Published online Jun 7, 2012. doi: 10.3748/wjg.v18.i21.2630
Revised: February 29, 2012
Accepted: March 20, 2012
Published online: June 7, 2012
AIM: To investigate the effect of high dose glargine on the expression profiles of microRNAs in human pancreatic cancer cells.
METHODS: Real-time polymerase chain reaction array (RT-PCR) was applied to investigate miRNAs differentially expressed in Sw1990 cells treated with or without 100 IU/L glargine. Stem-loop RT-PCR was used to confirm the results of the array assay in Sw1990 and Panc-1 cells. The effects of miR-95 on cell growth, apoptosis, invasion and migration abilities were respectively examined by CCK8 assay, apoptosis assay, Matrigel invasion and migration assay in Sw1990 and Panc-1 cells. Nude mice xenograft models with Sw1990 cells were built to investigate pancreatic cancer growth in vivo after transfection by the lentivirus pGLV3-GFP- miR-95.
RESULTS: Ten miRNAs were significantly up-regulated and 2 miRNAs down-regulated in glargine treated Sw1990 cells when compared with non-treated cells (2.48-fold changes on average, P < 0.01). miR-95, miR-134 and miR-34c-3p are the top three miRNAs regulated by glargine (3.65-fold, 2.67-fold and 2.60-fold changes respectively, P < 0.01) in Sw1990 cells. Stem-loop RT-PCR confirmed that high dose glargine up-regulated the expression of miR-95 and miR-134 in both Sw1990 and Panc-1 cells. The most obvious change is the apparent increase of miR-95. Forced expression of miR-95 significantly increased cell proliferation (Sw1990: 2.510 ± 0.129 vs 2.305 ± 0.187, P < 0.05; Panc-1: 2.439 ± 0.211 vs 2.264 ± 0.117, P < 0.05), invasion (Sw1990: 67.90 ± 12.33 vs 47.30 ± 5.89, P < 0.01; Panc-1: 37.80 ± 8.93 vs 30.20 ± 5.14, P < 0.01), migration (Sw1990: 101 ± 6.00 vs 51.20 ± 8.34, P < 0.01; Panc-1: 91.80 ± 9.22 vs 81.50 ± 7.47, P < 0.01) and inhibited cell apoptosis (Sw1990: 22.05% ± 1.92% vs 40.32% ± 1.93%, P < 0.05; Panc-1: 20.17% ± 0.85% vs 45.60% ± 1.43%, P < 0.05) when compared with paired negative controls, whereas knockdown of miR-95 obtained the opposite effect. Nude mice xenograft models confirmed that miR-95 promoted the growth of pancreatic cancer in vivo when compared with negative control (tumor volume: 373.82 ± 23.67 mL vs 219.69 ± 17.82 mL, P < 0.05).
CONCLUSION: These observations suggested that modulation of miRNA expression may be an important mechanism underlying the biological effects of glargine.
- Citation: Li WG, Yuan YZ, Qiao MM, Zhang YP. High dose glargine alters the expression profiles of microRNAs in pancreatic cancer cells. World J Gastroenterol 2012; 18(21): 2630-2639
- URL: https://www.wjgnet.com/1007-9327/full/v18/i21/2630.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i21.2630
Pancreatic cancer is the fourth leading cause of cancer-related deaths in Western countries and has the poorest survival rate (< 5%) among the common malignancies[1,2]. Recently, antidiabetic therapies have been shown to affect the risk of pancreatic cancer. Some observational studies in humans have linked glargine with a putative increased cancer risk, including pancreatic cancer[3,4].
Glargine (A21Gly, B31Arg, B32Arg human insulin) is a widely used insulin analog in which a 24-h action profile is achieved by altering the amino acid sequence of the alpha (α) and beta (β) chains of the C terminus[5]. It has been shown that glargine increases resistance to apoptosis in several tumor cell lines[6]. Given the increased affinity to the insulin-like growth factor-I receptor (IGF-IR) in vitro[7], glargine may increase the bioavailability of IGF-I by altering the levels of IGF-binding proteins[8,9]. IGF-I is a more potent growth factor than insulin, promoting proliferation and inhibiting apoptosis, and plays an important role in facilitating malignant cell survival and metastasis[10,11]. This may be the theoretical basis for the potential carcinogenicity of glargine. However, data regarding the effect of glargine on pancreatic cancer are inconsistent. Administration of glargine didn’t alter proliferation of Colo-357 pancreatic carcinoma cells and survival of patients with pancreatic carcinoma[12]. Thus, the role of glargine in pancreatic carcinogenesis deserves further investigation.
MicroRNAs (miRNAs) are endogenous, non-coding small RNAs, 19-25 nucleotides in length, which are now recognized as crucial post-transcriptional regulators of gene expression[13-15]. It has been demonstrated that miRNAs play important roles in biological processes that affect tumor progression including migration, invasion, epithelial to mesenchymal transition (EMT) and metastasis[16-18]. miRNAs are promising as early biomarkers, prognostic indicators and therapeutic targets for anticancer treatments[19-21]. Aberrant miRNA expression has also been frequently reported in pancreatic cancer[22]. However, very few compounds, not to mention glargine, which affect cell growth and/or development, have been shown to affect miRNA expression.
In this present study, we elucidated the miRNAs signature in response to glargine treatment in human pancreatic cancer cells. Our results indicated that glargine alters specific miRNA expression in human pancreatic cells, especially miR-95. The effect of miR-95 on apoptosis, proliferation, migration and invasion ability of pancreatic cancer cells were further investigated. Moreover, nude mice xenograft models were built to investigate pancreatic cancer growth in vivo after transfection by the lentivirus pGLV3-GFP-miR-95. It therefore appeared that miR-95-related changes were important effects of glargine.
Pancreatic ductal cancer cell lines Sw1990 and Panc-1 were conserved in our own laboratory and were cultured in Dulbecco’s modified Eagle’s medium (DMEM; GIBCO) with 10% fetal bovine serum (FBS; GIBCO) in a humidified incubator at 37 °C with an atmosphere of 5% CO2.
