Published online Aug 7, 2007. doi: 10.3748/wjg.v13.i29.3932
Revised: February 20, 2007
Accepted: April 4, 2007
Published online: August 7, 2007
AIM: To examine Krüppel-like factor 6 (KLF6) mutations in nonpolypoid-type tumors and alterations of K-ras, p53, and B-raf in relation between mutation and morphologic type, particularly nonpolypoid-type colorectal carcinomas.
METHODS: Fifty-five early nonpolypoid colorectal carcinomas were analyzed. Loss of heterozygosity (LOH) of KLF6 and p53 was determined by microsatellite assay. Mutations of KLF6, K-ras, and B-raf were examined by polymerase chain reaction-single-strand conformation polymorphism followed by direct sequencing. In LOH-positive and/or mutation-positive tumors, multiple (4-7) samples in each tumor were microdissected and examined for genetic alterations. p53 expression was evaluated by immunohistochemistry.
RESULTS: LOH of KLF6 and p53 was found in 14 of 29 (48.3%) and 14 of 31 (45.2%) tumors, respectively. In 10 of the 14 (71.4%) KLF6 LOH-positive tumors and 9 of the 14 (64.3%) p53 LOH-positive tumors, LOH was found in all of the microdissected samples. In 1 of the 10 (10.0%) KLF6 LOH-positive tumors, a single missense mutation was identified. K-ras and B-raf mutations were found in 5 of 55 (9.1%) and 6 of 55 (10.9%) tumors, respectively. However, these mutations were detected only in subsets of microdissected tumor samples.
CONCLUSION: These data suggest that KLF6 and p53 mutations are involved in the development of nonpolypoid colorectal carcinoma, whereas K-ras and B-raf mutations are not.
-
Citation: Mukai S, Hiyama T, Tanaka S, Yoshihara M, Arihiro K, Chayama K. Involvement of Krüppel-like factor 6 (
KLF6 ) mutation in the development of nonpolypoid colorectal carcinoma. World J Gastroenterol 2007; 13(29): 3932-3938 - URL: https://www.wjgnet.com/1007-9327/full/v13/i29/3932.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i29.3932
Two distinct types of colorectal carcinomas have been described, polypoid and nonpolypoid[1-4]. The concept of a precursor lesion led to the adenoma-carcinoma sequence hypothesis, based on the polypoid type, whereby a benign adenoma progresses to malignant adenocarcinoma. Nonpolypoid-type colorectal carcinoma was first described in detail by Muto et al[5]. These lesions show subtle morphologic differentiation and are often undetectable, posing diagnostic problems. Nonpolypoid-type tumors frequently invade the submucosal layer, even though they are less than 10 mm in diameter[6]. Because of the high malignancy potential of nonpolypoid-type tumors, these are considered important lesions. Nonpolypoid-type carcinomas are thought to be de novo tumors that develop directly from normal colorectal mucosa.
It has been reported that K-ras mutations and loss of heterozygosity (LOH) at chromosome 3p are more frequent in early polypoid colorectal carcinomas than in nonpolypoid-type tumors[2,3]. In addition, APC mutations are less frequent in nonpolypoid-type tumors than in polypoid-type tumors[2]. Thus, these two types of colorectal tumors may possess different genetic backgrounds.
Krüppel-like factors (KLFs) are core transcription factors that regulate numerous mammalian genes[7]. The KLF6 gene encodes one of these transcription factors, which has been identified as a tumor-suppressor gene involved in the regulation of cell proliferation and differentiation[7]. KLF6 mediates the inhibition of proliferation by upregulating the cell-cycle inhibitor CDKN1A (p21WAF1/CIP1) through an interaction with cyclin D1 in a p53-independent manner[8]. Recently, a high frequency (44%) of KLF6 somatic mutations has been reported in colorectal cancers[9]. However, the relation between these genetic changes and morphologic type, particularly nonpolypoid type, is unclear. In this study, examined KLF6 mutations in early nonpolypoid-type tumors. We also examined alterations of K-ras, p53, and B-raf genes. In addition, we examined genetic alterations in multiple samples from a given tumor obtained by microdissection.
