Tongji Medical College and Hospital.
| Clinicopathological features | n | ΔCtvalue of miR-373 | P value |
| Age (yr) | |||
| < 60 | 19 | 27.69 ± 3.76 | 0.059 |
| ≥ 60 | 29 | 25.65 ± 4.35 | |
| Gender | |||
| Male | 33 | 24.43 ± 2.43 | 0.877 |
| Female | 15 | 29.01 ± 3.76 | |
| Tumor size (cm) | |||
| < 2 | 29 | 25.92 ± 3.64 | 0.606 |
| ≥ 2 | 31 | 25.49 ± 2.59 | |
| Pathological type | |||
| Adenocarcinoma | 44 | 26.71 ± 3.18 | 0.390 |
| Mucocellulare carcinoma | 2 | 24.63 ± 3.57 | |
| Adenosquamous carcinoma | 1 | 23.94 | |
| Squamous carcinoma | 0 | ||
| Undifferentiated carcinoma | 1 | 27.91 | |
| Cell differentiation | |||
| Well | 14 | 19.09 ± 3.46 | 0.031a |
| Moderately | 23 | 21.97 ± 1.74 | |
| Poorly | 11 | 28.43 ± 4.09 | |
| Bismuth classification | |||
| Bismuth I | 6 | 24.66 ± 3.31 | 0.082 |
| Bismuth II | 13 | 27.65 ± 2.71 | |
| Bismuth III | 17 | 26.99 ± 4.02 | |
| Bismuth IV | 12 | 26.22 ± 3.96 | |
| Lymphatic node metastasis | |||
| Absent | 19 | 28.59 ± 2.53 | 0.224 |
| Present | 29 | 25.01 ± 2.64 | |
| Clinical stages | |||
| I + II | 13 | 18.35 ± 2.62 | 0.017a |
| III + IV | 35 | 27.95 ± 3.12 |
The QBC939 cell line, originating from human common bile duct adenocarcinoma, was kindly provided by Dr. Shuguang Wang from Southwest Hospital of the Third Military Medical University (China)[24]. The HEK293 cell line was purchased from the Cell Bank of Chinese Academy of Sciences (China). Cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (Gibco-BRL; Carlsbad, CA, United States). For the epigenetic study, QBC939 cells were treated with 5.0 μmol 5-Aza-2-CdR for 5 d, and 200 nmol trichostatin A (TSA; Sigma-Aldrich; St. Louis, MO, United States) was added on day 5.
RNA was extracted using the mirVana™ miRNA Isolation Kit (Applied Biosystems, Carlsbad, CA, United States). cDNA synthesis and analysis of miR-373 expression were performed according to the TaqMan MicroRNA Assay protocol (Applied Biosystems). U6 (RUN6B) was used as an endogenous control. Polymerase chain reaction (PCR) was conducted in three independent replicates for each sample. Expression of miR-373 was normalized to U6, and fold change was calculated based on the 2−ΔΔCt method.
Genomic DNA was isolated from tissues and cells using the DNeasy® Blood and Tissue Kit (Qiagen, Valencia, CA, United States). For DNA methylation detection, 1.5 μg of genomic DNA was modified with sodium bisulfite using the EpiTect Bisulfate Kit (Qiagen). CG Genome Universal Unmethylated DNA and CG Genome Universal Methylated DNA (mDNA) (Millipore, Darmstadt, Germany) were also modified for use as positive and negative controls, respectively (100% values).
Methylation-specific PCR (MSP) and MethySYBR[25,26] quantative methylation-specific PCR (qMSP) were performed with primers specific for fully methylated and fully unmethylated CpG island sequences (MmiR-373, UmiR-373). Primers for converted (ActB) and unconverted (ActG) β-actin special sequences containing no CpG sites were used as a control to correct CT values and for efficiency of bisulfite conversion, respectively. CT values of samples MmiR-373, UmiR-373, and ActB were calculated using corresponding standard curves, after which they were corrected to DNA amount with ActB values. The sum of percent of fully methylated reference (PMR) and percent of unmethylated reference (PUR) DNA sample amounts (MmiR-373 + UmiR-373 = 100%) was calculated.
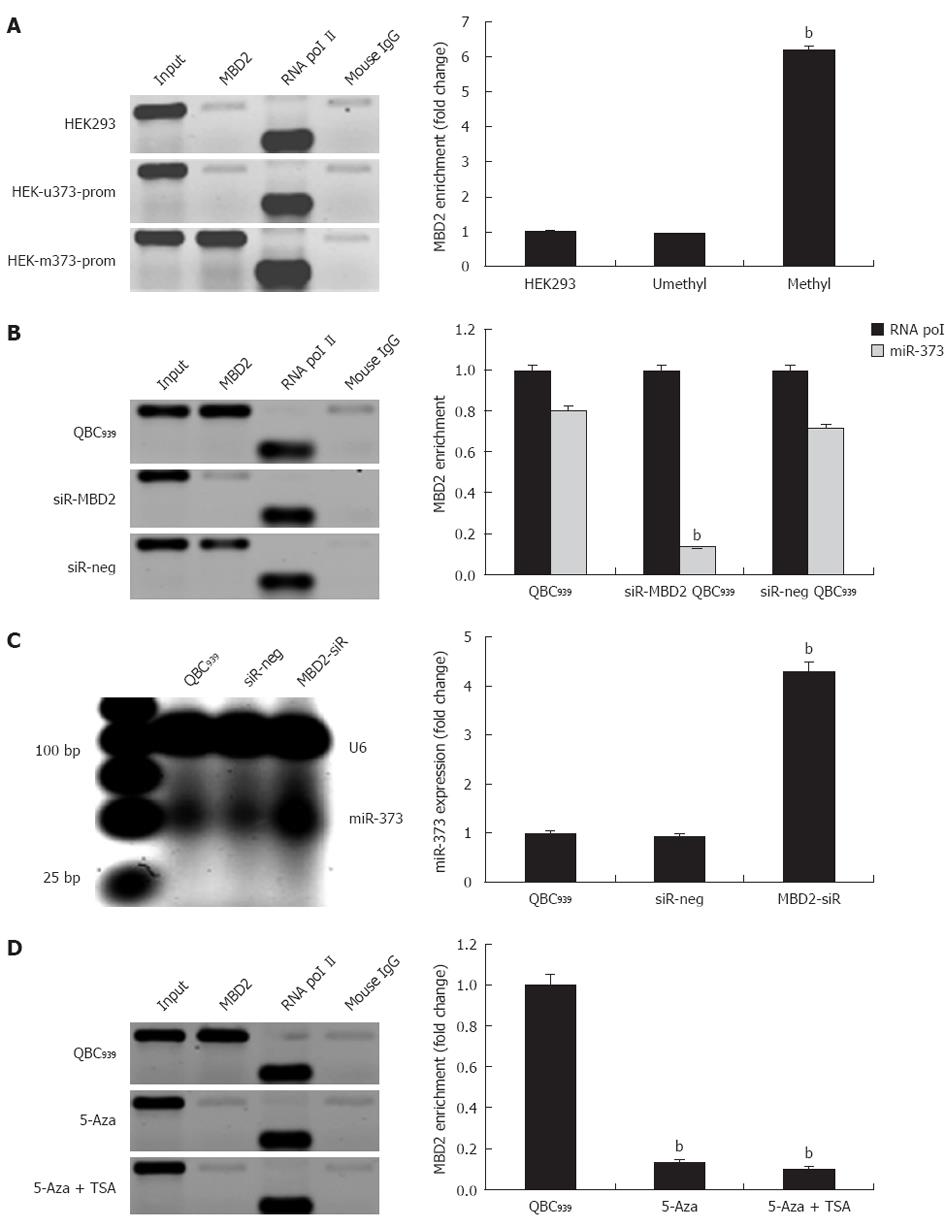
The Chromatin immunoprecipitation (ChIP) assay was performed with the ChIP-IT™ Express kit (Active Motif, Carlsbad, CA, United States) according to the manufacturer’s instructions using 2 μg ChIP-validated antibodies (Mecp2, MBD2, and mouse IgG, Active Motif; MBD1, Invitrogen, Carlsbad, CA, United States). The presence of target proteins at DNA segment were validated with primers (5’-AGATCGAGACCATCCTGGCTAACA-3’; 5’-TGAGAATGAGTCTTGCTCTGTCGC-3’) to a product of 201 base pairs (bp) in size, and the enrichment of RNA polymerase II on the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene (116 bp in size) was quantified as the endogenous control. ChIP-qPCR reactions were performed using SYBR® GreenER™ Assay (Invitrogen) in a genomic DNA model. Protein enrichment was expressed as the ratio of immunoprecipitated DNA (IP-DNA) to the total amount of DNA in the chromatin sample (input). Fold change was calculated as the ratio of IP-DNA/input (target) to IP-DNA/input (RNA polymerase II), and was then normalized to the control.
