Published online Sep 7, 2005. doi: 10.3748/wjg.v11.i33.5129
Revised: February 15, 2005
Accepted: February 18, 2005
Published online: September 7, 2005
AIM: To identify chromosomal translocations specific to gastric cancer (GC), spectral karyotyping (SKY) analysis was performed on established cell lines and cancerous ascitic fluids.
METHODS: SKY analysis of 10 established cell lines and seven cancerous ascitic fluid samples identified recurrent chromosomal breakpoints and translocations in GC, several of which involved chromosomal loci of oncogenes or tumor suppressor genes.
RESULTS: A total of 630 chromosomal breaks were identified. Chromosome no.8 was the most frequently involved in rearrangements (65 breaks), followed by chromosomes no.11 (53), no. 1 (49), no. 7 (46), no. 13 (37), no. 3 (36), no. 17 (33), and no. 20 (29). Frequent breakpoints were detected in 8q24.1 (30 breaks), 11q13 (29), 13q14 (16), 20q11.2 (14), 7q32 (13), 17q11.2 (13), 18q21 (12), 17q23 (9), 18q11.2 (9). SKY analysis identified a total of 242 chromosomal rearrangements including 190 reciprocal and non-reciprocal translocations. The recurrent combinations of chromosomal bands involved in translocations were 8q24.1 and 13q14 (3 cases), 8q24.1 and 11q13 (3), 11q13 and 17q11.2 (2), and 18q11.2 and 20q11.2 (2). Our study validated the ability of SKY to characterize in detail the chromosomal rearrangements in solid tumors and derived cell lines. Moreover, fluorescence in situ hybridization helped to identify the insertions, translocations, and homogeneously staining regions of MYC and CCND1 gene loci.
CONCLUSION: The non-random co-localization of certain cytogenetic bands suggests the importance of chromosomal translocations in gastric carcinogenesis, by serving as landmarks for the cloning of GC causing genes.
- Citation: Yamashita Y, Nishida K, Okuda T, Nomura K, Matsumoto Y, Mitsufuji S, Horiike S, Hata H, Sakakura C, Hagiwara A, Yamagishi H, Taniwaki M. Recurrent chromosomal rearrangements at bands 8q24 and 11q13 in gastric cancer as detected by multicolor spectral karyotyping. World J Gastroenterol 2005; 11(33): 5129-5135
- URL: https://www.wjgnet.com/1007-9327/full/v11/i33/5129.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i33.5129
Chromosomal aberrations are fundamental to cancer formation because they interfere with the function of oncogenes and tumor suppressor genes. Identification of recurrent chromosomal translocation may thus contribute to the cloning of cancer causing genes. Several genetic alterations associated with gastric cancer (GC) have been reported: (1) amplifications of MYC, HST1/FGF4, INT2/FGF3, ERBB2, MET, and KSAM/FGFR2 genes[1-3], and (2) mutations of APC, KRAS, TP53, E-cadherin (CDH1), β-catenin (CTNNB1) and RUNX3/PEBP2 genes[3-5]. These specific alterations have been implicated in multi-stage carcinogenesis of GC.
At the same time, previous cytogenetic studies of GC have demonstrated frequent aberrations of chromosome nos. 1, 3, 6, 7, 8, 13, 17, 20 and Y[6-9], while other chromosomes have also been found to be repeatedly involved in cytogenetic aberrations as demonstrated by recent comparative genomic hybridization (CGH) studies[10-13]. The chromosomal breakpoints identified were 1p22, 3p21, 3q23, 11p13-15, and 19p13 on primary gastric tumors[6,7,14], and 1q32, 5q11-22, 14q22, and 15q15 on human GC cell lines[15]. However, specific chromosomal translocations have not yet been identified in GC, because complete karyotypic analysis was precluded by the complicated and cryptic nature of the rearrangements as well as the poor banding of condensed chromosomes. To overcome these limitations, multiplex fluorescence in situ hybridization (FISH) has been successfully used for two patients with GC[16].
To identify chromosomal translocations specific to human GC, we used multicolor spectral karyotyping (SKY) to analyze established cell lines and cancerous ascitic fluid samples and were able to identify and characterize recurrent chromosomal breakpoints and translocations in GC, several of which involved chromosomal loci of oncogenes or tumor suppressor genes.
