Published online Nov 21, 2010. doi: 10.3748/wjg.v16.i43.5502
Revised: July 20, 2010
Accepted: July 27, 2010
Published online: November 21, 2010
AIM: To study the biological and clinical characteristics of transcription factor forkhead box protein 3 (FOXP3) in hepatocellular carcinoma (HCC).
METHODS: We analyzed the expression and localization of FOXP3 in HCC tissues and cell lines to evaluate its biological features. The relationship between FOXP3 staining and clinical risk factors of HCC was assessed to identify the clinical characteristics of FOXP3 in HCC.
RESULTS: The mRNA and protein expression of FOXP3 were found in some hepatoma cell lines. Immunohistochemical (IHC) analysis of HCC sections revealed that 48% of HCC displayed FOXP3 staining, but we did not find any FOXP3 staining in normal liver tissues and para-tumor tissues. IHC and Confocal analysis showed that the expressions of FOXP3 were mainly present in the nucleus and cytoplasm of tumor cells in tissues or cell lines. In HCC, the distribution of FOXP3 was similar to that of the cirrhosis, but not to the hepatitis B virus. Those findings implicate that FOXP3 staining seems to be associated with the high risk of HCC.
CONCLUSION: The clinical characteristics of FOXP3 in HCC warrants further studies to explore its functions and roles in the cirrhosis and development of HCC.
- Citation: Wang WH, Jiang CL, Yan W, Zhang YH, Yang JT, Zhang C, Yan B, Zhang W, Han W, Wang JZ, Zhang YQ. FOXP3 expression and clinical characteristics of hepatocellular carcinoma. World J Gastroenterol 2010; 16(43): 5502-5509
- URL: https://www.wjgnet.com/1007-9327/full/v16/i43/5502.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i43.5502
Hepatocellular carcinoma (HCC), which consists predominantly of primary liver cancer, is the fifth most common malignancy in men and the eighth one in women worldwide. The number of new cases of HCC is about 564 000 per year[1]. Cirrhosis and virus infection, such as hepatitis B virus (HBV) and hepatitis C virus (HCV), are the major known risk factors for HCC[2,3]. HCC has a poor prognosis and a low survival rate in the majority of patients[4]. The current treatment options of HCC include surgical resection, liver transplantation and local ablative therapy, which are effective only in limited tumors[5]. To improve the treatment of HCC will require a better understanding of the biological development and molecular events in the immune system of HCC.
Forkhead box protein 3 (FOXP3) is a member of the forkhead/winged-helix family of transcriptional regulators and is highly conserved in normal cells. The full-length protein contains 431 amino acids. Foxp3 is considered to be an important gene of thymically derived and naturally occurring regulatory T cells (Tregs)[6]. Mutations in human Foxp3 are associated with immune diseases, such as multi-organ autoimmune disorder, immune dysregulation, polyendocrinopathy, enteropathy and X-linked syndrome (IPEX)[7], in which Tregs from affected patients are greatly reduced in number and suppressive activity[8-10]. A high prevalence of Tregs is thought to be an unfavorable prognostic indicator for HCC[11].
Recent publications described the expression of FOXP3 in pancreatic carcinoma cells, melanoma cells and other tumor cells[12-14]. It has been found that FOXP3 expression was related to the regulation of several cytokines, such as IL-10 and TGF-β2, and FOXP3 might mediate the inhibiting efficacy of tumor cells to escape immune destruction. Those reports implicated that FOXP3 performs its functions in the regulation of tumor progression by expressing not only in Tregs, but also in tumor cells. We assumed that FOXP3 may also be functional in tumor cells of HCC.
This study was designed to investigate whether expression of FOXP3 transcripts and mature protein is related to HCC. We also evaluated the distribution of FOXP3 in human HCC tissues and its relationship with the diagnosis, differentiation and clinical risk factors of HCC.
Normal (8) and cancerous liver tissues (21) were obtained from HCC patients who underwent resection of liver. The circulating HBV markers and ultrasound examination were performed regularly. The quantitative cirrhosis score was derived from the ultrasonographic evaluation of the liver surface, liver parenchyma, caliber of intrahepatic blood vessels and spleen size. Cirrhosis scores of ≥ 7 were used to define cirrhosis[15]. Normal controls were histologically normal tissues obtained from patients who underwent partial hepatectomy for metastatic tumor or liver biopsy. Microarray tissues were obtained from Cybrdi, USA. The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki in a prior approval by the Fourth Military Medical University, China.