The promoter luciferase reporter plasmid harboring the CpG island of miR-373 gene was constructed according to a previously described approach[27] and designated pGL4-373-Prom. In brief, the miR-373 gene locus was screened in the database (http://genome.ucsc.edu/), a 5-kb fragment upstream of the pre-miR-373 sequence was analyzed (http://www.fruitfly.org/seq_tools/promoter) to figure out the putative promoter and transcriptional start site (TSS), and a CpG island was predicted using Methprimer software (http://www.urogene.org/ methprimer/). The results showed 402 bases of putative CpG islands spanning -251 to +150 bp and containing 26 CpG dinucleotides in the 5’-flank region of the human miR-373 gene (predicted TSS is recognized as +1).
A 726 bp fragment of the miR-373 gene (GenBank accession no. NR_029866) was amplified from QBC939 cell genomic DNA using the following PCR-specific primers: 5’-CGATGGTACCTGGAAAGTGCTGCGACATTT-3’ (sense), which contains an artificial Kpn I site, and 5’-TCATGCTAGCAGAGGTTGGCCTCCAATCAT-3’ (antisense), which contains an artificial Nhe I site and four protective bases. The PCR-amplified fragments were digested with Kpn I/Hne I and then inserted into the pGL4.22-basic plasmid (Promega, Madison,WI, United States) to generate the miR-373 gene promoter luciferase reporter plasmid designated pGL4-373-Prom.
Precursor miR-373 clones (miRNA Accession: MI 0000781), a scrambled control clone, wild-type MBD2 3’UTR vector pEZX-MBD2-3’UTR (Gene Accession: NM_004992.3), a scrambled control, and full-length cDNA expression vector of MBD2 (pDONR™_MBD2), were purchased from GeneCopoeia company (GeneCopoeia, Rockville, MD, United States). The potential binding sequences of miR-373 on the MBD2 3’UTR were mutated using the QuikChange™ Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, United States). Silencer® Select siRNA for MBD2 and the scrambled control were purchased from Applied Biosystems (siRNA ID: s17081, Target RefSeq Number: NM_003927). The recombinants were confirmed by full-length sequencing.
For protein knock-down by RNAi, 5 × 106 freshly elutriated HEK293 cells were transfected with targeting MBD2-siRNA or scrambled control at a final concentration of 200 nmol, using HiPerfect Transfection Kit (Qiagen) according to the manufacturer’s instructions. Seventy-two hours after transfection, the cells were harvested.
pGL4-373-prom was methylated in vitro using M. Sss I (4 U/μg DNA) (New England Biolab, Ipswich, MA, United States) in the presence of 160 μmol S-adenosylmethionine (SAM) for 16 h. The mDNA, re-designed as pGL4-m373-prom, was digested with either Sal I (blocked by M. Sss I) or Bam I (not blocked by M. Sss I) restriction enzymes. Sensitivity to Sal I and resistance to Bam I indicated efficient DNA methylation.
pGL4-m373-prom or pGL4-u373-prom were transfected into HEK293 cells in 35-mm dishes using Lipofectamine™ LTX and Plus Reagent (Invitrogen). Twenty-four hours post-transfection, cells were plated into 100-mm dishes at various densities and incubated in RPMI 1640 medium containing 2.0 μg/mL of puromycin for 7 d, followed by maintenance with 1.0 μg/mL puromycin. The stable cell line established was designated HEK-m373-prom and HEK-u373-prom.
For promoter luciferase activity assay, pGL4-m373-prom or pGL4-u373-prom were transfected into HEK293 cells; pRL-TK was cotransfected as an endogenous control, and methylated and unmethylated pGL4-control were also used as controls. For the 3’UTR luciferase reporter assay, vectors containing WT-3’UTR or MUT-3’UTR were co-transfected with precursor miR-373 or pre-miR-neg. Cells were harvested 72 h after transfection, and luciferase activity was quantified using the Dual-Luciferase® Reporter Assay System (Promega, Madison, WI, United States) according to the manufacturer’s protocol.
Proteins (50 μg) were analyzed by Western blotting with primary antibodies against Mecp2 (1:500, Sigma-Aldrich), MBD1 (1:1000, Millipore, Darmstadt, Germany) and MBD2 (1:1000, Millipore), which produced a signal of approximately 75 kDa, 61 kDa, and 50 kDa, respectively. GAPDH (1:5000, abcam, Cambridge, MA, United States) was used as a loading control.