Sw1990 cells (3 × 105 per well) were plated on 6-well plates in DMEM with 10% FBS. After 24 h of incubation at 37 °C, the cells were treated with or without 100 IU/L glargine. Glargine was replenished every 24 h. The cultures were incubated for 2 d, then the total RNA was isolated from cell samples using Trizol reagent (Invitrogen) following the manufacturer’s protocol. Then, cDNA synthesis was performed using Universal cDNA synthesis kit (Exiqon). The expression levels of 372 human mature miRNAs were examined using the miRCURY LNA™ Universal real time microRNA polymerase chain reaction system, Ready-to-use human panel I (Exiqon, kangchen, China).
Briefly, total RNA containing miRNA was polyadenylated, and cDNA was synthesized using a poly (T) primer with a 3’degenerate anchor and a 5’universal tag. Then, cDNA served as a template for microRNA quantitative real-time polymerase chain reaction (qPCR) using miRCURY LNA Universal RT miRNA PCR kit (Exiqon). The miRNA Ready-to-use human panel I is a 384-well PCR plate containing dried down LNA™ primer sets for one real-time PCR reaction per well. Three small RNA (U6snRNA, SNORD38B, SNORD49A) and three miRNA (miR-103, miR-191 and miR-423-5p) reference genes are included on the panel. The amplification profile was denatured at 95 °C for 10 min, followed by 40 cycles of 95 °C for 10 s and 60 °C for 60 s. At the end of the PCR cycles, melting curve analyses were performed. All reactions were conducted three times. Expression levels of mature miRNAs were evaluated using comparative CT method (2-ΔCT).
The miRNAs (miR-95, miR-134 and miR-34c-3p) were quantitated by stem-loop real time reverse transcription (RT)-PCR to confirm the reliability of the miRNA array assay. In brief, Sw1990 and Panc-1 cells (3 × 105 per well) were seeded on 6-well plates in DMEM with 10% FBS. After 24 h of incubation, the cells were treated with different concentrations of glargine (0-150 IU/L) for 48 h or treated with 100 IU/L glargine for different periods (24-72 h). Glargine was replenished every 24 h. Then the total RNA was isolated. 0.2-0.5 μg of total RNA was reverse transcribed to cDNA using a target-specific stem-loop primer indicated in Table 1. cDNA in water was added to 5 μL of the 2 × SYBR green master mix (Applied Biosystems Inc, Foster City, United States), 400 nmol/L of gene-specific primer and water used to make the solution up to 10 μL. The reactions were amplified at 95 °C for 10 min followed by 40 cycles at 95 °C for 15 s and 60 °C for 60 s. U6 small nuclear RNA (U6) served as the endogenous control. The relative amount of each miRNA to U6 was described using the formula 2-ΔCt where ΔCt = (Ct miRNA - CtU6). Each sample was run in triplicate.
| Gene name | Primer sequences (5’-3’) | |
| miRNA-95 | Stem-loop primer | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTGCTCAAT |
| Sense | CGGGTATTTATTGAGCA | |
| Antisense | AACTGGTGTCGTGGAG | |
| miRNA-134 | Stem-loop primer | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCCCTCTG |
| Sense | TGTGACTGGTTGACCAGAG | |
| Antisense | AACTGGTGTCGTGGAG | |
| miRNA-34c-3p | Stem-loop primer | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCTGGCCGTG |
| Sense | AATCACTAACCACACGG | |
| Antisense | AACTGGTGTCGTGGAG | |
| U6 | Stem-loop primer | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAAAAATAT |
| Sense | CAAGGATGACACGCAAAT | |
| Antisense | TGGTGTCGTGGAGTCG | |
The miR-95 sequence was constructed as follows: (forward) hsa-miR-95-BamH I: GATCCGTTCAACGGGTATTTATTGAGCATTCAAGAGATGCTCAATATACCCGTTGAACTTTTTTG; (reverse) hsa-miR-95-EcoR I: AATTCAAAAAAGTTCAACGGGTATATTGAGCATCTCTTGAATGCTCAATAAATACCCGTTGAACG. The sequence was amplified and cloned into the pGLV3-GFP vector (GenePhama) to generate pGLV3-GFP- miR-95. The negative control was pGLV3-shRNA-NC. Virus packaging was performed in HEK 293T cells after the co-transfection of 20 mg pGLV3-GFP-miR-95 vector with 15 mg of the packaging plasmid pHelper 1.0 Vector and 10 mg of the envelope plasmid pHelper 2.0 Vector using Lipofectamine 2000 (Invitrogen).Viruses were harvested 48 h after transfection, and viral titers were determined.
After glargine treatment, the expression of miR-95 was increased most obviously in Sw1990 cells and Panc-1 cells, so we further investigated the functional roles of miR-95 in pancreatic cancer cells. miR-95 mimics, miR-95 inhibitor and negative control siRNA oligonucleotides were chemosynthesized (Shanghai GenePhama Co. Ltd). The oligonucleotides used in these studies were has-miR-95 mimics: 5’-UUCAACGGGUAU UUAUUGAGCA-3’ and 5’-CUCAAUAAAUACCCGUUGAAUU-3’. Mimics negative control: 5’-UUCUCCGAACGUGUCACGUTT-3’ and 5’-ACGUGACACGUUCGGAGAA TT-3’, hsa-miR-95 inhibitor: 5’-UGCUCAAUAAAUACCCGUUGAA-3’. MicroRNA inhibitor negative control: 5’-CAGUACUUUUGUGUAGUACA A-3’.
Cells were cultured to 80% to 90% confluence after being seeded into 6-well plates and were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. For transient transfection, Sw1990 or Panc-1 cells in each well of a 6-well plate were transfected with 12.5 μL miRNA inhibitor or 7.5 μL miRNA mimic oligonucleotides. Transfection efficiency was evaluated by FAM in control vector or real-time PCR. For stably transfected cells, cells were transfected with lentivirus at 80%-90% confluency. Sw1990 cells (1 × 105) were infected with recombinant lentivirus in the presence of 5 μg/mL polybrene (GenePhama).
A total of 104 Sw1990 or Panc-1 cells per well were plated in 96-well plates before transfection and cultured for 24 h in normal conditions. They were then transfected with hsa-miR-95 mimics or hsa-miR-95 inhibitor along with paired negative controls. The cells were incubated at 37 °C for 48 h. Cell proliferation was assessed using Cell Counting Kit 8 (Dojindo, Tokyo, Japan) according to manufacturer’s protocol.