Samples from early nonpolypoid colorectal tumors resected surgically or endoscopically were collected at Hiroshima University Hospital (Hiroshima, Japan) during the period 2000 to 2002. Nonpolypoid-type colorectal carcinomas were diagnosed according to the criteria of Shimoda et al[10]. In brief, nonpolypoid-type tumors were tumors without intramucosal protuberant growth. Ten samples were mucosal adenocarcinomas, and 45 were adenocarcinomas that had invaded the submucosal layer. The present study was approved by the local ethical committee (No. I-Rin-Hi-107).
Tissue sections (4-μm thick) were prepared from formalin-fixed, paraffin-embedded colorectal tissues. The sections were stained with hematoxylin and eosin (HE) for histologic examination. Depth of invasion was classified as intramucosal (m) or submucosal (sm).
Tissue sections (10-μm thick) were stained with HE, dehydrated in a graded ethanol series, and then dried without a cover glass. Tissue samples from 55 tumors and corresponding normal tissues were cut with sterile needles, and the DNA was extracted with 20 μL extraction buffer (100 mmol/L Tris-HCl, pH 8.0, 2 mmol/L EDTA, 400 μg/mL proteinase K) at 55°C overnight. The tubes were boiled for 5 min to inactivate proteinase K, and 1-2 μL of each extract was used for polymerase chain reaction (PCR) amplification.
LOH analysis of KLF6 and p53 was performed by microsatellite assay. Primer pairs specific for microsatellites KLF6 (D10S591 and D10S594) and p53 (TP53) are shown in Table 1[11,12]. The microsatellite assay was performed as described previously[12]. In brief, each 15-μL reaction mixture containing 10-20 ng genomic DNA, 6.7 mmol/L Tris-HCl, pH 8.8, 6.7 mmol/L EDTA, 6.7 mmol/L MgCl2, 0.33 μmol/L primer labeled with [γ32-P]dATP, 0.175 μmol/L unlabeled primer, 1.5 mmol/L of each deoxynucleotide triphosphate, and 0.75 units AmpliTaq Gold DNA polymerase (Perkin-Elmer, Branchburg, NJ) was amplified with 40 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and elongation at 72°C for 30 s. PCR products were separated by electrophoresis on 6% polyacrylamide-8 M urea-32% formamide gels and subjected to autoradiography overnight at -80°C on Fuji RX film. LOH was identified when only one major band was detected in DNA isolated from cancerous tissue; two major bands were present in the normal tissue specimen from the same sample.
| Priming region | Primer sequence | |
| Loss of heterozygosity analysis | ||
| p53 (TP53) | 5′-AGGGATACTATTCAGCCCGAGGTG-3′ | |
| 5′-ACTGCCACTCCTTGCCCCATTC-3′ | ||
| KLF6 (D10S591) | 5′-ACCTCGAAGGTCTGTTCTCC-3′ | |
| 5′-GGCTTTATGGATCATATTAATCCAC-3′ | ||
| KLF6 (D10S594) | 5′-GGGCAGCGTTGCTGAGA-3′ | |
| 5′-GCACCCAGATAGGCATAGAGA-3′ | ||
| Mutation analysis | ||
| K-ras exon 1: | 5′-GGCCTGCTGAAAATGACTGA-3′ | |
| 5′-GGTGCAGGACCATTCTTTGAT-3′ | ||
| B-raf exon 11: | 5′-AAACACTTGGTAGACGGGAC-3′ | |
| 5′-AATGTGGTGACATTGTGACAAGT-3′ | ||
| B-raf exon 15: | 5′-CTTCATGAAGACCTCACAGT-3′ | |
| 5′-TCCACTGATTAAATTTTTGGCC-3′ | ||
| KLF6 | exon 1: | 5′-GCCTCGCCGCCCTCGC-3′ |
| 5′-AACTCCAAACAGCCGACCC-3′ | ||
| exon 2: | 5′-CAATCACGTGCCTTCTCTGG-3′ | |
| 5′-GAGAAAGTGAGGATTTGTCTG-3′ | ||
| exon 3 | 5′-GTCTGCGGGTCAGTGAAGTC-3′ | |
| 5′-GTCATCACATTCCCAAGGCC-3′ | ||
DNA samples were screened for mutations of KLF6, K-ras, and B-raf by PCR-single-strand conformation polymorphism (SSCP) analysis. The PCR primers were designed to amplify the exons including mutational hot spots of KLF6, K-ras, and B-raf and are listed in Table 1[11,13,14]. PCR-SSCP analysis was performed as described previously[12]. In brief, each 25 μL reaction mixture contained 1 × AmpliTaq Gold Buffer (8.0 mmol/L Tris-HCl, pH 8.3, 40 mmol/L KCl; Perkin-Elmer), 4 mmol/L MgCl2, 0.3 mmol/L of each deoxynucleotide triphosphate, 100 pmol of each primer, 10-20 ng genomic DNA, 2.5 mCi [γ32-P]dCTP (3000 Ci/mmol/L, 10 mCi/mL), and 1.25 U AmpliTaq Gold DNA polymerase. The reaction mixtures were heated to 95°C for 10 min, followed by 45 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 2 min, and strand elongation at 72°C for 2 min. After PCR, the samples were electrophoresed on 6% polyacrylamide gels (ratio of acrylamide:bis-acrylamide, 19:1) with 10% glycerol at 4°C. The gels were then subjected to autoradiography overnight at -80°C.