Data analysis was performed using SPSS for windows version 14.0 (Chicago, IL, United States. Student’s test, one-way analysis of variance, and Pearson were used according to the data characteristics. Duration of hilar cholangiocarcinoma recurrence and death measured from the date of surgery was referenced against disease-free survival and overall survival time. Survival duration was calculated via the Kaplan-Meier method. The log-rank test was employed for comparison of cumulative survival rate and disease-free survival in the patient group. P values < 0.05 were considered statistically significant.
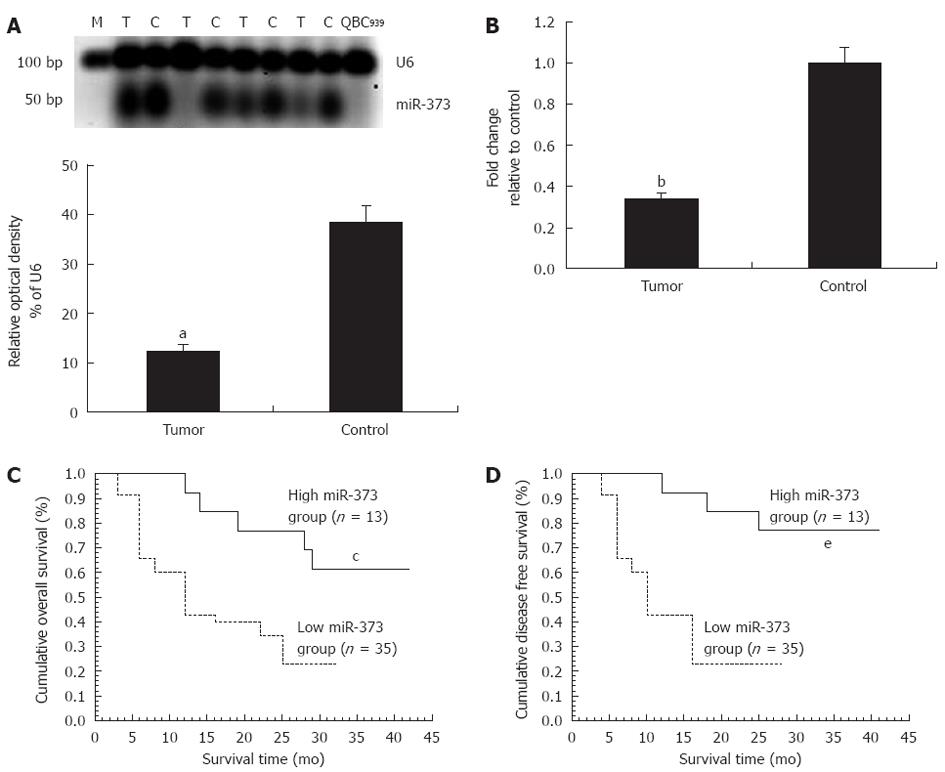
In patients with hilar cholangiocarcinoma, significant down-regulation of miR-373 was observed in QBC939 cells and 35 (72.92%) tumors, including seven undetectable samples (P < 0.01) (Figure 1A). Fold-change analysis showed a 2.94-fold decrease in the tumor group compared to the control (P < 0.01, Figure 1B). In regard to the correlation between miR-373 expression and clinicopathological factors, miR-373 showed low expression in specimens with poor cell differentiation (P = 0.031) and advanced clinical stages (stage III, IV vs I, II) (P = 0.017) (Table 1), while no association was observed with age, gender, tumor size, different pathological types, Bismuth classification, or lymphatic metastasis (P > 0.05). Further studies were conducted to evaluate the correlation between miR-373 expression and survival. Kaplan-Meier analysis showed that down-regulation of miR-373 correlated with decreased overall survival (Figure 1C, P < 0.05, log-rank test) and disease-free survival (Figure 1D, P < 0.05, log-rank).
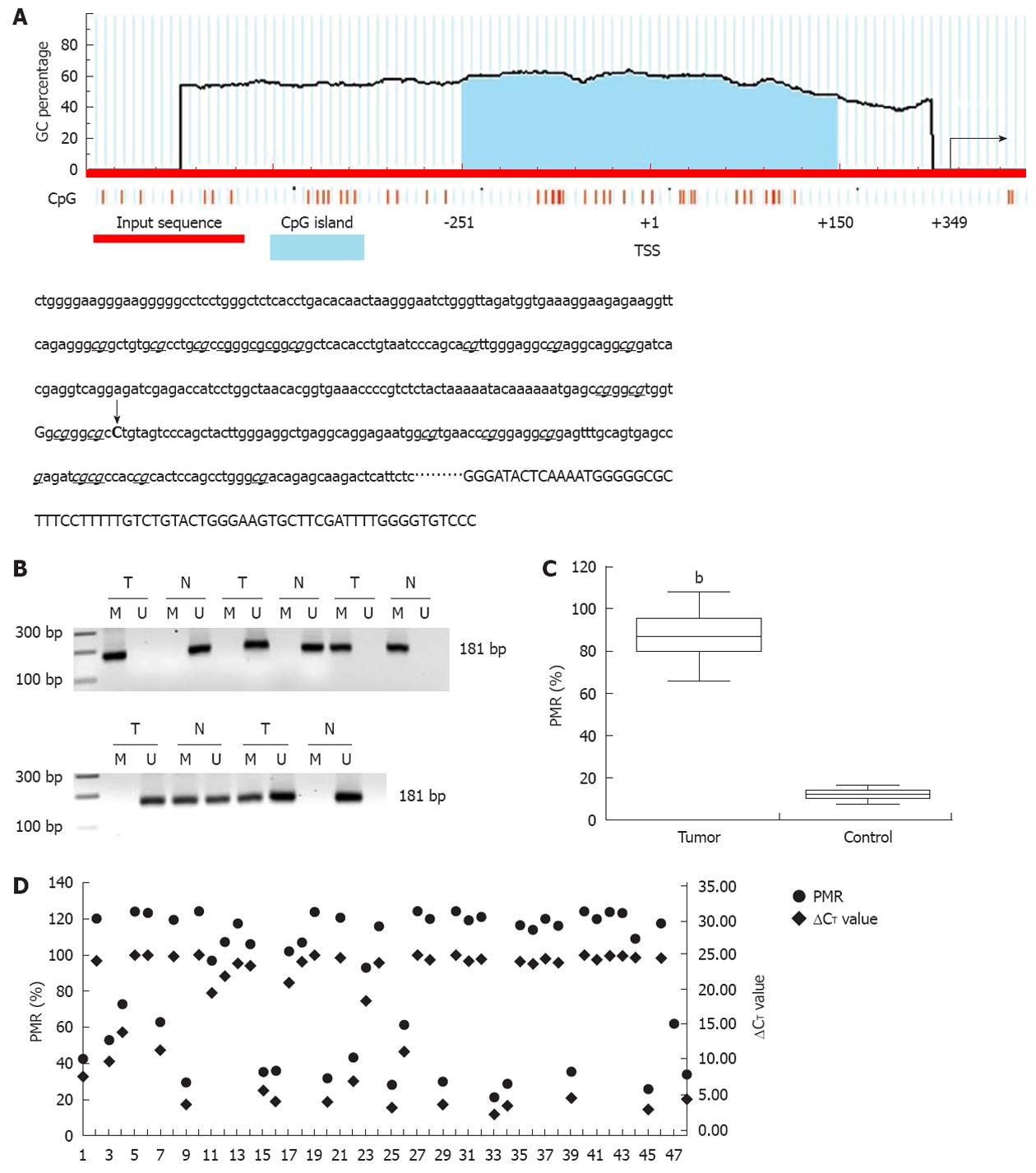
According to the literature, the CpG island is a region of at least 200 bp, a GC percentage greater than 50%, and an observed/expected CpG ratio greater than 60%. A 402 base canonical CpG island spanning -251 to +150 bp and containing 26 CpG dinucleotides encompasses the transcription start site (TSS, recognized as +1, Figure 2A). Methylation of the promoter-associated CpG island was investigated with standard MSP and MethylSYBR. In standard MSP, methylation was present in 38 (38/48, 79.17%) tumors, including 26 homozygous and 12 heterozygous samples, which are indicated by a single methylation band or by both methylation and unmethylation bands, respectively (Figure 2B). Heterozygous methylation was also observed in five control tissues. These results were validated by qMSP and a fluorescent signal was detected in same samples. The value of PMR and PUR was 87.4% and 14.7% in the tumor and control groups, respectively (Figure 2C, P < 0.01).