At 72 h after transfection, apoptosis was detected using Annexin V-FITC Apoptosis Detection Kit (Biovision, United States). Results were calculated by the percentage of apoptotic cells in all cells counted.
At 48 h after transfection, the invasive ability of the cells was assayed using Transwells (8 mm pore size, Corning Costar Corp). The Transwells were put into the 24-well plates. First, 0.1 mL Matrigel (50 mg/mL, BD Biosciences) was added onto the plate surface and incubated for 2 h, and then the supernatant was removed. Freshly trypsinized and washed Panc-1 or Sw1990 cells were suspended in DMEM containing 1% FBS. Then 0.1 mL of the cell suspension (1 × 105 cells) was added to the upper chamber of each insert that was coated with Matrigel. Next, 0.6 mL of DMEM containing 10% FBS was added into the lower compartment, and the cells were allowed to invade for 24 h at 37 °C in a 5% CO2 humidified incubator. After incubation, the cells were fixed with 95% absolute alcohol and followed by crystal violet stain. The number of migrated cells on the lower surface of the membrane was counted under a microscope in 10 fields with magnification of × 200. Each experiment was performed in triplicate.
At 48 h after transfection, the ability of Panc-1 or Sw1990 cells to migrate was detected using Transwells (8 mm pore size, Corning Costar Corp). The Transwells were put into the 24-well plates. Freshly trypsinized and washed cells were suspended in DMEM containing 1% FBS. 5 × 104 cells/well were placed in the top chamber of each insert (BD Biosciences, NJ), with the non-coated membrane. 0.6 mL of DMEM containing 10% FBS was added into the lower chambers. After incubating for 24 h at 37 °C in a 5% CO2 humidified incubator, the cells were fixed with 95% absolute alcohol and stained with crystal violet stain. The number of migrated cells on the lower surface of the membrane was counted under a microscope in 10 fields with magnification of × 200. Each experiment was performed in triplicate.
The stable cell line Sw1990 was harvested from tissue culture flasks after transfection with the pGLV3-GFP-miR-95 and control pGLV3-GFP vector using trypsin and washed three times with PBS. About 1 × 107 cells were implanted into the right flanks of female nu/nu mice (five in each group). Tumor volume (V) was measured with an external caliper every 4 d and it was calculated as V = 0.52 (length × width2). After four weeks, all the animals were sacrificed and tumors were removed.
Data were expressed as the mean ± SD unless otherwise noted. The differences between groups were analyzed using a two-tailed Student’s t-test when only two groups were present and the null hypothesis was rejected at the 0.05 level.
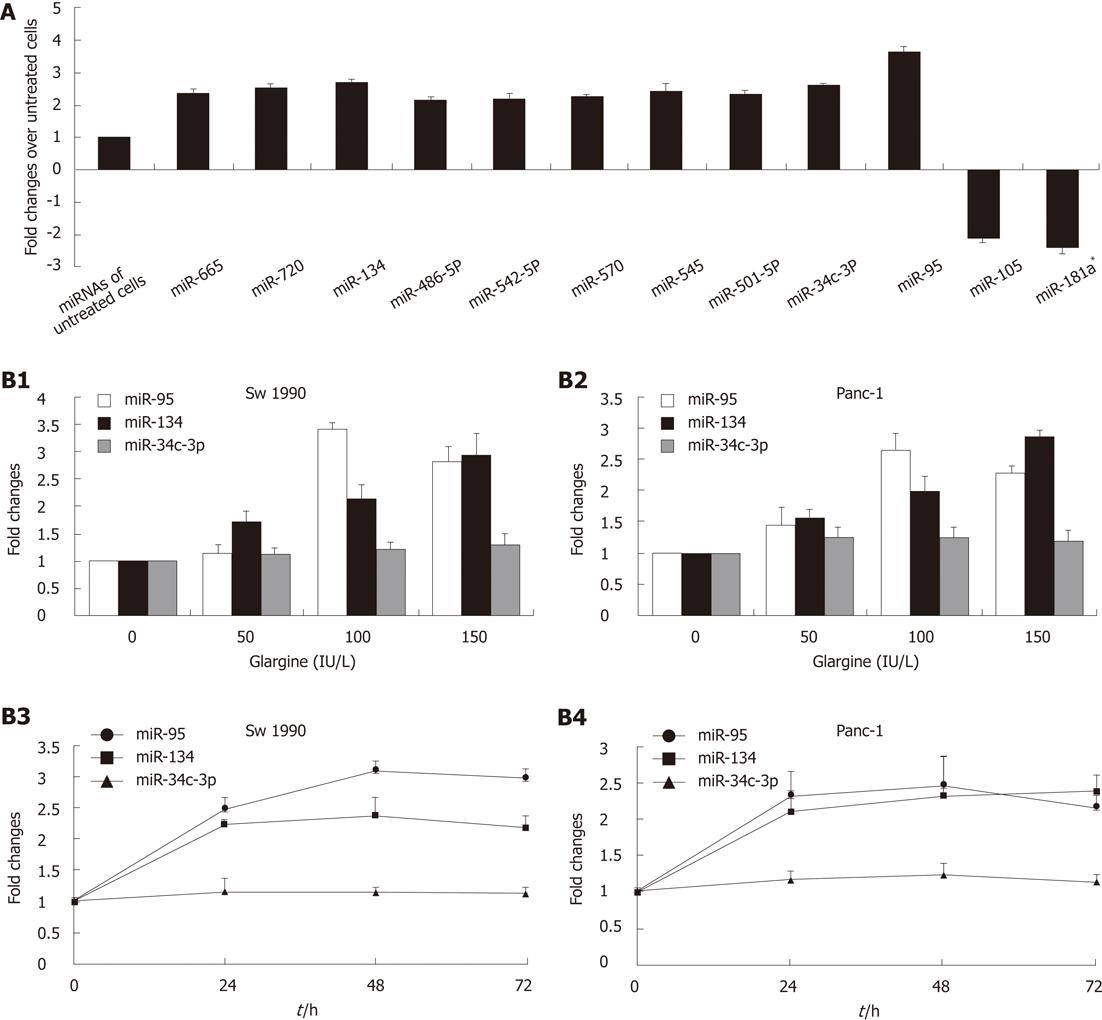
To study the responses of miRNAs to glargine, miRNA real time PCR array analysis of miRNA expression was conducted with total RNAs extracted from Sw1990 pancreatic cells treated with or without 100 IU/L glargine. Differential expression between glargine-treated and non-treated cells was defined using a cut off value of 2-fold change. We observed that 10 miRNAs were significantly up-regulated and 2 miRNAs were significantly down-regulated (2.48-fold on average, P < 0.01) in glargine treated Sw1990 cells when compared with non-treated cells. miR-95, miR-134 and miR-34c-3p are the top three miRNAs regulated by glargine (3.65-fold, 2.67-fold and 2.60-fold changes respectively, P < 0.01) in Sw1990 cells (Figure 1A).