To confirm KLF6, K-ras, and B-raf mutations, direct sequencing was performed as described previously[15]. The aberrantly migrating band on the SSCP gel was removed, reamplified, and directly sequenced on both strands with an ABI PRISM 310 Genetic Analyzer (Perkin-Elmer Applied Biosystems, Foster City, CA). For the sequencing reaction, a PRISM AmpliTaq DNA polymerase FS Ready Reaction Dye Terminator Sequencing Kit (Perkin-Elmer Applied Biosystems) was used.
For mutation- or LOH-positive tumors, multiple (4-7) samples of each tumor were cut out by microdissection with sterile needles. DNA was then extracted and examined for mutations of KLF6, K-ras, and B-raf and for LOH of KLF6 and p53.
p53 expression was evaluated by immunohistochemistry (IHC) using a monoclonal antibody (DO7, diluted 1:50; Novocastra, Newcastle, United Kingdom). Immunohistochemical staining was performed with the labeled streptavidin-biotin method (LSAB kit; Dako, Denmark). After deparaffinization and rehydration, the tissue sections were subjected to microwave treatment for 15 min. Endogenous peroxidase activity was blocked with hydrogen peroxide, and sections were incubated with goat serum at room temperature for 20 min to block nonspecific binding. Primary antibody was applied overnight at 4°C. This was followed by incubation with biotinylated secondary antibody for 20 min and then with a streptavidin-biotin-peroxidase complex reagent for 20 min at room temperature. The sections were washed thoroughly with phosphate-buffered saline between incubation steps. Diaminobenzidine tetrahydrochloride was used as the chromogen, and sections were counter-stained with hematoxylin. Cases with sporadic or no p53 staining were considered negative, whereas focal or diffuse immunoreactivity was considered positive. Immunohistochemical evaluations were performed without any knowledge of clinicopathologic data.
Chi-square and Fischer’s exact probability tests were used for comparisons of clinicopathologic and genetic factors. P < 0.05 was regarded as significant.
Clinicopathologic features of the 55 colorectal tumors are shown in Table 2. We first examined genetic alterations in tissue samples without microdissection. Individual data and summary of genetic analyses are shown in Tables 3 and 4, respectively. LOH of KLF6 was found in 14 of 29 (48.3%) nonpolypoid tumors, including 2 of 6 (33.3%) mucosal tumors and 12 of 23 (52.2%) submucosal tumors. Mutations of KLF6 were detected in 2 of 34 (5.9%) tumors, including none of 6 (0%) mucosal tumors and 2 of 34 (5.9%) submucosal tumors. LOH of p53 was found in 14 of 31 (45.2%) tumors, including 4 of 8 (50.0%) mucosal tumors and 10 of 23 (43.5%) submucosal tumors. p53 overexpression was found in 19 of 55 (34.5%) tumors, including 2 of 10 (20.0%) mucosal tumors and 17 of 45 (37.8%) submucosal tumors. K-ras mutations were found in 5 of 55 (9.1%) tumors, including 1 of 10 (10.0%) mucosal tumors and 4 of 45 (8.9%) submucosal tumors. All of the detected mutations were G13C. B-raf mutations were found in 6 of 55 (10.9%) tumors, including 1 of 10 (10.0%) mucosal tumors and 5 of 45 (11.1%) submucosal tumors. Three of the 6 B-raf mutations were A560C, and the others were A608C, A396T, and G378A. Two samples showed both K-ras and B-raf mutations. In each genetic alteration, there were no significant difference between the frequency in mucosal carcinomas and that in submucosal carcinomas. There were no significant associations between genetic alterations and clinicopathologic characteristics including patients’ sex, tumor location, histology and depth.