To determine the correlation between promoter methylation and miR-373 expression, we divided methylation into four groups according to the following PMR values: (PMR above 90.00%), hypermethylation (PMR range from 42.00% to 89.99%), standard methylation (PMR range from 20.00% to 41.99%), and hypomethylation (PMR below 20.00%). Compared to its counterparts, miR-373 expression distinctly decreased in 88.5% (23/26) supermethylated samples (P < 0.01). There was a comparative reduction in seven hypermethylated samples (P < 0.05), and a dramatic increase was seen in 10 hypomethylated tumors and 43 control tissues (P < 0.01); no difference was detected in five standard methylated samples (P > 0.05). Interestingly, three supermethylated tumors were characterized by relatively high miR-373 expression (samples 14, 18, and 44). Despite these three extra tumors, promoter methylation demonstrated a inverse relationship with miR-373 expression in hilar cholangiocarcinoma (Figure 2D).
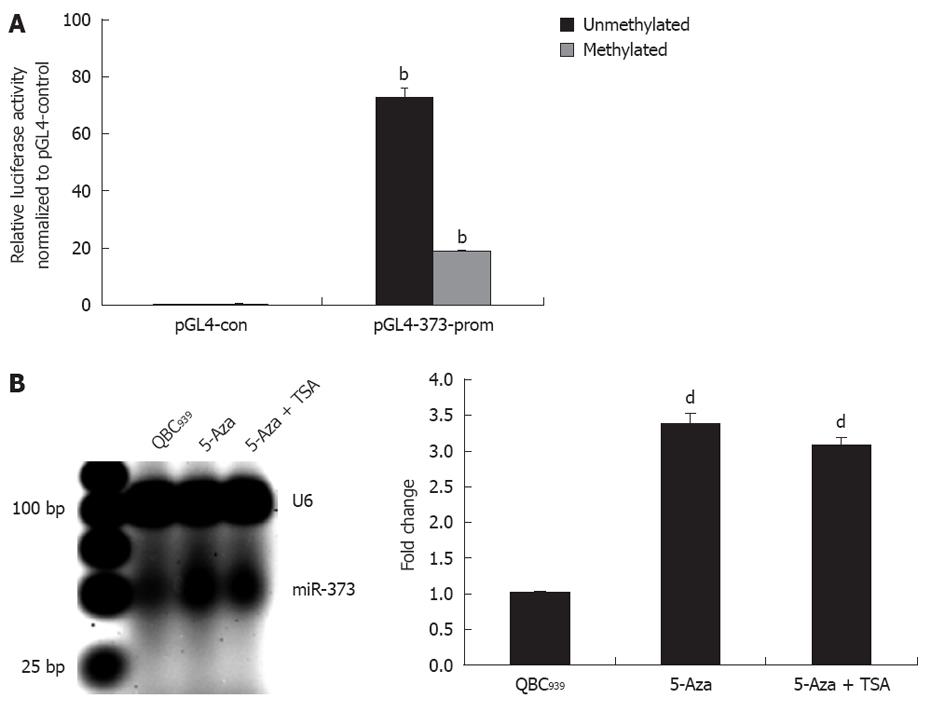
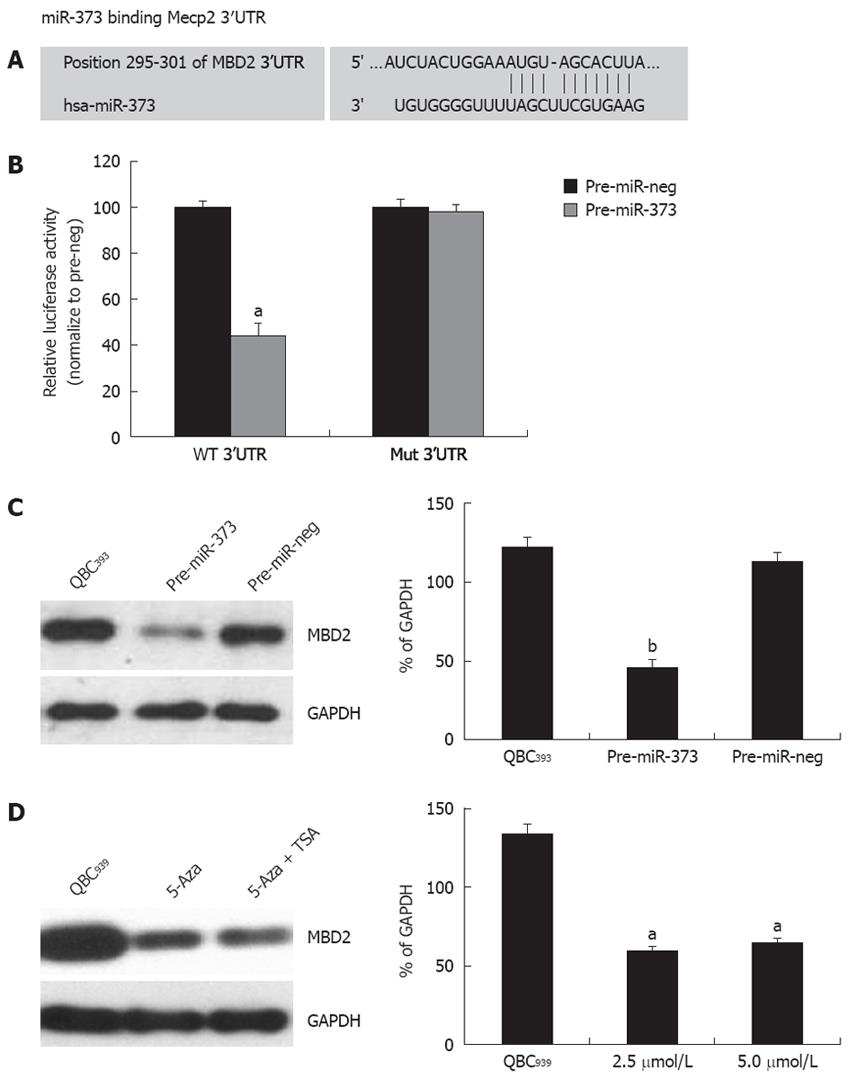
For further study on the contribution of promoter-associated CpG island methylation and inhibition of miR-373, pGL4-373-prom was constructed and methylated in vitro followed by transfection into HEK293 cells. As shown in Figure 3A, the relative luciferase activity in pGL4-u373-prom presented a higher level and decrease significantly in pGL4-m373-prom (P < 0.01).This phenomenon was also proven by data showing reactivation of miR-373 with epigenetic treatment of QBC939 cells. A 3.4-fold increase in miR-373 in cells treated with 