After treatment with increasing concentrations of glargine (50, 100, 150 IU/L) for 48 h, miR-95 was up-regulated by 1.18 fold (P > 0.05), 3.41 fold (P < 0.01) and 2.92 fold (P < 0.01) on average respectively in Sw1990 cells and 1.45 fold (P < 0.01), 3.41 fold (P < 0.01) and 2.92 fold (P < 0.01) on average respectively in Panc-1 cells, when compared with non-treated cells (0 IU/L). No obvious dose dependent responses were observed; miR-134 was up-regulated in a dose dependent manner (Sw1990: 1.69, 2.10 and 2.93 fold on average respectively, P < 0.01; Panc-1: 1.56, 1.99 and 2.88 fold on average respectively, P < 0.01) in both Sw1990 and Panc-1 cells; miR-34c-3p showed no significant changes (Sw1990: 1.03, 1.05 and 1.06 fold on average respectively, P > 0.05; Panc-1: 1.25, 1.25 and 1.19 fold on average respectively, P > 0.05) in both Sw1990 and Panc-1 cells (Figure 1B-1,2).
After treatment with 100 IU/L glargine for different periods (24, 48 and 72 h), miR-95 was up-regulated by 2.50 fold (P﹤0.01), 3.10 fold (P < 0.01) and 2.99 fold (P < 0.01) on average, respectively, in Sw1990 cells and 2.31 fold (P < 0.01), 2.46 fold (P < 0.01) and 2.16 fold (P < 0.01) on average, respectively, in Panc-1 cells, when compared with the cells at 0 h; miR-134 was up-regulated by 2.22 fold (P < 0.01), 2.37 fold (P < 0.01), 2.17 fold (P < 0.01) on average, respectively, in Sw1990 cells and 2.10 fold (P < 0.01), 2.31 fold (P < 0.01) and 2.37 fold (P < 0.01) on average, respectively, in Panc-1 cells; miR-34c-3p showed no significant changes (Sw1990: 1.16, 1.14 and 1.13 fold on average respectively, P > 0.05; Panc-1: 1.17,1.24 and 1.13 fold on average, respectively, P > 0.05) in both Sw1990 and Panc-1 cells (Figure 1B-3,4).
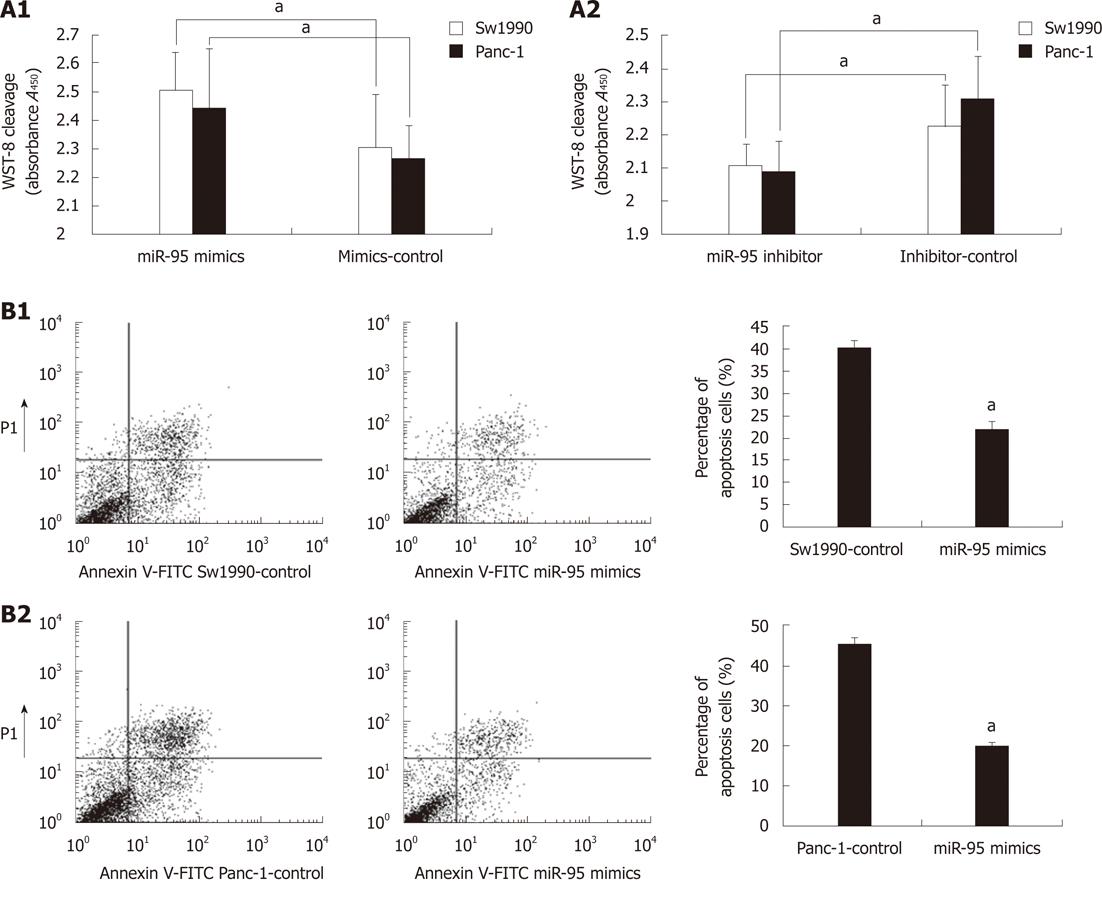
We investigated the potential oncogenic role of miR-95 in Sw1990 and Panc-1 cells. First, we tested miR-95 expression using stem-loop real-time PCR. It increased or decreased after transfected with miR-95 mimics or anti-miR-95 inhibitor. We observed a significant increase in proliferation (Sw1990: 2.51 ± 0.13 vs 2.31 ± 0.19, P < 0.05; Panc-1: 2.44 ± 0.21 vs 2.26 ± 0.12, P < 0.05) after transfection of miR-95 mimics (Figure 2A-1). In contrast, anti-miR-95 inhibitor significantly decreased cell proliferation (Sw1990: 2.11 ± 0.07 vs 2.23 ± 0.13, P < 0.05; Panc-1: 2.09 ± 0.09 vs 2.31 ± 0.13, P < 0.05) (Figure 2A-2). These data indicate that cell proliferation can be significantly promoted by increase of miR-95 expression.