Complete medium (RPMI-1640) contained RPMI-1640 supplemented with 2 mmol/L Glutamax, 100 U/mL penicillin, 100 μg/mL streptomycin, and 10 mmol/L HEPES (Invitrogen, USA) and 10% FCS (Thermo Trace, Australia). The following cell lines were obtained from the Biotechnology Center of the Fourth Military Medical University, such as SMMC-7721 and Hepa-G2. All tumor cell lines were maintained in complete RPMI-1640 and passaged using trypsin/EDTA (Invitrogen, USA). Foxp3 transiently transferred 293 cells were established. Melanocytes were freshly prepared when used (derived from normal human skins from the Department of Dermatology of Xijing Hospital of the Fourth Military Medical University).
Total RNA was isolated from HCC cells and melanocyte (as control) using Trizol reagent (Invitrogen, USA). A total of 500 ng RNA was reversely transcribed using the Kit from Takara, Japan. The polymerase chain reaction (PCR) was performed for Foxp3 fragment amplification. The following primers were used (5’-3’): Foxp3 sense: CACAACATGCGACCCCCTTTCACC; Foxp3 antisense: AGGTTGTGGCGGATGGCGTTCTTC. β-actin was used as an internal control for normalization (primer sequences available on request). Semi-quantitative PCR of Foxp3 transcripts was done by comparing the signal intensity of PCR product of Foxp3 gene with that of β-actin gene from the same RNA sample using agarose gel electrophoresis. The intensity of the product band was quantified by densitometric scanning of the gel (Pharmacia Biotech) using “Total image” 1D GEL ANALYSIS software. DNA marker (Takara, Japan) was run in each gel to confirm the size of PCR product.
To examine the protein expression level of FOXP3 in HCC, whole cell lysate was subjected to SDS-PAGE electrophoresis followed by blotting on a nitrocellulose (NC) membrane. During FOXP3 detection, membranes were probed with goat anti-human FOXP3 polyclonal antibody at 4°C overnight followed by incubation with a secondary horseradish peroxidase-conjugated antibody. The mouse anti-human β-actin monoclonal antibody was used as an internal control (R&D, USA). Chemiluminescent detection was done with the enhanced chemiluminescence detection kit (Anmei, China).
To determine the expression levels of FOXP3, hepatoma cell lines were stained for FOXP3 and analyzed by flow cytometry. Cells were washed in PBS containing 1% bovine serum albumin (BSA) and 0.1% NaN3 before antibody staining followed by fixation with 1% paraformaldehyde. Fluorescein isothiocyanate (FITC)-conjugated rat anti-human FOXP3 monoclonal antibody was purchased from eBiosciences, USA. A total of 105 events were collected using Becton Dickinson FACScaliber (Becton Dickinson, USA). Analysis was performed using the WinMDI 2.8 program (Purdue University Cytometry Laboratories, USA).
For double-label immunofluorescence, formalin-fixed cell line slides were treated in 3% hydrogen peroxide in methanol for 10 min. Following three rinses in PBS, the slides were treated in blocking solution (Zhongshan, China) for 1 h, and then incubated with fluorescein isothiocyanate (FITC)-conjugated rat anti-human FOXP3 monoclonal antibody (eBioscience, USA) for another 1 h at room temperature. After rinsed in PBS, the slides were treated with a 1:1000 dilution of diamidino-phenyl-indole (DAPI) (stock solution: 1 mg/mL) (Sigma-Aldrich, USA) for 30 min. DAPI staining was used to visualize nuclei. The slides were mounted with the anti-fade mounting medium. Slides were examined under a Leica TCS-SP laser scanning confocal microscope (Leica, German). All images were collected using a pinhole of 1 Airy.
Paraffin-embedded resected liver cancer specimens were provided by Xijing Hospital tissue bank. Rat anti-human FOXP3 monoclonal antibody was applied to paraffin-embedded sections after microwave antigen retrieval for 10 min in citrate buffer (pH 6.0). Specimens were treated with 0.3% hydrogen peroxide in methanol for 15 min after incubation with the primary antibody to block endogenous peroxidase activity. The secondary antibody of horseradish peroxidase-labeled goat anti-rat antibody (Zhongshan, China) was incubated for 1 h. These slides were examined systematically using an image analyzer system (Olympus BH-2 microscope, Japan).
Slides were reviewed under light microscopy by two pathologists separately. Semi-quantitative analysis of FOXP3 staining was assessed as 0, 1+, 2+, and 3+ as previously established[16,17]. Grade 0 was defined as the complete absence or weak FOXP3 staining in < 1% of the tumor cells; grade 1+ was focal FOXP3 staining in 1%-10% of tumor cells; grade 2+ was positive FOXP3 staining in 11%-50% of tumor cells; and grade 3+ was positive FOXP3 staining in > 50% of tumor cells. A global assessment of the entire tumor was made without selection for the invasive front or areas of active tumor growth. The frequency and semi-quantitative analysis of positive tumors for all regions were calculated for statistical comparisons.