| KLF6 | p53 | K-ras | B-raf | |||
| Case | LOH1 | Mutation | IHC2 | LOH4 | Mutation | Mutation |
| Intramucosal cancer | ||||||
| 16 | NI3 | Wild | (-) | NI | GGC to GCC [13, Gly to Ala (1/4)] | Wild |
| 17 | (-) | (-) | Wild | Wild | ||
| 29 | LOH (3/3) | Wild | (-) | Wild | Wild | |
| 30 | LOH (2/4) | Wild | (-) | (-) | Wild | Wild |
| 31 | Wild | (-) | (-) | Wild | Wild | |
| 32 | Wild | (++) | LOH (3/3) | Wild | Wild | |
| 33 | (-) | (+) | LOH (1/3) | Wild | AAA to CAA [560, Lys to Gln (1/3)] | |
| 34 | (-) | (+++) | Wild | Wild | ||
| 35 | Wild | (-) | LOH (1/3) | Wild | Wild | |
| 36 | (-) | (-) | LOH (1/4) | Wild | Wild | |
| Submucosal cancer | ||||||
| 1 | LOH (1/4) | Wild | (-) | Wild | Wild | |
| 2 | Wild | (-) | (-) | Wild | Wild | |
| 3 | NI | Wild | (+) | GGC to GCC [13, Gly to Ala (1/4)] | CAT to CTT [396, His to Leu (1/4)] | |
| 4 | (-) | (+++) | LOH (1/5) | Wild | Wild | |
| 5 | Wild | (+++) | (-) | Wild | Wild | |
| 6 | NI | Wild | (-) | LOH (2/3) | Wild | Wild |
| 7 | NI | (+++) | (-) | Wild | Wild | |
| 8 | LOH (2/3) | Wild | (++) | (-) | Wild | Wild |
| 9 | NI | Wild | (+++) | Wild | Wild | |
| 10 | NI | Wild | (-) | LOH (2/3) | Wild | Wild |
| 11 | NI | (+++) | (-) | Wild | Wild | |
| 12 | NI | (+++) | LOH (2/4) | Wild | Wild | |
| 13 | LOH (3/7) | Wild | (+++) | (-) | Wild | Wild |
| 14 | NI | Wild | (-) | (-) | Wild | Wild |
| 15 | LOH (3/4) | Wild | (+++) | Wild | Wild | |
| 18 | (-) | (+) | Wild | Wild | ||
| 19 | NI | (-) | Wild | Wild | ||
| 20 | Wild | (-) | GGC to GCC [13, Gly to Ala (1/4)] | CAG to CCG [608, Glu to Pro (1/4)] | ||
| 21 | NI | Wild | (++) | (-) | Wild | GTG to ATG [378, Val to Met (1/3)] |
| 22 | LOH (2/4) | CTG to GCT [735, Leu to Ala (2/4)] | (++) | Wild | Wild | |
| 23 | NI | CTG to GCT [735, Leu to Ala (2/4)] | (+) | (-) | Wild | Wild |
| 24 | NI | Wild | (-) | (-) | GGC to GCC [13, Gly to Ala (1/5)] | Wild |
| 25 | (+) | Wild | AAA to CAA [560, Lys to Gln (1/5)] | |||
| 26 | (-) | Wild | (-) | Wild | AAA to CAA [560, Lys to Gln (2/6)] | |
| 27 | (-) | (-) | NI | Wild | Wild | |
| 28 | Wild | (-) | Wild | Wild | ||
| 37 | (-) | (-) | LOH (4/4) | Wild | Wild | |
| 38 | (-) | (+++) | LOH (4/4) | Wild | Wild | |