Ten GC cell lines (KMK2, KHM14K, MKN1, MKN28, MKN45, MKN74, GT3TKB, SNU1, SNU5, and SNU16) and seven cancerous ascitic fluid samples from advanced GC (patient nos. 1-7) were the subjects of our study. SNU1, SNU5, and SNU16 were obtained from the Korean Research Institute of Bioscience and Biotechnology (Taejon, South Korea), and MKN1, MKN28, MKN45, MKN74, and GT3TKB from the RIKEN Cell Bank (Tsukuba, Japan). Two cell lines, KMK2 and KHM14K, were established at the Kumamoto University School of Medicine (Kumamoto, Japan) and their clinico-pathological features are summarized in Tables 1 and 2. Six cell lines were derived from poorly differentiated adenocarcinoma (MKN45, SNU1, SNU5, SNU16, KMK2, and KHM14K), two from moderately differentiated adenocarcinoma (MKN28 and MKN74), one from undifferentiated mucin-producing adenocarcinoma (GT3TKB), and one from adenosquamous cell carcinoma (MKN1). According to Lauren’s classifications[17], seven cell lines (MKN45, SNU1, SNU5, SNU16, KMK2, KHM14K, and GT3TKB) were categorized as diffuse type, and two cell lines (MKN28 and MKN74) as intestinal type.
| Cell line | Histologic grade1 | Site2 | Representative karyotype3 |
| MKN45 | Poor | LT | 85,XX,del (1) (q32), del (1) (q32), +der (1) t (1;2) (p34;q21), +der (1) t (1;2) (p34;q21), der (2) t (2;22) (p23;q11.2), +del |
| (2) (q23), +der (3) t (3;8) (p21.3;q24.1), +der (4) H (9qter→9q22::4p12→4q35::11p11.2→11pter), +der (4) (9qter→9 | |||
| q22::4p12→4q35::11p11.2→11pter), +der (5) t (5;8) (q31;q22), +der (5) t (5;8) (q31;q22), | |||
| der (6) t (1;6) (q25;q27), +der (7) t (7;12) (q32;q21), +der (7) t (7;11) (q11.1;q13), +der (7) t (7;11) (q11.1;q13), | |||
| +der (7) t (7;11) (q11.1;q13), +der (7) t (7;9) (q11.2;q34),-8,-8, +9, +10, +der (10) t (8;10) (q23;p15), | |||
| +der (10) t (8;10) (q23;p15), +del (11) (p11.2), +der (12) t (9;12) (p22;q24.1), +der (12) t (10;12) (q11.2;q12), +13, +13, | |||
| +der (14) t (13;14) (q22;p11.2), +der (14) t (13;14) (q22;p11.2), +16, +16, +16, +der (17) t (12;17) (q22;p13), | |||
| +i (17) (p11.1), +der (19) (19pter→19q13.4::7? | |||
| →7?::11q23→11qter), +der (19) (19pter→19q11::8q21.2→8q24.1::20q13.1→20qter), +20, +20, +der (20) t (7;20) | |||
| (q11.2;p12), +der (20) t (7;20) (q11.2;p12), +der (20) t (8;20) (p11.2;q11.2), | |||
| +der (21) t (20;21) (p11.2;q11.2), +der (21) t (12;21) | |||
| (p11.2;q22.3), +22, +22, +mar (6pter→6q21::20p13→20q11.2::7p11.2→7p15::8q22?8q24.1::18q21→18qter) | |||
| SNU1 | Poor | AS | 85,XXYY, +del (1) (p22), +del (1) (p22), -4, -5,-5, -11,-11, -13, -14, -17, -18, +20, +20, +20, +mar |
| SNU5 | Poor | AS | 74,XX,der (1) t (1;8) (p36.3;q24.1),der (1) t (X;1) (p11.4;p32), +der (1) t (X;1) (p11.4;p32), +der (2) t (2;11) (p21;q13), |
| +der (2) t (2;11) (p21;q13), +2,-3,der (4) t (4;17) (q33;q23), +der (4) t (4;13) (q31;q32), +der (4) (4pter→4q12::13q14 | |||
| →13q22::10q24→10qter), +del (5) (q15), +del (5) (p12), +5, +6, der (7) t (7;11) (q36;q13), +del (7) (q32), +del (7) (q32), | |||
| +der (7) (7pter→7q22::11q13→11q23::13q12→13qter), +der (7) t (7;8) (q36;q24.1), +der (8) t (8;13) (q24.1;q14), + | |||