5-Aza-2’-CdR and a 3.1-fold increase in cells treated with 5-Aza-2’-CdR and TSA were detected (Figure 3B). These results suggest that the promoter-associated CpG island acts as a cis-element of miR-373 gene transcription, and its function can be abrogated by methylation in hilar cholangiocarcinomas and QBC939 cells.
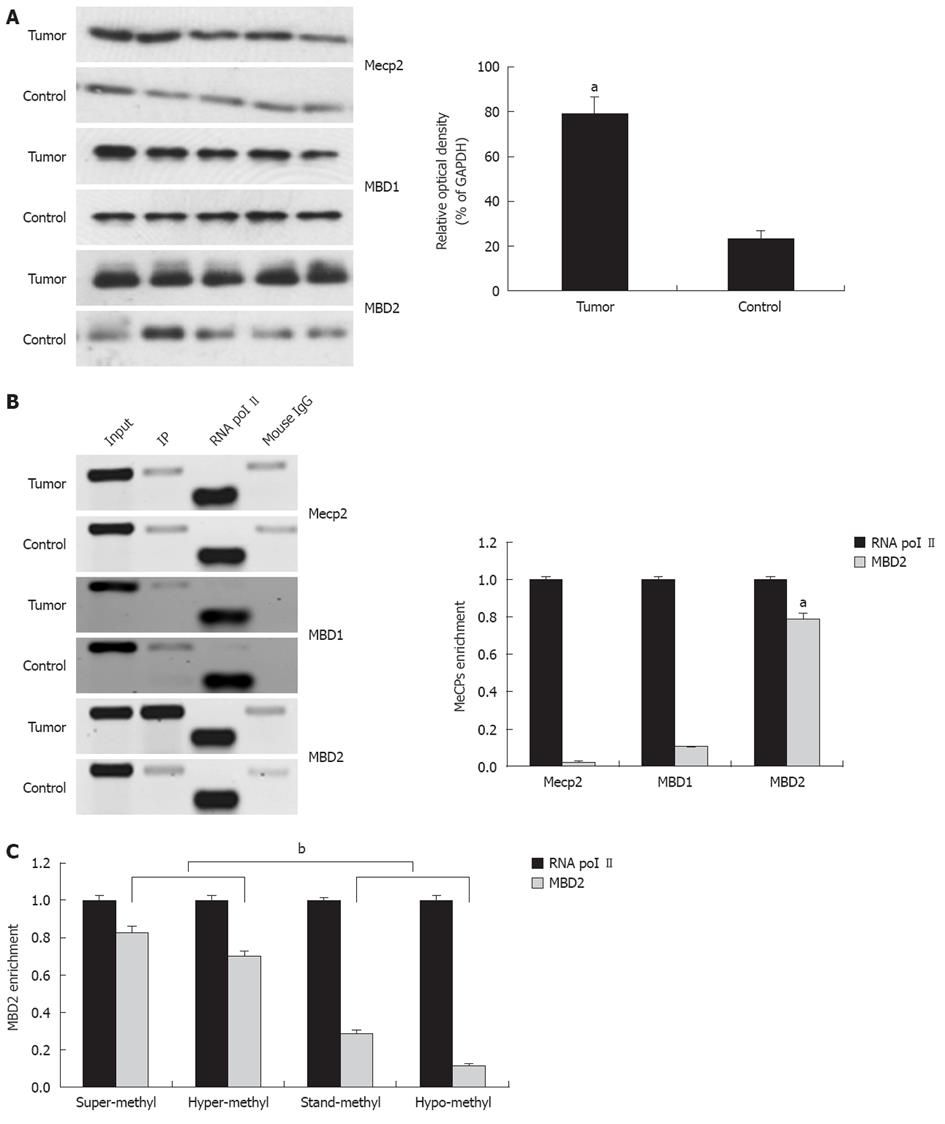
Among the MeCPs, Mecp2, MBD1, and MBD2 have been established to be involved in the methylation-dependent repression of transcription. Therefore, we explored protein expression using antibodies directed against Mecp2, MBD1, and MBD2. Compared to the control, a 2.9-fold increase in MBD2 expression was found in tumors while no difference in MBD1 and Mecp2 expression were detected (Figure 4A).
The presence of Mecp2, MBD1, and MBD2 in the region of the promoter-associated CpG island was assessed by the ChIP assay using ChIP-validated antibodies (Figure 4B). In hilar cholangiocarcinoma, the amount of CpG island fragment immunoprecipitated by MBD2 was greater than the input. ChIP-qPCR analysis showed 0.03-fold, 0.11-fold, and 0.79-fold for Mecp2, MBD1, and MBD2 compared to the endogenous control of RNA polymerase II enrichment of GAPDH, respectively (P < 0.01, Figure 4B, bottom panel). These findings indicate that the fraction of promoter-associated CpG island is selectively immunoprecipitated by MBD2, but not by Mecp2 and MBD1. In addition, fold enrichment analysis showed a remarkable correlation between miR-373 promoter methylation and MBD2 enrichment in four groups with different frequencies of methylation (P < 0.01, Figure 4C).