| 39 | (-) | (-) | Wild | Wild | ||
| 40 | (-) | (++) | (-) | Wild | Wild | |
| 41 | Wild | (-) | Wild | Wild | ||
| 42 | (-) | Wild | Wild | |||
| 43 | (-) | (++) | Wild | Wild | ||
| 44 | LOH (3/3) | Wild | (-) | NI | Wild | Wild |
| 45 | LOH (1/5) | Wild | (+++) | LOH (5/5) | Wild | Wild |
| 46 | (-) | (+++) | (-) | Wild | Wild | |
| 47 | LOH (3/3) | Wild | (-) | (-) | Wild | Wild |
| 48 | LOH (2/4) | Wild | (-) | Wild | Wild | |
| 49 | Wild | (-) | (-) | GGC to GCC [13, Gly to Ala (1/4)] | Wild | |
| 50 | Wild | (-) | LOH (3/5) | Wild | Wild | |
| 51 | (-) | (-) | (-) | Wild | Wild | |
| 52 | LOH (3/4) | Wild | (-) | Wild | Wild | |
| 53 | (-) | LOH (1/4) | Wild | Wild | ||
| 54 | LOH (1/5) | Wild | (-) | Wild | Wild | |
| 55 | LOH (1/4) | Wild | (+++) | LOH (3/4) | Wild | Wild |
| Genetic alteration | Frequency | ||
| Mucosal cancer | Submucosal cancer | Total | |
| KLF6, LOH1 (+) | 2/6 (33.3%) | 12/23 (52.2%) | 14/29 (48.3%) |
| KLF6, mutation | 0/6 (0%) | 2/28 (7.1%) | 2/34 (5.9%) |
| p53, LOH (+) | 4/8 (50.0%) | 10/23 (43.5%) | 14/31 (45.2%) |
| p53, overexpression | 2/10 (20.0%) | 17/45 (37.8%) | 19/55 (34.5%) |
| K-ras, mutation | 1/10 (10.0%) | 4/45 (8.9%) | 5/45 (9.1%) |
| B-raf, mutation | 1/10 (10.0%) | 5/45 (11.1%) | 6/55 (10.9%) |
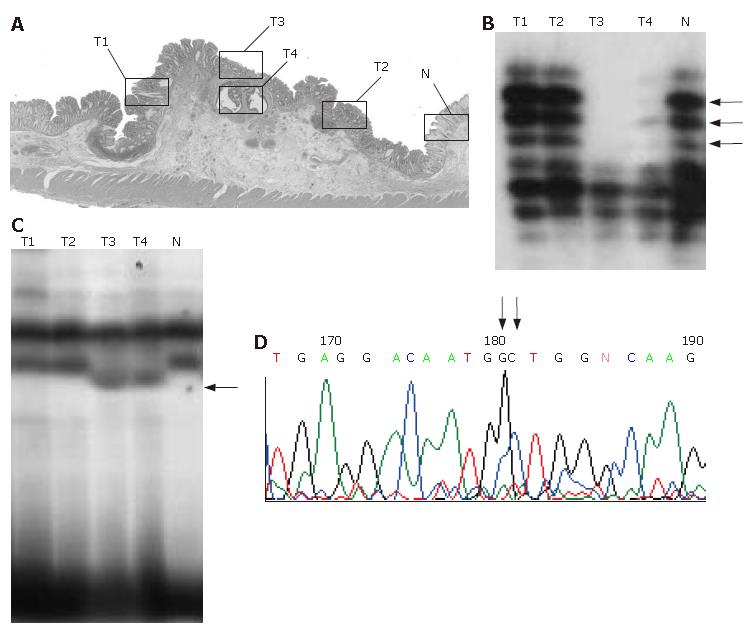
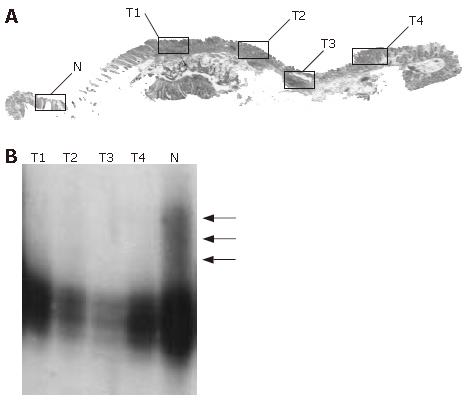
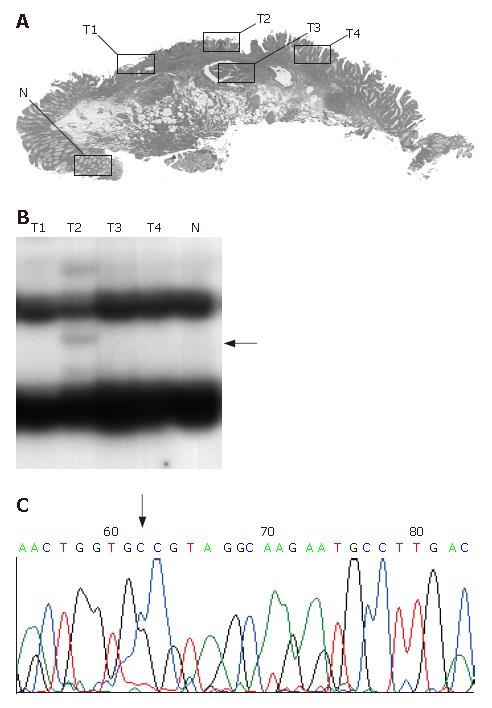
In mutation- or LOH-positive tumors, we next examined genetic alterations by microdissection. In the 10 of the 14 (71.4%) KLF6 LOH-positive tumors, LOH was found in all of the microdissected tumor samples. In 1 of 14 KLF6 LOH-positive tumors, case 22, a missense mutation (exon 3, codon 735, CTG to GCT) was detected (Figure 1). In 9 of the 14 (64.3%) p53 LOH-positive tumors, LOH was found in all of the microdissected tumor samples (Figure 2). In both of the K-ras and B-raf mutation-positive cases, the mutations were detected in only a subset of microdissected tumor samples (Figure 3).