Differences in proportions were compared by a Pearson Chi-square test or Fisher’s exact test as appropriate. The statistical correlation between the grades of HCC differentiation and the staining level of FOXP3 was analyzed by the Cochran-Mantel-Haenszel test. The dependability between the distribution of cirrhosis or HBV infection and FOXP3 expression was evaluated by t test. Differences with a P value less than 0.05 were considered to be statistically significant. All analyses were done using SAS statistical software version 9.1 (Cary, USA).
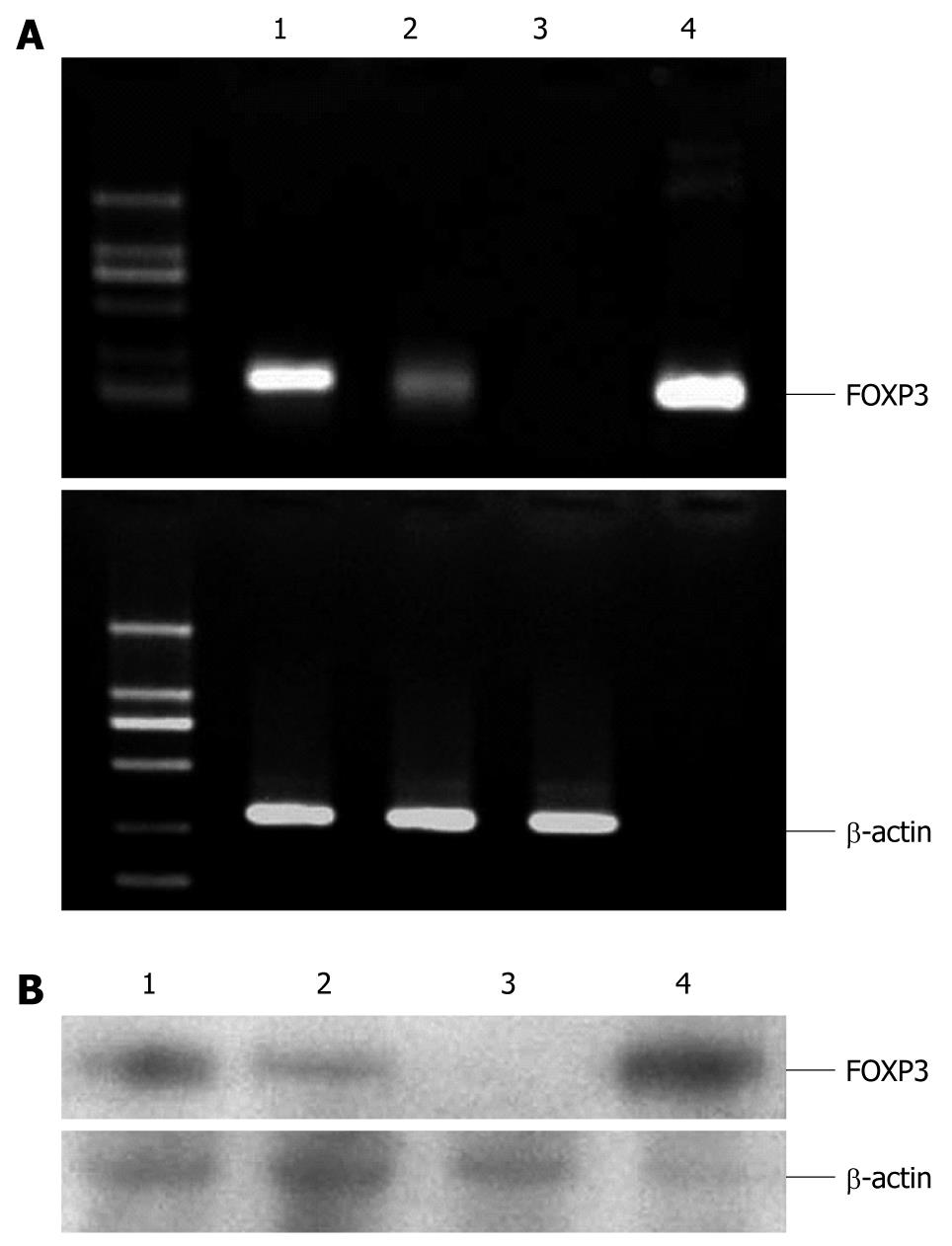
In RT-PCR analysis, Foxp3 was found in some hepatoma cell lines. To further validate the changes of Foxp3, semi-quantitative RT-PCR analysis from two hepatoma cell lines and melanocytes were conducted. The results revealed a significant overexpression of Foxp3 in HCC specimens as compared with nonmalignant melanocytes (Figure 1A). The cell lines shown in Figure 1A (SMMC-7721 and Hepa-G2) were uniformly positive for Foxp3 transcription. But we did not find any Foxp3 transcription in melanocytes as expected. Foxp3 full length plasmid was constructed and performed as a positive control.
