Published online Nov 14, 2005. doi: 10.3748/wjg.v11.i42.6620
Revised: April 15, 2005
Accepted: April 18, 2005
Published online: November 14, 2005
AIM: To detect the germline mutations of hMLH1 and hMSH2 based on mRNA sequencing to identify hereditary non-polyposis colorectal cancer (HNPCC) families.
METHODS: Total RNA was extracted from peripheral blood of 14 members from 12 different families fulfilling Amsterdam criteria II. mRNA of hMLH1 and hMSH2 was reversed with special primers and heat-resistant reverse transcriptase. cDNA was amplified with expand long template PCR and cDNA sequencing analysis was followed.
RESULT: Seven germline mutations were found in 6 families (6/12, 50%), in 4 hMLH1 and 3 hMSH2 mutations (4/12, 33.3%); (3/12, 25%). The mutation types involved 4 missense, 1 silent and 1 frame shift mutations as well as 1 mutation in the non-coding area. Four out of the seven mutations have not been reported previously. The 4 hMLH1 mutations were distributed in exons 8, 12, 16, and 19. The 3 hMSH2 mutations were distributed in exons 1 and 2. Six out of the 7 mutations were pathological, which were distributed in 5 HNPCC families.
CONCLUSION: Germline mutations of hMLH1 and hMSH2 can be found based on cDNA sequencing so as to identify HNPCC family, which is highly sensitive and has the advantages of cost and time saving.
-
Citation: Wang CF, Zhou XY, Zhang TM, Sun MH, Shi DR. Detection of germline mutations of
hMLH1 andhMSH2 based on cDNA sequencing in China. World J Gastroenterol 2005; 11(42): 6620-6623 - URL: https://www.wjgnet.com/1007-9327/full/v11/i42/6620.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i42.6620
hMLH1 and hMSH2 are the two most important genes for HNPCC, which is the most common hereditary colon syndrome accounting for 10% of all colorectal cancers. It is autosomally dominant with a penetrance rate of 80-90%. HNPCC occurrence is closely associated with deficiency or loss of function of mismatch repair (MMR) genes. Affected individuals have an approximately 70% lifetime risk of colon cancer with a mean onset age of 44 years and an approximately 40% lifetime risk of endometrial cancer in females. At least 5 MMR genes, hMLH1, hMSH2, hMSH6, hPMS1, and hPMS2, have been implicated in HNPCC[1,2]. Information of genetic linkage analysis shows that germline mutations of hMLH1 and hMSH2 account for nearly 90% of all germline mutations found in HNPCC[3]. Germline mutations in MMR genes predispose to colorectal and other HNPCC associated epithelial cancers. Identification of MMR gene germline mutations has direct clinical implications in counseling and management of HNPCC.
Methods such as microsatellite instability (MSI), immunohistochemistry (IHC)[4-6], and sequencing of genes are employed to screen HNPCC. The most specific method is to detect the germline mutations of MMR. Its cost and sensitivity limitations can be overcome at least in part by RNA-based analysis[7]. It is the first time in China that we identified HNPCC families by detecting germline mutations of hMLH1 and hMSH2 genes based on cDNA sequencing with special primers and heat-resistant reverse transcriptase.
Fourteen anticipants from 12 unrelated families fulfilling Amsterdam criteria II for HNPCC were studied. Personal and family cancer history was obtained from the patients and their relatives. Pathological diagnosis and death were confirmed by review of medical records, pathological reports or death certificates.
Three microliters of peripheral blood was taken from each participant. Total RNA was extracted using TRIzol (Sigma Company) according to the manufacturer’s instructions.
cDNA was synthesized with transcriptor reverse tran-scriptase (Roche Diagnostics) using 0.5 μg of total RNA and specific primers complementary to the 3’ end of hMLH1 (2484-TATGTTAAGACACATCTATTTATTTA-2459) and to the 3’ end of hMSH2 (3145-CCACCAAACTACATGATTTTATTTATAAAATTC-3114). RT was performed at 60 °C for 60 min.
cDNA of hMLH1 and hMSH2 was amplified in two overlapping fragments using primers (Table 1) to generate products of ~2 000 bp. PCR was performed using expand long template PCR (Roche Diagnostics) at 94 °C for 5 min; then 10 cycles at 94 °C for 30 s, at 59 °C for 30 s, at 68 °C for 3 min; 32 cycles at 94 °C for 30 s, at 57 °C for 30 s, at 68 °C for 3 min with a final elongation at 68 °C for 7 min.