Results of this study suggest that KLF6 and p53 mutations are involved in the development of nonpolypoid colorectal carcinoma, whereas K-ras and B-raf mutations are not. KLF6 encodes a nuclear protein with three zinc fingers at the C terminus a serine/threonine-rich central region, and an acidic domain in the N-terminal region. The zinc fingers are responsible for specific binding to guanine-rich core promoter elements. The central region may be involved in activation or posttranslational regulatory pathways, and the acidic N-terminal domain may be involved in transcriptional activation. The DNA binding and transcriptional activity of this protein, in conjunction with its expression pattern, suggests that it may participate in the regulation and/or maintenance of the basal expression of pregnancy-specific glycoprotein genes and possibly other TATA box-less genes[7-9].
K-ras encodes a 21-kD plasma membrane-bound guanosine triphosphate-binding protein that is a key regulatory component of signal transduction pathways that transmit growth stimulatory signals from cell surface receptors to intracellular targets[16]. K-ras mutations are found in approximately 30% of colorectal cancers[17]. The majority of the mutations involve a single amino acid substitution at codon 12 or 13, which decreases the intrinsic guanosine triphosphatase activity, which leads to the constitutive activation of the K-ras signaling pathway. Interestingly, K-ras mutations are associated with polypoid-type colorectal tumors.
The raf family, which includes A-raf, B-raf, and raf-1, comprises serine/threonine kinases that are regulated by binding to ras. K-ras mutations lead to activation of this signaling pathway and to malignant transformation[18]. B-raf mutations are observed in malignant melanoma, colorectal cancers, and ovarian borderline tumors. They are mutually exclusive with ras mutations[14,19]. B-raf mutations are observed in approximately 10% of colorectal carcinomas[19]. Mutations occur in two regions of the kinase domain, in exon 15 (the activation segment, which protects the substrate-binding site), and less commonly, in exon 11 (the G loop, which mediates ATP binding). Mutated B-raf protein shows increased kinase activity and can transform NIH3T3 cells[14]. B-raf mutations are also associated with polypoid-type colorectal tumors.