We further investigated the effect of miR-95 on apoptosis and found that apoptosis decreased dramatically (Sw1990: 22.05% ± 1.92% vs 40.32% ± 1.93%, P < 0.05; Panc-1: 20.17% ± 0.85% vs 45.60% ± 1.43%, P < 0.05) in Sw1990 and Panc-1 cells 72 h after transfection with miR-95 mimics (Figure 2B). It suggested that miR-95 may function as a strong apoptotic suppressor in human pancreatic cancer cells.
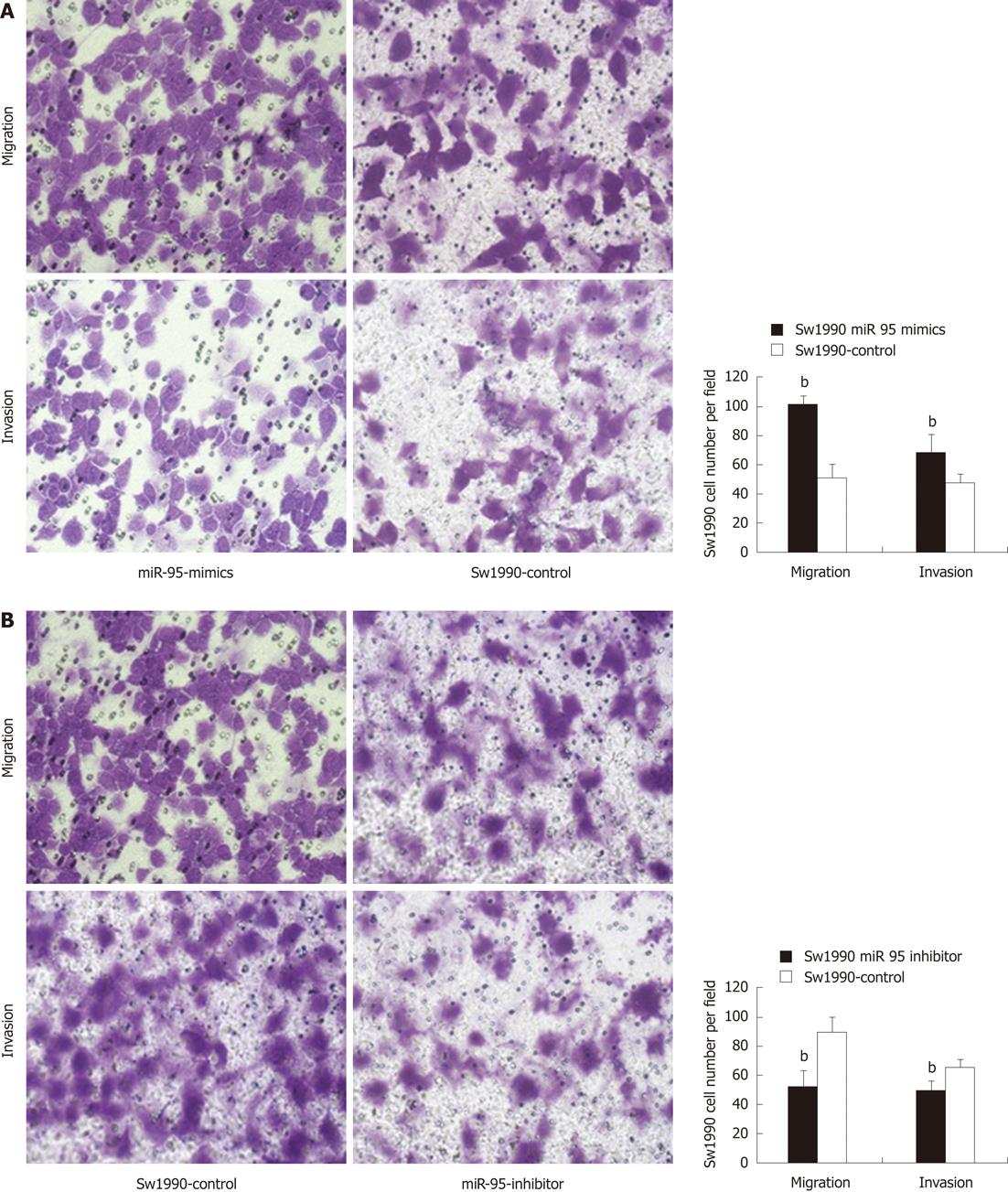
In the cell invasion and migration assay, we observed that depletion of miR-95 significantly impaired the ability of Sw1990 cells to migrate and invade through the matrigel-coated membranes or the non-matrigel-coated membranes towards serum-containing medium (invasion: 49.40 ± 6.59 vs 65.80 ± 5.09; migration: 52.30 ± 10.87 vs 88.90 ± 10.46, P < 0.01), when compared with a paired negative control (Figure 3B); Increased expression of miR-95 significantly promoted the ability of Sw1990 cells to migrate and invade through matrigel-coated membranes or non-matrigel-coated membranes towards serum-containing medium (invasion: 67.90 ± 12.33 vs 47.30 ± 5.89; migration: 101.00 ± 6.00 vs 51.20 ± 8.34, P < 0.01), when compared with the paired negative control (Figure 3A). Similar results were found in Panc-1 cells (depletion of miR-95, invasion: 57.90 ± 10.55 vs 73.80 ± 11.95, migration: 66.40 ± 10.1 vs 99.50 ± 8.85, P < 0.01; forced expression of miR-95, invasion: 37.80 ± 8.93 vs 30.20 ± 5.14; migration: 91.80 ± 9.22 vs 81.50 ± 7.47, P < 0.01) (Figure 4A and B). These results indicated that miR-95 may be important in the progression of pancreatic cancer through increasing cell invasion and migration.