| Sense | Antisense |
| hMLH1-1F(1-18) | hMLH1-5R(2198-2175) |
| CTTGGCTCTTCTGGCGCC | GAGCGCAAGGCTTTATAGACAATG |
| hMLH1-4F(1333-1353) | hMLH1-6R(2484-2459) |
| GCTGAAGTGGCTGCCAAAAAT | TATGTTAAGACACATCTATTTATTTA |
| hMSH2-1F(1-21) | hMSH2-7R(2753-2732) |
| GGCGGGAAACAGCTTAGTGGG | GGGCATTTGTTTCACCTTGGAC |
| hMSH2-6F(1898-1920) | hMSH2-8R(3145-3114) |
| CGTGTCAAATGGAGCACCTGTTC | CCACAAACTACATGATTTTATTTATAAAATTC |
PCR products were size fractionated by agarose gel electrophoresis and analyzed by ethidium bromide staining.
Purified PCR fragments were sequenced directly using a DNA sequencing kit according to Applied Biosystems from USA with BigDye Terminators on an ABI3700 automated DNA sequencer.
cDNA of hMLH1 (2 484 bp) was sequenced in six overlapping fragments and cDNA of hMSH2 (3 145 bp) was sequenced in eight overlapping fragments using primers (Table 2).
| Sense | Antisense |
| hMLH1-1F CCTGGCTCTTCTGGCGCC | hMLH1-1R CTTTTCTCCTCGTGGCTATGTTGT |
| hMLH1-2F ATGTGCTGGCAATCAAGGGA | hMLH1-2R GGTGCACATTAACATCCACATTCT |
| hMLH1-3F CCAAAAACACACACCCATTCCT | hMLH1-3R CCTTTGTTGTATCCCCCTCCA |
| hMLH1-4F GCTGAAGTGGCTGCCAAAAAT | hMLH1-4R CATCTTCCTCTGTCCAGCCACTC |
| hMLH1-5F TTGCCATGCTTGCCTTAGATAGTC | hMLH1-5R GAGCGCAAGGCTTTATAGACAATG |
| hMLH1-6F GCTCCATTCCAAACTCCT | hMLH1-6R TATGTTAAGACACATCTATTTATTTA |
| hMSH2-1F GGCGGGAAACAGCTTAGTGGG | hMSH2-1R CTCTGGCCATCAACTGCGGAC |
| hMSH2-2F GGCTTCTCCTGGCAATCTCTCTCA | hMSH2-2R CTTGATTACCGCAGACAGTGATGAAAC |
| hMSH2-3F GCAAAAAGGGAGAGCAGATGAATAGTG | hMSH2-3R GGCAAGTCGGTTAAGATCTGGGAAT |
| hMSH2-4F AGATGCAGAATTGAGGCAGACTTTACA | hMSH2-4R GGACTTTTTCTTCCTTACAGGTTACACG |
| hMSH2-5F CAGAGATCTTGGCTTGGACCCT | hMSH2-5R TTCAACACAAGCATGCCTGGAT |
| hMSH2-6F CGTGTCAAATGGAGCACCTGTTC | hMSH2-6R GATTGGCCAAGGCAGTAAGTTCAT |
| hMSH2-7F AATCATAGATGAATTGGGAAGAGGAACT | hMSH2-7R GGGCATTTGTTTCACCTTGGAC |
| hMSH2-8F CTATCTGGAAAGAGAGCAAGGTGAA | hMSH2-8R CCACAAACTACATGATTTTATTTATAAAATTC |
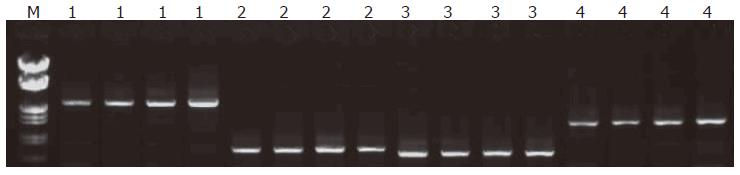
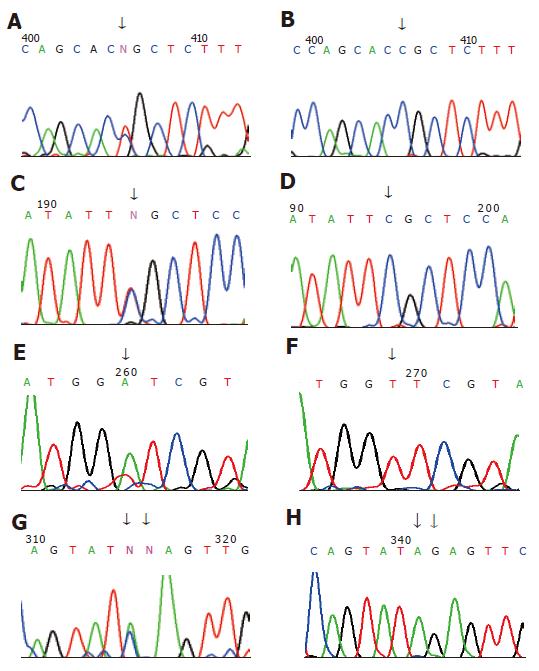
The sizes of amplified hMLH1 and hMSH2 segments were respected (Figure 1). Seven germline mutations were found in 6 out of 12 families, 4 hMLH1 and 3 hMSH2 mutations (4/12, 33.3%); (3/12, 25%). The mutation types involved 4 missense, 1 silent and 1 frame shift mutations as well as 1 mutation in non-coding area, including hMLH1 mutation in family H2 at 649 codon 217 exon 8: CGC→TGC; hMLH1 mutation in family H31 at 1742 codon 581 exon 16: CCG→CTG; hMLH1 missense mutation in family H114 at 1151 codon 384 exon 12: GTT→GAT; family H111 hMLH1 non-coding area at 2438 exon 19: A→C; family H11 hMSH2 at 14 codon 5: CCG→CAG; family H38 hMSH2 mutations at 295 and 296 codon 99 exon 2: 295: A→C, 296:del.G (Table 3, Figure 2).