Furthermore, as shown in Figure 1B, FOXP3 protein expression could be detected in hepatoma cell lines by Western blotting. Because it was the first time to demonstrate FOXP3 expression in liver cells, we confirmed the staining results in Western blotting with two different anti-FOXP3 antibodies (eBioscience and R&D, USA). We found that the intensity of FOXP3 expression varied in different cell lines. In this experiment, Foxp3 transiently transferred 293 cells were established as a positive control cell line.
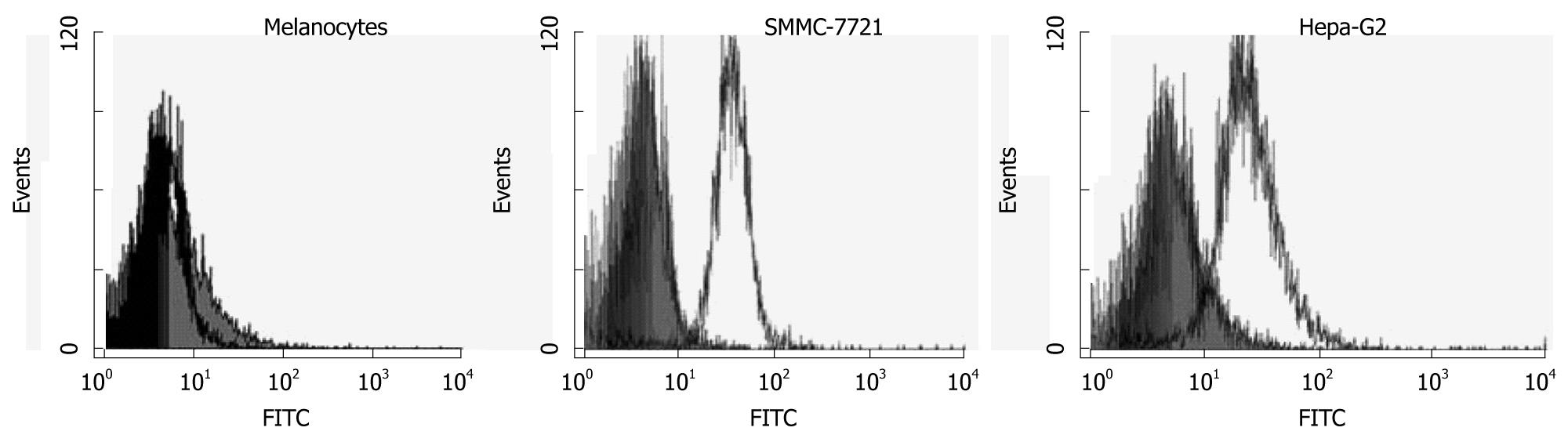
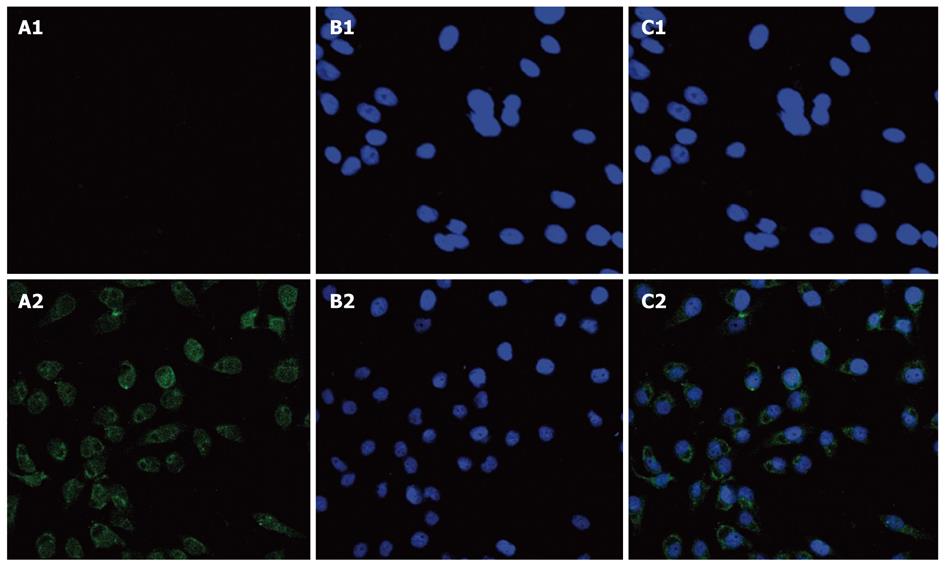
FOXP3 FITC-staining resulted in a shift in the fluorescence of the entire population of hepatoma cell lines compared with the isotype control in the results of flow cytometry. In contrast, melanocytes did not express FOXP3, as shown in Figure 2. Confocal microscopy was used to examine the distribution of FOXP3 in hepatoma cell lines and melanocytes. In these experiments, the nuclei were stained with DAPI to facilitate analysis. The results showed that SMMC-7721 cells exhibited intense nucleic and less cytoplasmic FOXP3 expression (Figure 3).