In QBC939 cells and hilar cholangiocarcinoma, we evaluated whether inhibition of miR-373 was closely related to hypermethylation and enrichment of MBD2 at the promoter-associated CpG island. Specifically, to unravel whether the enrichment of MBD2 is critical for inactivation of miR-373, exogenous MBD2 expression was induced in stable cell lines of HEK-u373-prom or HEK-m373-prom. The ChIP assay was performed 72 h post-transfection, and fold change analysis showed a 6.2-fold enrichment of MBD2 in pGL4-m373-prom compared to wild-type HEK293 cells, while no change was found in the pGL4-u373-prom group (Figure 5A, P < 0.01).
Further knock-down studies in QBC939 were performed to reduce endogenous MBD2 by a specific siRNA. A 4.3-fold increase in miR-373 expression was observed in knock-down cells compared to wild-type QBC939 cells (Figure 5C). Sequential ChIP assay revealed that knock-down of MBD2 resulted in depletion of MBD2 enrichment at the CpG island (Figure 5B). Furthermore, knock-down of MBD2 was not compensated by the binding of MBD1 or Mecp2. In addition, in MBD2 knock-down QBC939 cells, 5-Aza-2-CdR and TSA treatments had an innocent effect on MBD2 enrichment (Figure 5D). These findings suggest that the enrichment of MBD2 is specific to the methylated region of miR-373 promoter-associated CpG island, MBD2 likely mediates CpG methylation-dependent inhibition of miR-373 in hilar cholangiocarcinomas.
One putative miR-373 binding site was predicted to have greater specificity to MBD2 3’UTR, ranging from dinucleotide 295 to 301 bp, as predicted by four algorithms (TargetScan, PicTar, miRanda, miRbase Target) (Figure 6A). To investigate whether the 3’UTR of MBD2 is a functional target of miR-373, wild-type MBD2-3’UTR vector was transfected into HEK293 cells with pre-miR-373, which led to a decrease of 55.8% reporter activity compared to the pre-miR-neg (Figure 6B, P < 0.001). After the conserved targeting regions for miR-373 recognition were mutated, relative luciferase activity of the reporter gene was also restored (Figure 6B). These observations suggest that the predicted complementary sequence in MBD 3’UTR is a functional element of miR-373.
On the contrary, enhanced expression of miR-373 by transfecting pre-miR-373 into QBC939 cells resulted in a significant reduction of MBD2 protein (Figure 6C). Furthermore, reactivation of miR-373 expression in epigenetic-treated QBC939 cells (Figure 3B) led to an inhibition of MBD2 protein (Figure 6D). Taken together, these findings suggest that MBD2 3’UTR carries a miR-373 regulatory site, and miR-373 can negatively regulate MBD2 through binding to the miRNA locus of MBD2 3’UTR.
In previous studies, miR-373 has displayed controversial characteristics in different cancers. In testicular germ cell tumors[27], esophageal cancer[28], and breast cancer[29], miR-373 behaves as a novel oncogene. Whereas in prostate cancer[30] and malignant cholangiocytes, including the extrahepatic cholangiocarcinoma cell line[31], miR-373 shows characteristics of a tumor suppressor. Regardless of this divergence, it has been well established that miR-373 participates in tumorigenesis, invasion, and metastasis by mediating gene expression.
In this study, we show that miR-373 is dramatically down-regulated in hilar cholangiocarcinoma, and correlates closely with poor cell differentiation, advanced clinical stages, shorter overall survival, and disease-free survival. Our findings are in agreement with the last two reports. Although it is difficult to definitively explain these directly opposing results, the expression pattern of individual miRNAs with strict tissue- and clinical-feature-specificity[32], and the different target genes involved in the unique regulation network of various cancers, could lead to these discrepancies.
Epigenetic dysregulation of miRNAs in human cancer constitutes an emerging mechanism implicated in the development of cancer[33]. Great effort has been devoted to understanding the relevance of aberrant CpG methylation patterns, and their roles in gene transcription. The inverse correlation between miR-373 expression and hilar cholangiocarcinoma progression prompted us to study the molecular mechanisms underlying miR-373 gene inhibition. In the present study, miR-373 promoter-associated CpG island was found to be hypermethylated in tumor tissues and QBC939 cells. Reciprocal assays were performed with demethylation of the CpG island by treatment of QBC939 cells with 5-aza-CdR in the absence or presence of combination with TSA, which contributes to the reactivation of miR-373. In addition, pre-methylation of pGL4-373-prom in vitro inhibited luciferase activity. Together, the results presented here provide evidence that promoter-associated CpG island methylation is a major cause of miR-373 gene suppression in hilar cholangiocarcinoma.
Promoter-associated CpG-methylation, along with MBPs and HDACs, has been identified as a major epigenetic event in the loss of gene expression during tumor progression[34]. In this study, we further investigated the involvement of MBPs in methylation-mediated suppression of miR-373 in hilar cholangiocarcinoma. MBD2 is an exclusive MBP that is aberrantly expressed in hilar cholangiocarcinoma. ChIP assays showed that MBD2 selectively bound to methylated CpG sequences. Moreover, silencing conferred by DNA methylation and MBD2 enrichment in QBC939 cells was reversed by treatment with 5-aza-CdR and TSA. These findings indicate that MBD2 plays an important role in recruiting transcription-repressive machinery to the methylated promoter, thereby suppressing transcriptional activation of miR-373. Confirming these findings, siRNA knock-down of MBD2 triggered a stimulation of miR-373 in QBC939 cells. Taken together, these data suggest that MBD2 is an important factor in methylation-mediated inhibition of miR-373.
The reciprocal correlation of expression between miR-373 and MBD2 encouraged us to explore whether miR-373 is a negative regulator of MBD2. To this end, four algorithms were used to predict the alignment of miR-373 with MBD2 3’UTR. In consensus, the seed region of miR-373 matched nucleotides 295-301 of the MBD2 3’UTR, suggesting the ability of miR-373 to directly bind to MBD2 mRNA. However, not all miRNAs identified in this manner are likely to be functional since factors such as steric hindrance may render them inaccessible to the mRNA[35]. Hence, functional validation experiments including transfection of miR-373 precursors showed that miR-373 can down-regulate the relative luciferase activity of MBD 3’UTR reporterer vectors and MBD2 protein in QBC939 cell lines. These findings indicate that miR-373 functionally regulates the expression of MBD2 by targeting the 3’UTR.