| Families | Genes | Exon | Codons affected | DNA change | Amino acid change | Mutation types |
| H31 | hMLH1 | 16 | 581 | T>C, at 1742 | Pro→Leu | Missense |
| H111 | hMLH1 | 19 | Non-coding area | A>T, at 2438 | ||
| H114 | hMLH1 | 12 | 384 | T→A, at 1151 | Val→Asp | Missense |
| H2 | hMLH1 | 8 | 217 | T->C, at 649 | Arg→Cyc | Missense |
| H11 | hMLH1 | 1 | 5 | A>C, at 14 | Pro→His | Missense |
| H38 | hMLH1 | 2 | 99 | A>C, at 295 | Arg→Arg | Silent |
| H38 | hMLH1 | 2 | 99 | Del G, at 296 | Frame shift | Frame shift |
Colorectal cancer (CRC) is one of the most common malignant tumors and its incidence is increasing gradually. According to the different molecular mechanism, CRC is divided into sporadic and genetic types. The latter type HNPCC is characterized by its early onset[8-10], location in the proximal colon and an increased risk of neoplasms in extracolonic organs including endometrium, stomach, urothelium, small intestine, ovary and multiple metachronous CRCs[9,11-14]. Its prognosis is better than sporadic type of CRC[15]. HNPCC is closely associated with the deficiency or loss of MMR gene function. Iden-tification of MMR gene germline mutations has direct clinical implications in counseling and management of HNPCC.
Methods are available for the identification of HNPCC. The most specific method is to detect the ger-mline mutations of MMR genes. Up to now, the germline mutations are mainly detected by genomic DNA-based sequencing (gDNA). A lot of information shows that the germline mutations of MMR genes associated with HNPCC are mainly localized in exons[3,5,7,13]. The gDNA-based sequencing is invariably affected by introns. cDNA-based sequencing of MMR genes has been reported recently. The new technique utilizes specific primers and heat-resistant reverse trancriptase to specifically synthesize cDNA of MMR genes, then full-length cDNA is amplified in two over fragments using specific primers followed by sequencing analysis of cDNA. The technique can successfully avoid the influence of introns. Additionally, it is well known that RNA is easily decayed. If RNA samples are stored too long, reverse transcription with random primers and common reverse transcription enzyme often fails, while the new technique employs specific primers and heat-resistant reverse trancriptase, the limitations can be overcome at least in part, thus improving the specificity and efficiency. Anna et al[7] compared the two techniques and found that cDNA-based sequencing not only has the advantage of specificity and efficiency, but also a lower cost, being 2.5-3 times less expensive than gDNA-based sequencing. We used 35 pair primers to amplify the two genes in the past, and only 4 pair primers were used in the present study, the procedure is greatly simplified.
We detected 7 germline mutations in 14 anticipants with HNPCC from 12 different families employing the new technique. The 3 mutations, at sites 1151, 14, and 217 in hMLH1 reported, the first two have been verified to be pathological. Moreover, the mutation at 1151 in hMLH1 has been found only in Japan and Korea, which is likely to be a hot mutation site in East Asia. The mutation at site 217 in hMLH1 occurs at a less conserved region is in 80 healthy Japanese. Whether it is pathological or not needs further study. None of the 4 unreported mutations belongs to polymorphism[17]. The 6 pathological mutations (2 reported, 4 unreported) were distributed in 5 HNPCC families in our study.