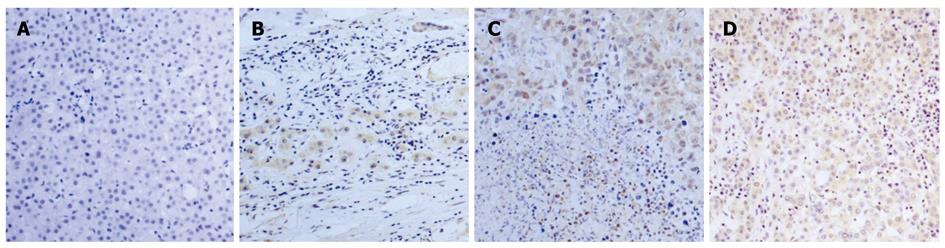
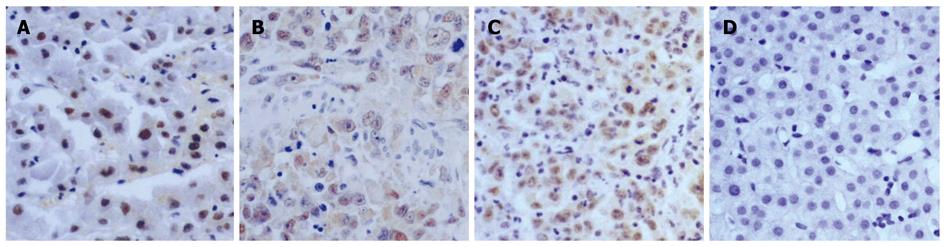
IHC was used to analyze the protein expression and localization of FOXP3 in HCC tissues. FOXP3 staining was done on a set of 29 tissue sections (Table 1) and a tissue array containing 154 cores (Table 2). Those samples were selected from normal and cancerous HCC tissues. Pathologists evaluated the expression level of FOXP3 in different HCC tissue samples according to the percentage of FOXP3 staining (Figure 4). Ten of 21 (48%) HCC tissue sections displayed FOXP3 staining. Interestingly, we noticed that FOXP3 was mainly localized in the nucleus of well differentiated HCC tissues and cytoplasm of moderately and poorly differentiated HCC tissues in tissue sections and array. To confirm the validity of the observation, we used two different anti-human FOXP3 antibodies (eBioscience and R&D, USA) to repeat the experiments. Both antibodies gave similar patterns about FOXP3 localization in HCC tissues.
| Demographic or clinical characteristic | No. of tumorspecimens (n = 21) | FOXP3 | P | FOXP3 immunohistochemistry intensity score | P | ||||
| Positive | Negative | 0 | 1 | 2 | 3 | ||||
| Gender | |||||||||
| Male | 20 | 10 (50) | 10 (50) | 1.00001 | 10 (50) | 7 (35) | 3 (15) | 0 (0) | 1.00001 |
| Female | 1 | 0 (0) | 1 (100) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | ||
| Age (yr) | |||||||||
| > 60 | 4 | 3 (75) | 1 (25) | 0.31081 | 1 (25) | 1 (25) | 2 (50) | 0 (0) | 0.10311 |
| ≤ 60 | 17 | 7 (41) | 10 (59) | 10 (59) | 6 (35) | 1 (6) | 0 (0) | ||
| Differentiation grade | |||||||||
| Well | 7 | 4 (57) | 3 (43) | 1.00001 | 3 (43) | 3 (43) | 1 (14) | 0 (0) | 1.00001 |
| Moderate | 7 | 3 (43) | 4 (57) | 4 (57) | 2 (29) | 1 (14) | 0 (0) | ||
| Poor | 7 | 3 (43) | 4 (57) | 4 (57) | 2 (29) | 1 (14) | 0 (0) | ||
| Tumor | 21 | 10 (48) | 11 (52) | 0.02651a | 11 (52) | 7 (33) | 3 (15) | 0 (0) | |
| Normal | 8 | 0 (0) | 8 (100) | 8 (100) | 0 (0) | 0 (0) | 0 (0) | ||
| Demographic or clinical characteristic | No. of tumorspecimens (n = 140) | FOXP3 | P | FOXP3 immunohistochemistry intensity score | P | ||||
| Positive | Negative | 0 | 1 | 2 | 3 | ||||
| Gender | |||||||||
| Male | 119 | 28 (24) | 91 (76) | 0.61932 | 91 (76) | 20 (17) | 7 (6) | 1 (1) | 0.48951 |
| Female | 21 | 6 (28) | 15 (72) | 15 (72) | 3 (14) | 3 (14) | 0 (0) | ||
| Age (yr) | |||||||||
| > 60 | 30 | 9 (30) | 21 (70) | 0.41032 | 21 (70) | 4 (13) | 5 (17) | 0 (0) | 0.15421 |
| ≤ 60 | 110 | 25 (23) | 85 (77) | 85 (77) | 19 (17) | 5 (5) | 1 (1) | ||
| Differentiation grade | |||||||||
| Well | 32 | 11 (34) | 21 (66) | 0.24102 | 21 (66) | 8 (25) | 2 (6) | 1 (3) | 0.28871 |
| Moderate | 73 | 14 (19) | 59 (81) | 59 (81) | 8 (11) | 6 (8) | 0 (0) | ||
| Poor | 35 | 9 (26) | 26 (74) | 26 (74) | 7 (20) | 2 (6) | 0 (0) | ||
| Tumor | 140 | 34 (24) | 106 (76) | 0.04051a | 106 (76) | 23 (16) | 10 (7) | 1 (1) | |
| Normal | 14 | 0 (0) | 14 (100) | 14 (100) | 0 (0) | 0 (0) | 0 (0) | ||
The expression of FOXP3 at each histopathological grade were examined and recorded in all the patients. It is interesting that many cancerous patients were found to be FOXP3 positive, whereas the intensity of FOXP3 staining was low. The proportion of FOXP3 staining cells had no differences among the histopathological grades in HCC tissue array. In this experiment, 11 of 32 grade I (34%), 14 of 73 grade II (19%), and 9 of 35 grade III (26%) carcinoma cores (Figure 5) were positive in FOXP3 staining (P = 0.2410, Pearson χ2 test; Table 2). This result implicated that FOXP3 staining might be a useful prognostic factor, but not a clue in differentiation of HCC patients. We also found that normal liver tissues were devoid of FOXP3 expression (Figures 4 and 5). In addition, there was no relationship between FOXP3 expression and gender or age, no matter in tissue section or array (Tables 1 and 2).