In this study, several interesting observations were made. First, the heterozygous methylation of miR-373, as indicated by both methylated and unmethylated bands in MSP, was detected in 12 cancers and five normal bile duct tissues. This is contrary to the ‘all-or-none’ manner of DNA methylation in regulating gene expression. Although the mechanism is unclear, there are several points to keep in mind: (1) miR-373 gene exhibits allele-specific DNA methylation (ASM) which means that only one allele is methylated and the other one is unmethylated[36]. ASM has been documented in a number of cancer cases except imprinted regions and X chromosomes; (2) samples contain normal and malignant cells although multiple efforts have been adapted to obtain purified tissues; and (3) miR-373 displays discrepant methylation in various differentiated cells. Secondly, in primary samples, three supermethylated tumors were characterized with relatively high miR-373 expression. The relevant mechanism underlying transcription that escapes methylation-mediated suppression is unknown, but whether MBPs bind effectively to the methylated CpG dinucleotides may determine the expression level.
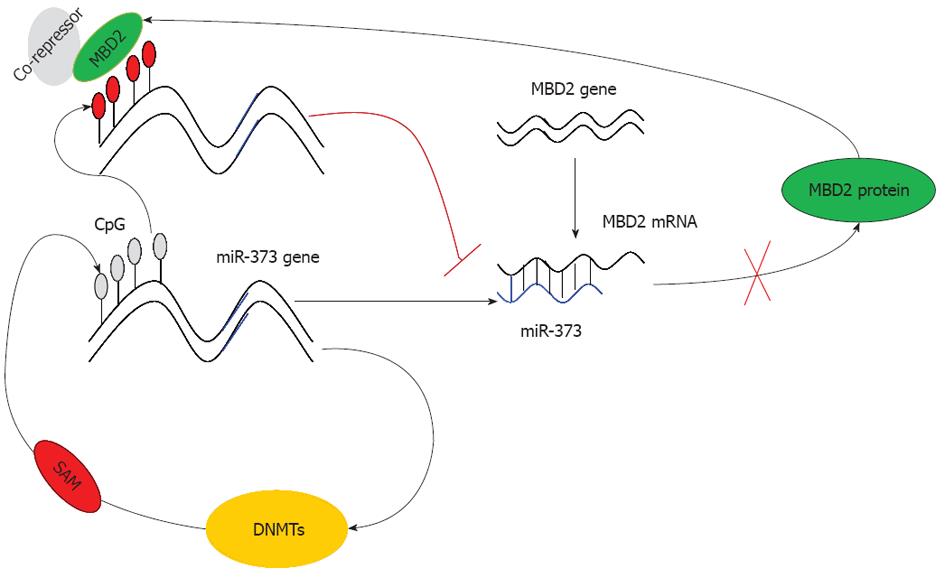
Based on these findings, we conclude that due to the hypermethylation of the promoter-associated CpG island and enrichment of MBD2, the function of miR-373 is restrained rendering it unable to inhibit MBD2. As a result, the expression of MBD2 is predominantly enhanced, leading to a strong inhibitory effect on miR-373 (Figure 7). This dysregulation ultimately results in the promotion of tumorigenesis and the development of hilar cholangiocarcinoma. In conclusion, our study proves that miRNA-373 behaves as a direct transcriptional target and negative regulator of MBD2 through a feedback loop of CpG methylation in hilar cholangiocarcinoma.
Both DNA methylation and microRNAs (miRNAs) are epigenetic and play vital roles in tumorigenesis and development of human malignance. DNA methylation represses transcription by impeding recognition of transcriptional activators to DNA sequences or recruiting methyl-CpG binding domain proteins (MBPs) to modify chromatin compaction and control gene silencing. miRNAs regulate gene expression mainly by binding to the three prime untranslated region of target mRNAs, leading to mRNA degradation or translation inhibition. Many studies have reported that the expression of miRNAs gene is regulated by DNA methylation, and in addition, DNA methyltransferases and MBPs are regulated by miRNAs.
Hilar cholangiocarcinoma displays highly aggressive malignancy. Many studies have reported miRNA expression and DNA methylation in hilar cholangiocarcinoma. In this study, the authors report evidence of the role of miR-373 in hilar cholangiocarcinoma. In particular, they show that miR-373 behaves as a direct transcriptional target and negative regulator of methyl-CpG-binding domain protein (MBD)2 through a feedback loop of CpG methylation in hilar cholangiocarcinoma.
It has been well established that miR-373 participates in tumorigenesis, invasion, and metastasis by mediating gene expression. In this study, the authors demonstrate that miR-373 is dramatically down-regulated in hilar cholangiocarcinoma, and closely correlates with poor cell differentiation, advanced clinical stage, shorter overall survival, and disease-free survival. The authors show that due to the hypermethylation of the promoter-associated CpG island and enrichment of MBD2, function of miR-373 is restrained, rendering it unable to inhibit MBD2. As a result, MBD2 expression is predominantly enhanced and has a strong inhibitory effect on miR-373. This dysregulation finally promotes the tumorigenesis and development of hilar cholangiocarcinoma.
This study provides the first evidence showing that miR-373 behaves as a direct transcriptional target and negative regulator of MBD2 through a feedback loop of CpG methylation in hilar cholangiocarcinoma. These results shed light on the mutual regulation between miRNA-373 and MBD2, which may eventually serve as useful biomarkers as well as therapeutic targets.
This study demonstrates the role of miR-373 in cholangiocarcinoma. In particular, the authors showed that miR-373 acts through a feedback loop of CpG methylation. This study is well designed and performed, and is of great interest for its novelty and impact in the field.