Of course, all mutations cannot be detected by the improved technique. For example, mutations in the promoter and 3'-untranslated regions of hMLH1 and hMSH2 cannot be detected. Sequencing of individual exons of gDNA also has such limitations.
Up to now, there is no optimal method to screen HNPCC patients or their families. The new technique can be utilized to screen HNPCC patients and their families, which may achieve a better result.
Science Editor Wang XL and Guo SY Language Editor Elsevier HK
| 1. | Huang D, Chen C, Sun W, Strom CM, Bender RA. High-throughput gene sequencing assay development for hereditary nonpolyposis colon cancer. Clin Colorectal Cancer. 2004;4:275-279. [RCA] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 2. | Wijnen J, de Leeuw W, Vasen H, van der Klift H, Møller P, Stormorken A, Meijers-Heijboer H, Lindhout D, Menko F, Vossen S. Familial endometrial cancer in female carriers of MSH6 germline mutations. Nat Genet. 1999;23:142-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 268] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 3. | Peltomäki P, Vasen H. Mutations associated with HNPCC predisposition -- Update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20:269-276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 317] [Cited by in RCA: 331] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 4. | Shin KH, Shin JH, Kim JH, Park JG. Mutational analysis of promoters of mismatch repair genes hMSH2 and hMLH1 in hereditary nonpolyposis colorectal cancer and early onset colorectal cancer patients: identification of three novel germ-line mutations in promoter of the hMSH2 gene. Cancer Res. 2002;62:38-42. [PubMed] |
| 5. | Peltomäki P, Gao X, Mecklin JP. Genotype and phenotype in hereditary nonpolyposis colon cancer: a study of families with different vs. shared predisposing mutations. Fam Cancer. 2001;1:9-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 48] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 6. | Wahlberg SS, Schmeits J, Thomas G, Loda M, Garber J, Syngal S, Kolodner RD, Fox E. Evaluation of microsatellite instability and immunohistochemistry for the prediction of germ-line MSH2 and MLH1 mutations in hereditary nonpolyposis colon cancer families. Cancer Res. 2002;62:3485-3492. [PubMed] |
| 7. | Jakubowska A, Górski B, Kurzawski G, Debniak T, Hadaczek P, Cybulski C, Kladny J, Oszurek O, Scott RJ, Lubinski J. Optimization of experimental conditions for RNA-based sequencing of MLH1 and MSH2 genes. Hum Mutat. 2001;17:52-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 8. | Anwar S, Hall C, White J, Deakin M, Farrell W, Elder JB. Hereditary non-polyposis colorectal cancer: an updated review. Eur J Surg Oncol. 2000;26:635-645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 9. | Jass JR. HNPCC and sporadic MSI-H colorectal cancer: a review of the morphological similarities and differences. Fam Cancer. 2004;3:93-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 108] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 10. | Aarnio M, Sankila R, Pukkala E, Salovaara R, Aaltonen LA, de la Chapelle A, Peltomäki P, Mecklin JP, Järvinen HJ. Cancer risk in mutation carriers of DNA-mismatch-repair genes. Int J Cancer. 1999;81:214-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 11. | Lucci-Cordisco E, Zito I, Gensini F, Genuardi M. Hereditary nonpolyposis colorectal cancer and related conditions. Am J Med Genet A. 2003;122A:325-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 66] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 12. | Park YJ, Shin KH, Park JG. Risk of gastric cancer in hereditary nonpolyposis colorectal cancer in Korea. Clin Cancer Res. 2000;6:2994-2998. [PubMed] |
| 13. | Ericson K, Halvarsson B, Nagel J, Rambech E, Planck M, Piotrowska Z, Olsson H, Nilbert M. Defective mismatch-repair in patients with multiple primary tumours including colorectal cancer. Eur J Cancer. 2003;39:240-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 14. | Lynch HT, de la Chapelle A. Genetic susceptibility to non-polyposis colorectal cancer. J Med Genet. 1999;36:801-818. [PubMed] |
| 15. | Watson P, Lin KM, Rodriguez-Bigas MA, Smyrk T, Lemon S, Shashidharan M, Franklin B, Karr B, Thorson A, Lynch HT. Colorectal carcinoma survival among hereditary nonpolyposis colorectal carcinoma family members. Cancer. 1998;83:259-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 16. | Wang Y, Friedl W, Lamberti C, Nöthen MM, Kruse R, Propping P. A novel missense mutation in the DNA mismatch repair gene hMLH1 present among East Asians but not among Europeans. Hum Hered. 1998;48:87-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 17. | Peltomaki P, Vasen HF. Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology. 1997;113:1146-1158. [RCA] [DOI] [Full Text] [Cited by in Crossref: 487] [Cited by in RCA: 459] [Article Influence: 16.4] [Reference Citation Analysis (0)] |