HCC is a heterogeneous tumor. Cirrhosis, HBV and HCV infections represent the major known risk factors for HCC. Those factors could cooperate or act alone to promote the incidence of HCC depending on the different pathways and molecules.
We detected the circulating HBV markers and liver cirrhosis levels to analyze the relationship between the expression of FOXP3 and HBV/cirrhosis to evaluate the potential clinical characteristics of FOXP3.
As shown in Table 3, among the HCC patients, 42.9% HBV infection and 33.3% cirrhosis of the section samples were FOXP3 positive. Statistical results revealed that FOXP3 expression coincided with the occurrence of cirrhosis (P = 0.092), while it is highly significant with HBV infection (P = 0.021) in HCC patients.
Although the factors and molecular events associated with the progression of HCC are complex and not well established, FOXP3 has been shown to play an important role in Tregs in HCC invasion[1,5,11]. In this study, we assessed the expression and subcellular localization of FOXP3 in hepatoma cell lines and HCC tissues to identify the fact that FOXP3 is expressed by tumor cells. We also identified some clinical characteristics of FOXP3 in HCC.
Few studies assessing FOXP3 expression in tumor tissues and cell lines have been reported. Hinz et al[12] described for the first time the expression and function of FOXP3 in pancreatic ductal adenocarcinoma cells and tissues. They detected FOXP3 expression in tumor cells of 24/39 patients with pancreatic carcinoma. Although they were unable to find a correlation between FOXP3 expression and tumor stage or survival rate, their findings indicate that pancreatic carcinoma cells share growth suppressive effects with Tregs depending on the function of FOXP3. Their results also suggest that FOXP3 may promote tumor cells to mimic characters of Tregs to represent an immune evasion function in microenvironment. Ebert et al[13] and Karanikas et al[14] reported that FOXP3 transcription factor was expressed by melanoma cells and lots of tumor cells. These evidences suggest that FOXP3 is related to tumor escape and can be used as a potential tumor antigen.
In agreement with their findings, Foxp3 transcription and protein expression were found in hepatoma cell lines (SMMC-7721 and Hepa-G2) in this study. After Confocal analysis, we noticed that nuclear and less cytoplasm FOXP3 expression was more prevalent in hepatoma cell lines. To our knowledge, this study provides the first evidence of nuclear and cytoplasm localization of FOXP3 in hepatoma cell lines. In addition, we found that FOXP3 was mainly expressed in the nucleus in well differentiated HCC tissues and cytoplasm in moderately and poorly differentiated HCC tissues. It implicates that the factors (such as cytokines, immune cells, ligand and receptor of tumor cells) coming from microenvironment and differentiation of tumor might induce a change in the subcellular localization of FOXP3 between cytoplasmic and nuclear expression patterns, which would result from post-translational modifications[18]. Therefore, the heterogeneous subcellular localization of FOXP3 in hepatoma cell lines and HCC tissues may reflect the different post-translationally modified forms of FOXP3. In particular, previous reports revealed the interaction of FOXP3 with nuclear factor of activated T cells (NFAT, a transcription factor) in Tregs[19]. It revealed that FOXP3 plays a key role in the formation of nuclear complexes that are important to regulate the transcription of functional genes[20].