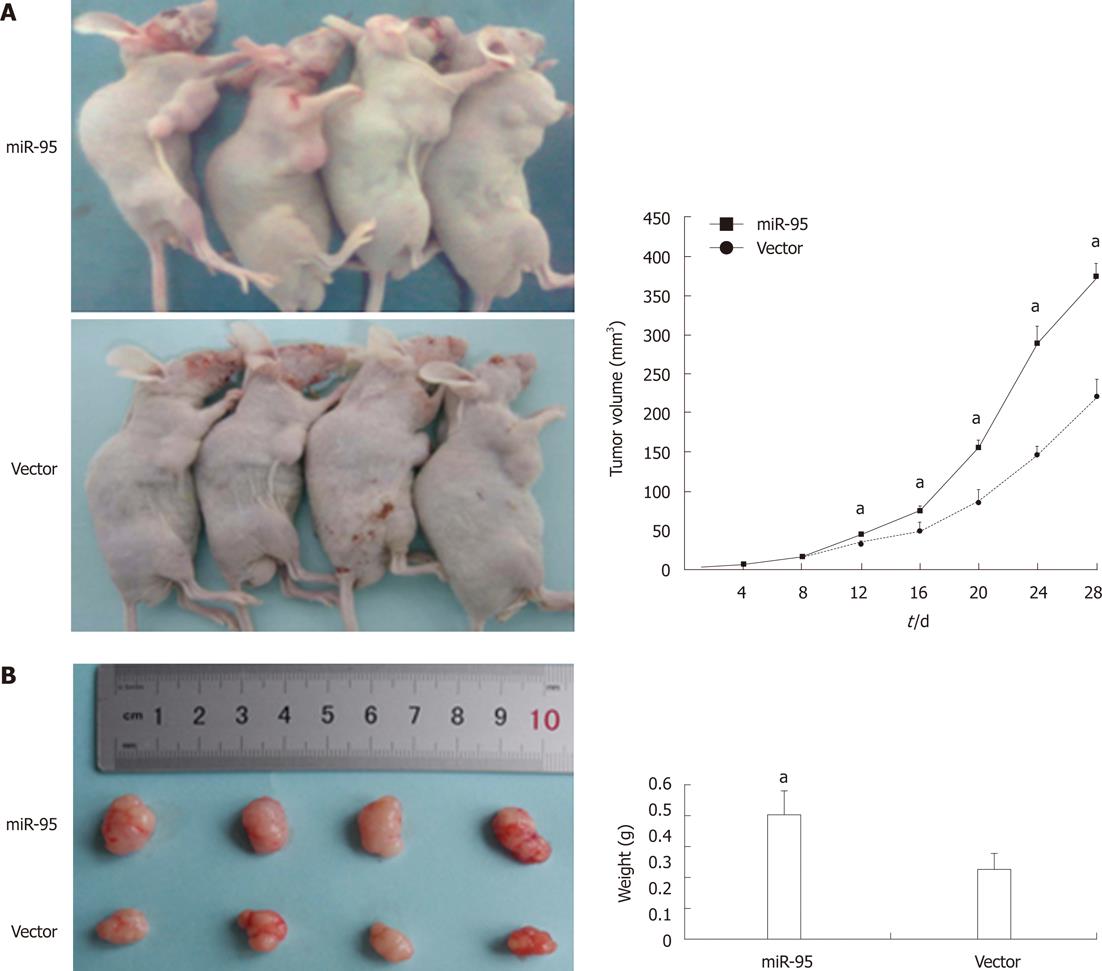
Sw1990 cells transfected with pGLV3-GFP-miR-95 or negative control pGLV3-GFP were injected into the right flank of nude mice. Four weeks later, the tumor volumes of xenografts were 373.82 ± 23.67 mm3 in the miR-95 transfected group and 219.69 ± 17.82 mm3 in the negative control group. The weight of xenografts was 0.40 ± 0.08 g in the miR-95 transfected group and 0.23 ± 0.05 g in the negative control group. miR-95 significantly increased the growth of the Sw1990 xenografts (Figure 5, P < 0.05).
In the present study we demonstrated that glargine altered specific miRNA expression in human pancreatic cells at 50-150 IU/L, which is equivalent to 300-900 nmol and is much higher than the physiological concentration of insulin (0.1-1 nmol). Our miRNA real time PCR array showed that high dose glargine (100 IU/L) up-regulated the expression of 10 miRNAs and down-regulated 2 miRNAs in Sw1990 pancreatic cancer cells. The most obvious change was the apparent increase of miR-95. Stem-loop real-time PCR confirmed the aberrant changes of miR-95 after treatment of high dose glargine in Sw1990 and Panc-1 pancreatic cancer cells. Then miR-95 showed significant anti-apoptotic and growth-promoting effects in vitro and in vivo. Ectopic expression and siRNA knockdown of miR-95 confirmed its invasion-promoting activity in vitro. Therefore, these results highlighted the miR-95-related changes as important effects of glargine.