In the present study of nonpolypoid tumors, frequent alterations of KLF6 and p53 were found. In more than 60% of the cases, these alterations were found in all of the microdissected tumor samples. On the contrary, the frequencies of K-ras and B-raf mutations were low, and these mutations were detected only in subsets of microdissected tumor samples. Alterations were detected homogeneously or heterogeneously in the microdissected samples for each gene. Homogeneous alterations were considered to be the result of expansion of a single clone. Heterogeneous alterations were considered to occur as the tumor progressed or diverged genetically. During genetic progression, in addition to the early homogeneous genetic events, alterations are detected only in subsets of microdissected tumor samples, indicating linear genetic progression of a single neoplastic clone. During genetic divergence, in addition to early homogeneous genetic events, different alterations of several genes are detected in different parts of the tumor, indicating that a single neoplastic clone has diverged in two or more directions[20]. Thus, alterations of KLF6 and p53 may be early and important events in the development of nonpolypoid-type tumors, whereas K-ras and B-raf mutations may not. However, morphologically mixed-type tumors, such as polypoid lesions in depressed tumors, are occasionally found. Tumors with mixed morphology may be the result of recently mutated K-ras. Microdissection is useful in the examination of genetic heterogeneity of tumors. Use of this method to examine genetic heterogeneity in tumors of mixed morphology will provide information regarding the genetic events involved.
Although LOH of KLF6 was detected in 14 of 31 (45.2%) tumors, only a single KLF6 missense mutation was detected in the present study. We examined the reported hot spot region of the gene. Current reports are all from Western countries. There are no studies on the frequency of KLF6 mutation in Asian countries. The mutation spectrum may differ between Western countries and Asian countries, including Japan. Further examination is necessary to clarify this issue.
Recently, Konishi et al[21] reported B-raf mutation in 9% of nonpolypoid tumors, and Noda et al[22] reported that 10% of tumors showed this mutation. They concluded that B-raf mutation may be involved in the development of nonpolypoid colorectal carcinoma. The mutation frequency of this gene was similar to that in the present study. However, mutation was detected only in subsets of microdissected tumor samples, indicating little or no role in the development of nonpolypoid tumors. Microdissection was not used in previous studies. Thus, genetic clonality was not assessed.
In conclusion, results of the present study suggest that KLF6 and p53 mutations are involved in the development of nonpolypoid colorectal carcinoma, whereas K-ras and B-raf mutations are not. Polypoid-type tumors were not examined in the present study; therefore, the present study had limitations. The frequency of KLF6 mutation in polypoid-type tumors is of great interest. A high frequency of KLF6 somatic mutation has been reported in colorectal carcinoma, and the adenoma-carcinoma sequence is predominant in the development of colorectal carcinoma. Therefore, KLF6 mutation in polypoid-type tumors may be frequent. In this relation, the frequency of KLF-6 mutation in adenomatous polyps is of great interest. However, there were no reports on the frequency. Future research will examine mutations in polypoid-type tumors and adenomatous polyps.
This study was supported in part by a Grant from the Japanese Society of Gastrointestinal Endoscopy, Chugoku Branch.
Supported in part by a Grant from the Japanese Society of Gastrointestinal Endoscopy, Chugoku Branch
S- Editor Liu Y L- Editor Rippe RA E- Editor Lu W
| 1. | Watanabe T, Muto T. Colorectal carcinogenesis based on molecular biology of early colorectal cancer, with special reference to nonpolypoid (superficial) lesions. World J Surg. 2000;24:1091-1097. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 2. | Umetani N, Sasaki S, Masaki T, Watanabe T, Matsuda K, Muto T. Involvement of APC and K-ras mutation in non-polypoid colorectal tumorigenesis. Br J Cancer. 2000;82:9-15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 42] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 3. | Sakamoto N, Terai T, Ajioka Y, Abe S, Kobayasi O, Hirai S, Hino O, Watanabe H, Sato N, Shimoda T. Frequent hypermethylation of RASSF1A in early flat-type colorectal tumors. Oncogene. 