In this study, we also found some clinical characteristics of FOXP3 in HCC. The statistical results revealed that HBV infection had different distribution patterns compared with the expression of FOXP3 in HCC. In our previous study, FOXP3 was not found in the hepatitis and normal liver cells. Therefore, we may draw a conclusion that FOXP3 of tumor cells has little relationship with the inflammation induced by HBV infection alone. But because of the limited number of patients, this still needs further studies to validate. On the other hand, 70% of FOXP3 positive HCC cases were companied with liver cirrhosis. This indicates that the pathogenesis and molecules of cirrhosis may have some effects on the regulation of FOXP3 expression. FOXP3 may be involved in the progression of cirrhosis-induced HCC.
On the contrary, Zou et al[21,22] reported that functional somatic mutations and down-regulation of the Foxp3 gene were commonly found in human breast cancer tissues. The expression of FOXP3 was correlated with HER-2/ErbB2 and SKP-2 overexpression. So the function and mechanism of FOXP3 may be different in various tumor cells, which still remains to be elucidated.
In conclusion, this is the first report of FOXP3 staining in hepatoma cell lines and HCC tissues, creating a new focus that FOXP3 is widely expressed in tumor cells and tissues. As the molecular mechanisms of FOXP3 still remain unclear, the real function of FOXP3 in different tumors need to be further studied so as to understand the critical molecular events associated with tumor progression.
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors. It is highly resistant to many conventional treatments and has a poor prognosis. forkhead box protein 3 (FOXP3) is considered to play an important role in naturally occurring regulatory T cells (Tregs). Recent reports have implicated that FOXP3 might have different effects on tumor cells. But the expression and mechanism of FOXP3 in HCC cells remain unclear.
Few reports showed the expression of FOXP3 in pancreatic carcinoma cells, melanoma cells and other tumor cells. The reports found that FOXP3 expression was related to the regulation of several cytokines, such as IL-10 and TGF-β2. FOXP3 might mediate the immune inhibiting efficacy of tumor cells to escape immune destruction. The authors assumed that FOXP3 may also be functional in HCC.
Interestingly, the authors found that hepatoma cell lines expressed FOXP3 mainly in nuclear and cytoplasm. But FOXP3 was almost expressed in nuclear in well differentiated HCC tissues and cytoplasm in moderately and poorly differentiated HCC tissues. It suggests that the microenvironment and differentiation of tumor might induce a change in the subcellular localization of FOXP3 between cytoplasmic and nuclear expression patterns. Therefore, the heterogeneous subcellular localization of FOXP3 in hepatoma cell lines and tissues may reflect the different post-translationally modified forms of FOXP3. The authors also found that the distribution of FOXP3 was similar to that of the cirrhosis, but not to that of HBV infection in HCC. Although the function of FOXP3 in HCC is not clear, the result suggests that FOXP3 may be involved in the cirrhosis-induced HCC. Altogether, those findings implicate that FOXP3 staining seems to be associated with a high risk in HCC.
This study is creating a new focus that FOXP3 is widely expressed in HCC cells and tissues. However, the real function of FOXP3 in HCC needs to be further studied so as to understand the critical molecular events in HCC progression.
The manuscript by Wang et al provides evidences supporting FOXP3 expression both in human hepatoma cells and in HCC tumors. Since this is the first time that expression of FOXP3 has been reported in liver cancer, the information is of interest.