Peer reviewers: Pradyumna Kumar Mishra, Professor, Translational Research, Tata Memorial Centre, Kharghar, Navi Mumbai 410210, India; Dr. Pietro Invernizzi, Center for Autoimmune Liver Diseases, Division of Internal Medicine, IRCCS Istituto Clinico Humanitas, Via Manzoni, 113, 20089 Rozzano, Italy
S- Editor Gou SX L- Editor A E- Editor Zheng XM
| 1. | Klatskin G. Adenocarcinoma of the hepatic duct at its bifurcation within the porta hepatis. An unusual tumor with distinctive clinical and pathological features. Am J Med. 1965;38:241-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 577] [Cited by in RCA: 499] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 2. | Akoad M, Jenkins R. Proximal biliary malignancy. Surg Clin North Am. 2008;88:1409-1428, x-xi. [PubMed] |
| 3. | Jarnagin WR, Fong Y, DeMatteo RP, Gonen M, Burke EC, Bodniewicz BS J, Youssef BA M, Klimstra D, Blumgart LH. Staging, resectability, and outcome in 225 patients with hilar cholangiocarcinoma. Ann Surg. 2001;234:507-517; discussion 517-519. [PubMed] |
| 4. | Aljiffry M, Walsh MJ, Molinari M. Advances in diagnosis, treatment and palliation of cholangiocarcinoma: 1990-2009. World J Gastroenterol. 2009;15:4240-4262. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 186] [Cited by in RCA: 193] [Article Influence: 12.1] [Reference Citation Analysis (1)] |
| 5. | Tischoff I, Tannapfe A. DNA methylation in hepatocellular carcinoma. World J Gastroenterol. 2008;14:1741-1748. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 128] [Cited by in RCA: 134] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 6. | Watt F, Molloy PL. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988;2:1136-1143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 407] [Cited by in RCA: 418] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 7. | Bird AP, Wolffe AP. Methylation-induced repression--belts, braces, and chromatin. Cell. 1999;99:451-454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1349] [Cited by in RCA: 1294] [Article Influence: 49.8] [Reference Citation Analysis (0)] |
| 8. | Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 457] [Cited by in RCA: 470] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 9. | Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471-481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 908] [Cited by in RCA: 959] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 10. | Fujita N, Takebayashi S, Okumura K, Kudo S, Chiba T, Saya H, Nakao M. Methylation-mediated transcriptional silencing in euchromatin by methyl-CpG binding protein MBD1 isoforms. Mol Cell Biol. 1999;19:6415-6426. [PubMed] |
| 11. | Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 636] [Cited by in RCA: 648] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 12. | Saito M, Ishikawa F. The mCpG-binding domain of human MBD3 does not bind to mCpG but interacts with NuRD/Mi2 components HDAC1 and MTA2. J Biol Chem. 2002;277:35434-35439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 134] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 13. | Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766-770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1342] [Cited by in RCA: 1383] [Article Influence: 69.2] [Reference Citation Analysis (0)] |
| 14. | Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857-866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5705] [Cited by in RCA: 6032] [Article Influence: 317.5] [Reference Citation Analysis (0)] |
| 15. | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14460] [Cited by in RCA: 16089] [Article Influence: 1005.6] [Reference Citation Analysis (2)] |
| 16. | Sevignani C, Calin GA, Siracusa LD, Croce CM. Mammalian microRNAs: a small world for fine-tuning gene expression. Mamm Genome. 2006;17:189-202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 239] [Cited by in RCA: 266] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 17. | Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051-4060. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2902] [Cited by in RCA: 2997] [Article Influence: 142.7] [Reference Citation Analysis (0)] |
| 18. | Wu L, Zhou H, Zhang Q, Zhang J, Ni F, Liu C, Qi Y. DNA methylation mediated by a microRNA pathway. Mol Cell. 2010;38:465-475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 441] [Cited by in RCA: 431] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 19. | Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805-15810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1300] [Cited by in RCA: 1282] [Article Influence: 71.2] [Reference Citation Analysis (0)] |
| 20. | Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, Schwind S, Pang J, Yu J, Muthusamy N. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411-6418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 624] [Cited by in RCA: 614] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 21. | Li D, Chen J, Gao Z, Li X, Yan X, Xiong Y, Wang S. 67-kDa laminin receptor in human bile duct carcinoma. Eur Surg Res. 2009;42:168-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 22. | Li XQ, Guo YY, De W. DNA methylation and microRNAs in cancer. World J Gastroenterol. 2012;18:882-888. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 22] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 23. | Stutes M, Tran S, DeMorrow S. Genetic and epigenetic changes associated with cholangiocarcinoma: from DNA methylation to microRNAs. World J Gastroenterol. 2007;13:6465-6469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in CrossRef: 16] [Cited by in RCA: 13] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 24. | Hattermann K, Mehdorn HM, Mentlein R, Schultka S, Held-Feindt J. A methylation-specific and SYBR-green-based quantitative polymerase chain reaction technique for O6-methylguanine DNA methyltransferase promoter methylation analysis. Anal Biochem. 2008;377:62-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 25. | Lo PK, Watanabe H, Cheng PC, Teo WW, Liang X, Argani P, Lee JS, Sukumar S. MethySYBR, a novel quantitative PCR assay for the dual analysis of DNA methylation and CpG methylation density. J Mol Diagn. 2009;11:400-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 26. | Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2463] [Cited by in RCA: 2548] [Article Influence: 115.8] [Reference Citation Analysis (0)] |
| 27. | Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169-1181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 954] [Cited by in RCA: 952] [Article Influence: 50.1] [Reference Citation Analysis (0)] |
| 28. | Lee KH, Goan YG, Hsiao M, Lee CH, Jian SH, Lin JT, Chen YL, Lu PJ. MicroRNA-373 (miR-373) post-transcriptionally regulates large tumor suppressor, homolog 2 (LATS2) and stimulates proliferation in human esophageal cancer. Exp Cell Res. 2009;315:2529-2538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 125] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 29. | Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 740] [Cited by in RCA: 745] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 30. | Yang K, Handorean AM, Iczkowski KA. MicroRNAs 373 and 520c are downregulated in prostate cancer, suppress CD44 translation and enhance invasion of prostate cancer cells in vitro. Int J Clin Exp Pathol. 2009;2:361-369. [PubMed] |
| 31. | Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, Jiang J, Schmittgen TD, Patel T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113-2129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 777] [Cited by in RCA: 782] [Article Influence: 41.2] [Reference Citation Analysis (0)] |
| 32. | Christodoulou F, Raible F, Tomer R, Simakov O, Trachana K, Klaus S, Snyman H, Hannon GJ, Bork P, Arendt D. Ancient animal microRNAs and the evolution of tissue identity. Nature. 2010;463:1084-1088. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 231] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 33. | Saito Y, Jones PA. Epigenetic activation of tumor suppressor microRNAs in human cancer cells. Cell Cycle. 2006;5:2220-2222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 193] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 34. | Fraga MF, Esteller M. Towards the human cancer epigenome: a first draft of histone modifications. Cell Cycle. 2005;4:1377-1381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 108] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 35. | Lai KW, Koh KX, Loh M, Tada K, Subramaniam MM, Lim XY, Vaithilingam A, Salto-Tellez M, Iacopetta B, Ito Y. MicroRNA-130b regulates the tumour suppressor RUNX3 in gastric cancer. Eur J Cancer. 2010;46:1456-1463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 132] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 36. | Tycko B. Allele-specific DNA methylation: beyond imprinting. Hum Mol Genet. 2010;19:R210-R220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 122] [Article Influence: 8.1] [Reference Citation Analysis (0)] |