Recent studies linked the use of glargine with increased risk of cancer. Hemkens et al[4] published a registry study that demonstrated a significantly increased risk of cancer diagnosis associated with high dosages of glargine. However, the Scottish study found a non-significant increased risk for specifically breast cancers[23]. The UK study found no link between glargine and cancer[3]. In addition, although glargine has been shown to increase resistance to apoptosis in several tumor cell lines, administration of glargine didn’t alter proliferation of Colo-357 pancreatic carcinoma cells in vitro[12]. Therefore, all epidemiological and laboratory evidence remains inconclusive and new indicators are needed to determine the role of glargine in carcinogenesis.
miRNAs have been recognized as promising diagnostic and prognostic markers for cancer diagnosis or treatment. For example , miR-34a family members were found to be directly regulated by TP53 and act as tumor suppressors[24]; miR-217 inhibited pancreatic cancer cell growth through targeting KRAS[25]; miR-10b promoted pancreatic cancer invasiveness and correlates with a poor prognosis[26]; The miRNA-200 family (miR-200a, miR-200b, miR-200c, miR-141 and miR-429) were found to inhibit tumor invasion and metastasis by regulating EMT[27]; miR-126 can inhibit cell adhesion, migration and invasion through the suppression of CRK[28]. Our study confirmed, for the first time, that miR-95 and miR-134 were primary glargine-responsive miRNAs.
miR-95 has been shown to be involved in carcinogenesis. A highly characterized example is colorectal carcinoma, in which miR-95 can promote cell proliferation by regulating sorting Nexin 1[29]. In pancreatic cancer, miR-95 is significantly upregulated in most tissues and cell lines[30]. In HeLa cells, inhibition of miR-95 caused a decrease in cell growth[31]. miR-134 gene is located at 14q32, and is involved in several physiological and pathological processes. For example, miR-134 plays an important role in translation-dependent guidance of nerve growth cones[32]; miR-134 is regarded as a potential plasma biomarker for the diagnosis of acute pulmonary embolism[33]; plasma miR-134 in bipolar disorder serves as a potential peripheral marker that can respond to acute manic episodes and is associated with effective mood stabilizer treatment[34]. Interestingly, recent studies indicated that miR-134 may also be involved in carcinogenesis. p53/p63/p73, the tumor suppressors, were believed to be regulators of the miR-134 processing complex[35]. Our study showed that high dose glargine can significantly upregulate the expression of miR-95 (no time or dose dependent manner) and miR-134 (dose dependent manner) in vitro, and for the first time investigated the role of miR-95 in pancreatic cancer.
In conclusion, our study demonstrated that alterations of specific miRNAs and miRNA-related changes were important effects of glargine, suggesting an important and novel new mechanism by which glargine mediates its potent effects on cell growth and apoptosis.
Glargine is widely used in the treatment of type 1 and type 2 diabetes mellitus. Recently, this insulin analogue has been suspected to be associated with an increased risk of cancer, including pancreatic cancer, but available evidence remains inconclusive.
Anti-diabetic therapies have been shown to affect the risk of pancreatic cancer. Metformin use was associated with reduced risk, and insulin or insulin analogue use was suspected to be associated with increased risk of pancreatic cancer in diabetic patients. However, data regarding the effect of glargine, one of the long-acting insulin analogues, on pancreatic cancer are inconsistent. Several researchers believed glargine to be a motigen, not a carcinogen. New biomarkers are needed to determine the role of glargine in the carcinogenesis of pancreatic cancer.
Recent reports have highlighted that miRNAs play important roles in biological processes that affect tumor progression and are promising biomarkers for cancer diagnosis or treatment. This is the first study to show the effects of high dose glargine on miRNA expression in pancreatic cancer cells. miR-95 was proved to be affected by high dose glargine and to be involoved in the carcinogenesis of pancreatic cancer.
By understanding the effects of glargine on pancreatic cancer cells, this study may help to clarify the role of glargine in the progress of pancreatic cancer.
Glargine is a widely used insulin analog in which a 24 h action profile is achieved. MicroRNAs (miRNAs) are endogenous, non-coding small RNAs, 19-25 nucleotides in length. It has been demonstrated that miRNAs play important roles in biological processes that affect tumor progression including migration, invasion and metastasis.
The authors examined the effects of high dose glargine on pancreatic cancer cells in vitro; miR-95 is up-regulated significantly by glargine. miR-95 can significantly increase pancreatic cancer cell proliferation, invasion and migration and inhibit cell apoptosis. Moreover, miR-95 is proved to inhibit pancreatic cancer growth in vivo. The results are interesting and may represent the role of glargine in pancreatic carcinogenesis.
Peer reviewer: Parimal Chowdhury, Professor, Department of Physiology and Biophysics, University of Arkansas for Medical Sciences, 4301 W Markham Street, AR 72205, United States
S- Editor Gou SX L- Editor O’Neill M E- Editor Xiong L
| 1. | Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8283] [Cited by in RCA: 8226] [Article Influence: 483.9] [Reference Citation Analysis (0)] |
| 2. | Warshaw AL, Fernández-del Castillo C. Pancreatic carcinoma. N Engl J Med. 1992;326:455-465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1056] [Cited by in RCA: 1015] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 3. | Currie CJ, Poole CD, Gale EA. The influence of glucose-lowering therapies on cancer risk in type 2 diabetes. Diabetologia. 2009;52:1766-1777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 859] [Cited by in RCA: 848] [Article Influence: 53.0] [Reference Citation Analysis (1)] |
| 4. | Hemkens LG, Grouven U, Bender R, Günster C, Gutschmidt S, Selke GW, Sawicki PT. Risk of malignancies in patients with diabetes treated with human insulin or insulin analogues: a cohort study. Diabetologia. 2009;52:1732-1744. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 463] [Cited by in RCA: 446] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 5. | Gerich JE. Insulin glargine: long-acting basal insulin analog for improved metabolic control. Curr Med Res Opin. 2004;20:31-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 6. | Weinstein D, Simon M, Yehezkel E, Laron Z, Werner H. Insulin analogues display IGF-I-like mitogenic and anti-apoptotic activities in cultured cancer cells. Diabetes Metab Res Rev. 2009;25:41-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 134] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 7. | Kurtzhals P, Schäffer L, Sørensen A, Kristensen C, Jonassen I, Schmid C, Trüb T. Correlations of receptor binding and metabolic and mitogenic potencies of insulin analogs designed for clinical use. Diabetes. 2000;49:999-1005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 574] [Cited by in RCA: 518] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 8. | Li G, Barrett EJ, Wang H, Chai W, Liu Z. Insulin at physiological concentrations selectively activates insulin but not insulin-like growth factor I (IGF-I) or insulin/IGF-I hybrid receptors in endothelial cells. Endocrinology. 2005;146:4690-4696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 116] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 9. | Strasser-Vogel B, Blum WF, Past R, Kessler U, Hoeflich A, Meiler B, Kiess W. Insulin-like growth factor (IGF)-I and -II and IGF-binding proteins-1, -2, and -3 in children and adolescents with diabetes mellitus: correlation with metabolic control and height attainment. J Clin Endocrinol Metab. 1995;80:1207-1213. [PubMed] [DOI] [Full Text] |