Peer reviewer: Fernando J Corrales, Associate Professor of Biochemistry, Division of Hepatology and Gene Therapy, Proteomics Laboratory, CIMA, University of Navarra, Avd. Pío XII, 55, Pamplona 31008, Spain
S- Editor Wang JL L- Editor Ma JY E- Editor Ma WH
| 1. | Bosch FX, Ribes J, Díaz M, Cléries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127:S5-S16. |
| 3. | Fattovich G, Stroffolini T, Zagni I, Donato F. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology. 2004;127:S35-S50. |
| 4. | Korangy F, Ormandy LA, Bleck JS, Klempnauer J, Wilkens L, Manns MP, Greten TF. Spontaneous tumor-specific humoral and cellular immune responses to NY-ESO-1 in hepatocellular carcinoma. Clin Cancer Res. 2004;10:4332-4341. |
| 5. | Ormandy LA, Hillemann T, Wedemeyer H, Manns MP, Greten TF, Korangy F. Increased populations of regulatory T cells in peripheral blood of patients with hepatocellular carcinoma. Cancer Res. 2005;65:2457-2464. |
| 6. | Hori S, Sakaguchi S. Foxp3: a critical regulator of the development and function of regulatory T cells. Microbes Infect. 2004;6:745-751. |
| 7. | Tanswell P, Garin-Chesa P, Rettig WJ, Welt S, Divgi CR, Casper ES, Finn RD, Larson SM, Old LJ, Scott AM. Population pharmacokinetics of antifibroblast activation protein monoclonal antibody F19 in cancer patients. Br J Clin Pharmacol. 2001;51:177-180. |
| 8. | Scott AM, Wiseman G, Welt S, Adjei A, Lee FT, Hopkins W, Divgi CR, Hanson LH, Mitchell P, Gansen DN. A Phase I dose-escalation study of sibrotuzumab in patients with advanced or metastatic fibroblast activation protein-positive cancer. Clin Cancer Res. 2003;9:1639-1647. |
| 9. | Yagi H, Nomura T, Nakamura K, Yamazaki S, Kitawaki T, Hori S, Maeda M, Onodera M, Uchiyama T, Fujii S. Crucial role of FOXP3 in the development and function of human CD25+CD4+ regulatory T cells. Int Immunol. 2004;16:1643-1656. |
| 10. | Allan SE, Passerini L, Bacchetta R, Crellin N, Dai M, Orban PC, Ziegler SF, Roncarolo MG, Levings MK. The role of 2 FOXP3 isoforms in the generation of human CD4+ Tregs. J Clin Invest. 2005;115:3276-3284. |
| 11. | Kobayashi N, Hiraoka N, Yamagami W, Ojima H, Kanai Y, Kosuge T, Nakajima A, Hirohashi S. FOXP3+ regulatory T cells affect the development and progression of hepatocarcinogenesis. Clin Cancer Res. 2007;13:902-911. |
| 12. | Hinz S, Pagerols-Raluy L, Oberg HH, Ammerpohl O, Grüssel S, Sipos B, Grützmann R, Pilarsky C, Ungefroren H, Saeger HD. Foxp3 expression in pancreatic carcinoma cells as a novel mechanism of immune evasion in cancer. Cancer Res. 2007;67:8344-8350. |
| 13. | Ebert LM, Tan BS, Browning J, Svobodova S, Russell SE, Kirkpatrick N, Gedye C, Moss D, Ng SP, MacGregor D. The regulatory T cell-associated transcription factor FoxP3 is expressed by tumor cells. Cancer Res. 2008;68:3001-3009. |
| 14. | Karanikas V, Speletas M, Zamanakou M, Kalala F, Loules G, Kerenidi T, Barda AK, Gourgoulianis KI, Germenis AE. Foxp3 expression in human cancer cells. J Transl Med. 2008;6:19. |
| 15. | Kuniholm MH, Lesi OA, Mendy M, Akano AO, Sam O, Hall AJ, Whittle H, Bah E, Goedert JJ, Hainaut P. Aflatoxin exposure and viral hepatitis in the etiology of liver cirrhosis in the Gambia, West Africa. Environ Health Perspect. 2008;116:1553-1557. |
| 16. | Iwasa S, Jin X, Okada K, Mitsumata M, Ooi A. Increased expression of seprase, a membrane-type serine protease, is associated with lymph node metastasis in human colorectal cancer. Cancer Lett. 2003;199:91-98. |
| 17. | Ariga N, Sato E, Ohuchi N, Nagura H, Ohtani H. Stromal expression of fibroblast activation protein/seprase, a cell membrane serine proteinase and gelatinase, is associated with longer survival in patients with invasive ductal carcinoma of breast. Int J Cancer. 2001;95:67-72. |
| 18. | Chen C, Rowell EA, Thomas RM, Hancock WW, Wells AD. Transcriptional regulation by Foxp3 is associated with direct promoter occupancy and modulation of histone acetylation. J Biol Chem. 2006;281:36828-36834. |
| 19. | Wu Y, Borde M, Heissmeyer V, Feuerer M, Lapan AD, Stroud JC, Bates DL, Guo L, Han A, Ziegler SF. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126:375-387. |
| 20. | Marson A, Kretschmer K, Frampton GM, Jacobsen ES, Polansky JK, MacIsaac KD, Levine SS, Fraenkel E, von Boehmer H, Young RA. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445:931-935. |
| 21. | Zuo T, Wang L, Morrison C, Chang X, Zhang H, Li W, Liu Y, Wang Y, Liu X, Chan MW. FOXP3 is an X-linked breast cancer suppressor gene and an important repressor of the HER-2/ErbB2 oncogene. Cell. 2007;129:1275-1286. |
| 22. | Zuo T, Liu R, Zhang H, Chang X, Liu Y, Wang L, Zheng P, Liu Y. FOXP3 is a novel transcriptional repressor for the breast cancer oncogene SKP2. J Clin Invest. 2007;117:3765-3773. |