2004;23:8900-8907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 39] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 4. | Akiyama Y, Arai T, Nagasaki H, Yagi OK, Nakahata A, Nakajima T, Ohkura Y, Iwai T, Saitoh K, Yuasa Y. Frequent allelic imbalance on chromosome 18q21 in early superficial colorectal cancers. Jpn J Cancer Res. 1999;90:1329-1337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 5. | Muto T, Kamiya J, Sawada T, Konishi F, Sugihara K, Kubota Y, Adachi M, Agawa S, Saito Y, Morioka Y. Small "flat adenoma" of the large bowel with special reference to its clinicopathologic features. Dis Colon Rectum. 1985;28:847-851. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 310] [Cited by in RCA: 264] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 6. | Kudo S, Tamura S, Hirota S, Sano Y, Yamano H, Serizawa M, Fukuoka T, Mitsuoka H, Nakajima T, Kusaka H. The problem of de novo colorectal carcinoma. Eur J Cancer. 1995;31A:1118-1120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 80] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 7. | Narla G, Heath KE, Reeves HL, Li D, Giono LE, Kimmelman AC, Glucksman MJ, Narla J, Eng FJ, Chan AM. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science. 2001;294:2563-2566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 351] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 8. | Chen C, Hyytinen ER, Sun X, Helin HJ, Koivisto PA, Frierson HF, Vessella RL, Dong JT. Deletion, mutation, and loss of expression of KLF6 in human prostate cancer. Am J Pathol. 2003;162:1349-1354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 114] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 9. | Reeves HL, Narla G, Ogunbiyi O, Haq AI, Katz A, Benzeno S, Hod E, Harpaz N, Goldberg S, Tal-Kremer S. Kruppel-like factor 6 (KLF6) is a tumor-suppressor gene frequently inactivated in colorectal cancer. Gastroenterology. 2004;126:1090-1103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 142] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 10. | Shimoda T, Ikegami M, Fujisaki J, Matsui T, Aizawa S, Ishikawa E. Early colorectal carcinoma with special reference to its development de novo. Cancer. 1989;64:1138-1146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 11. | Jeng YM, Hsu HC. KLF6, a putative tumor suppressor gene, is mutated in astrocytic gliomas. Int J Cancer. 2003;105:625-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 75] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 12. | Hiyama T, Yokozaki H, Shimamoto F, Haruma K, Yasui W, Kajiyama G, Tahara E. Frequent p53 gene mutations in serrated adenomas of the colorectum. J Pathol. 1998;186:131-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 13. | Bos JL. The ras gene family and human carcinogenesis. Mutat Res. 1988;195:255-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 564] [Cited by in RCA: 549] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 14. | Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949-954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7459] [Cited by in RCA: 7630] [Article Influence: 331.7] [Reference Citation Analysis (0)] |
| 15. | Yokozaki H, Shitara Y, Fujimoto J, Hiyama T, Yasui W, Tahara E. Alterations of p73 preferentially occur in gastric adenocarcinomas with foveolar epithelial phenotype. Int J Cancer. 1999;83:192-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 16. | Hoffman M. Getting a handle on Ras activity. Science. 1992;255:159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 17. | Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, Young J, Walsh T, Ward R, Hawkins N. Kirsten ras mutations in patients with colorectal cancer: the 'RASCAL II' study. Br J Cancer. 2001;85:692-696. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 632] [Cited by in RCA: 666] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 18. | Kolch W. Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem J. 2000;351 Pt 2:289-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 283] [Cited by in RCA: 243] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 19. | Chan TL, Zhao W, Leung SY, Yuen ST. BRAF and KRAS mutations in colorectal hyperplastic polyps and serrated adenomas. Cancer Res. 2003;63:4878-4881. [PubMed] |
| 20. | Shima H, Hiyama T, Tanaka S, Yoshihara M, Arihiro K, Chayama K. Genetic progression and divergence in superficial esophageal squamous cell carcinoma by loss of heterozygosity analysis. Oncol Rep. 2006;16:685-691. [PubMed] |
| 21. | Konishi K, Takimoto M, Kaneko K, Makino R, Hirayama Y, Nozawa H, Kurahashi T, Kumekawa Y, Yamamoto T, Ito H. BRAF mutations and phosphorylation status of mitogen-activated protein kinases in the development of flat and depressed-type colorectal neoplasias. Br J Cancer. 2006;94:311-317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 22. | Noda H, Kato Y, Yoshikawa H, Arai M, Togashi K, Nagai H, Konishi F, Miki Y. Frequent involvement of ras-signalling pathways in both polypoid-type and flat-type early-stage colorectal cancers. J Exp Clin Cancer Res. 2006;25:235-242. [PubMed] |