| 10. | Michell NP, Dent S, Langman MJ, Eggo MC. Insulin-like growth factor binding proteins as mediators of IGF-I effects on colon cancer cell proliferation. Growth Factors. 1997;14:269-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 11. | Baserga R. The insulin-like growth factor I receptor: a key to tumor growth? Cancer Res. 1995;55:249-252. [PubMed] |
| 12. | Erbel S, Reers C, Eckstein VW, Kleeff J, Büchler MW, Nawroth PP, Ritzel RA. Proliferation of colo-357 pancreatic carcinoma cells and survival of patients with pancreatic carcinoma are not altered by insulin glargine. Diabetes Care. 2008;31:1105-1111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 13. | Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11:252-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 958] [Cited by in RCA: 926] [Article Influence: 61.7] [Reference Citation Analysis (0)] |
| 14. | Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1689] [Cited by in RCA: 1864] [Article Influence: 116.5] [Reference Citation Analysis (0)] |
| 15. | Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27:5959-5974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 546] [Cited by in RCA: 594] [Article Influence: 34.9] [Reference Citation Analysis (0)] |
| 16. | Bandres E, Agirre X, Ramirez N, Zarate R, Garcia-Foncillas J. MicroRNAs as cancer players: potential clinical and biological effects. DNA Cell Biol. 2007;26:273-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 50] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 17. | Baranwal S, Alahari SK. miRNA control of tumor cell invasion and metastasis. Int J Cancer. 2010;126:1283-1290. [PubMed] |
| 18. | Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs--the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009;9:293-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 601] [Cited by in RCA: 623] [Article Influence: 38.9] [Reference Citation Analysis (0)] |
| 19. | Waldman SA, Terzic A. Translating MicroRNA discovery into clinical biomarkers in cancer. JAMA. 2007;297:1923-1925. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 43] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 20. | Bartels CL, Tsongalis GJ. MicroRNAs: novel biomarkers for human cancer. Clin Chem. 2009;55:623-631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 412] [Cited by in RCA: 413] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 21. | Wiggins JF, Ruffino L, Kelnar K, Omotola M, Patrawala L, Brown D, Bader AG. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70:5923-5930. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 543] [Cited by in RCA: 523] [Article Influence: 34.9] [Reference Citation Analysis (0)] |
| 22. | Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901-1908. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 873] [Cited by in RCA: 915] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 23. | Colhoun HM. Use of insulin glargine and cancer incidence in Scotland: a study from the Scottish Diabetes Research Network Epidemiology Group. Diabetologia. 2009;52:1755-1765. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 292] [Cited by in RCA: 276] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 24. | Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745-752. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1580] [Cited by in RCA: 1576] [Article Influence: 87.6] [Reference Citation Analysis (0)] |
| 25. | Zhao WG, Yu SN, Lu ZH, Ma YH, Gu YM, Chen J. The miR-217 microRNA functions as a potential tumor suppressor in pancreatic ductal adenocarcinoma by targeting KRAS. Carcinogenesis. 2010;31:1726-1733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 194] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 26. | Nakata K, Ohuchida K, Mizumoto K, Kayashima T, Ikenaga N, Sakai H, Lin C, Fujita H, Otsuka T, Aishima S. MicroRNA-10b is overexpressed in pancreatic cancer, promotes its invasiveness, and correlates with a poor prognosis. Surgery. 2011;150:916-922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 125] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 27. | Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2843] [Cited by in RCA: 3084] [Article Influence: 181.4] [Reference Citation Analysis (0)] |
| 28. | Crawford M, Brawner E, Batte K, Yu L, Hunter MG, Otterson GA, Nuovo G, Marsh CB, Nana-Sinkam SP. MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem Biophys Res Commun. 2008;373:607-612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 221] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 29. | Huang Z, Huang S, Wang Q, Liang L, Ni S, Wang L, Sheng W, He X, Du X. MicroRNA-95 promotes cell proliferation and targets sorting Nexin 1 in human colorectal carcinoma. Cancer Res. 2011;71:2582-2589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 114] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 30. | Zhang Y, Li M, Wang H, Fisher WE, Lin PH, Yao Q, Chen C. Profiling of 95 microRNAs in pancreatic cancer cell lines and surgical specimens by real-time PCR analysis. World J Surg. 2009;33:698-709. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 243] [Cited by in RCA: 254] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 31. | Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290-1297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1089] [Cited by in RCA: 1158] [Article Influence: 57.9] [Reference Citation Analysis (0)] |
| 32. | Han L, Wen Z, Lynn RC, Baudet ML, Holt CE, Sasaki Y, Bassell GJ, Zheng JQ. Regulation of chemotropic guidance of nerve growth cones by microRNA. Mol Brain. 2011;4:40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 33. | Xiao J, Jing ZC, Ellinor PT, Liang D, Zhang H, Liu Y, Chen X, Pan L, Lyon R, Liu Y. MicroRNA-134 as a potential plasma biomarker for the diagnosis of acute pulmonary embolism. J Transl Med. 2011;9:159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 84] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 34. | Rong H, Liu TB, Yang KJ, Yang HC, Wu DH, Liao CP, Hong F, Yang HZ, Wan F, Ye XY. MicroRNA-134 plasma levels before and after treatment for bipolar mania. J Psychiatr Res. 2011;45:92-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 139] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 35. | Boominathan L. The tumor suppressors p53, p63, and p73 are regulators of microRNA processing complex. PLoS One. 2010;5:e10615. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 151] [Article Influence: 10.1] [Reference Citation Analysis (0)] |