| der (8) t (8;13) (q24.1;q14), +9, +10, +der (10) t (2;10) (p21;q24), +der (10) t (10;15) (q22;q15), +11, +11, +11, +12, | |||
| der (13) (14qter→14q13::13p13→13q32::17q23→17qter), | |||
| +14, +14, +der (15) t (15;22) (p13;q11.2), +16, +i (17q) (p10), +19, +20, +20, +21,-22, +hsr, +dmin | |||
| SNU16 | Poor | AS | 74,XX,del (1) (p13), +hsr (1) (pter→q21::hsr::q21→qter), +2, +3, +6, +6, +6, +del (7) (q22), +del (7) (q22), del (7) (q32), |
| der (7) t (7;8) (q22;q24.1), | |||
| +der (8) t (8;11) (q24.1;q13), +9,del (10) (q22), +11, +der (11) (5pter→5p13::11p11.2→11q25::17q23→17qter), + | |||
| 12, +12, +13, +14, +der (15) (6pter→6p11.2→::hsr::15p11→15qter), +16, +17, +17, +18, +18, +19, +20, +20, | |||
| +22, 30-50dmin | |||
| KMK2 | Poor | AS | 66,X,del (X) (p11.2) (q22),der (1) (20qter→20q13.1::1p36.1→1q23::8q22→8qter), |
| der (1) (17qter→17q21::1p32→1q42::8q24.1→8qter), +der (1) (4qter→4q33::5q35→5q11.2::1p13?→q21::8q | |||
| 13→8qter), +der (1) t (1;5) (q21;q32), | |||
| +der (2) t (2;3) (q33;q25),t (3;5) (q25-26.2;q31), +der (3) (3pter→3q24::11q13→11q14::4p12→4pter), | |||
| der (5) del (5) (p12) del (5) (q21), +der (5) (12qter→12q24.1::5p15.1→5q22::13q12→13qter), | |||
| +der (5) t (5;7) (q13;q32), del (6) (q12), del (6) (q12), +del (6) (q25), +der (6) t (6;14) (q21;q22), | |||
| del (7) (q22), der (7) t (7;15) (p22;q13), +der (7) t (6;7) (q15;p15), +der (7) t (7;20) (p11.2;q13), der (8) t (8;17) (p23;q11.2), | |||
| der (9) (Xpter→Xp11.2::8q24.3→8q24.1::9p13→9qter), +der (9) t (3;9) (?;?), | |||
| del (10) (q22), +der (10) (14q?ter→14q?24::10p13→10q21::1p13→1p34::8q24.1→8q24.3::11q13.1→11q13.5 | |||
| ::7p15→7p22::15q13→15qter),der (12) (12pter→12q22::11q13.3→11q21::3q21→3qter), +der (12) (3pter→ | |||
| 3p21::12p13→12q24.3::Xq13→Xq22::6p21.3→6pter), +der (12) (17qter→17q11.2::12p11.2→12q24.1::13q12 | |||
| → 13qter),-13,der (13) (13pter→13q14::8q24.1→8q24.3::11q13.3→11q23::12q24.1→12qter),del (14) (q22), | |||
| del (15) (q15), der (16) t (1;16) (p36.1;q22),der (17) (17pter→17q11.2:: | |||
| 11q13.3→11q21::3q26.2→3qter), +der (17) t (16;17) (q22;q11.2), | |||
| der (18) (22qter→22q11.2::11q21→11q13.3:: | |||
| 8q24.1→8q22::18p11.2→18qter), +der (18) t (14;18) (q24;q21.3), | |||
| +der (20) t (12;20) (q22;q13.1), +der (20) (20pter→20q11.2::13q12→13q14::8q24.1→8q24.3::11q13.3→11qter), | |||
| +der (20) (20pter→20q11.2::13q12→13q14::8q24.1→8q24.3::11q13.3→11qter), +21, | |||
| +der (22) t (5;22) (p13;q11.2) | |||
| KHM14K | Poor | LN | 56,X,-Y, +1, +der (3) del (3) (p?) del (3) (q?), +der (3) (9qter?9q13::3q21→3p11::22q11.2→22qter), +der (5) (5qter |
| →5p15::3?→3?::22q11.2→22qter),i (5p) (q10), +der (6) (6pter→6q11::5?→5?::17q23→17qter), | |||
| t (8;16) (p23;q22), +der (8) t (8;20) (p11.2;q13.1), der (9) t (9;16) (p13;q12.1), +der (9) t (3;9) (q25;q13), +11, | |||
| der (12) t (5;12) (q13;p13),der (13) t (3;13) (p13;q14), (14;15) (q32;q15), -15, del (15) (q24), | |||
| +der (18) t (3;18) (p21.3;q21.1), +20, +20, +20,der (21) t (15;21) (q11.2-q13;p11.1),-22 | |||
| GT3TKBU | Ndiffer. mucin-producing | AS | 49,der (X) (Xpter→Xq28::HSR::1q32→1qter), +der (X) (Xpter→Xq28::HSR::1q32→1qter),-Y,-1, |
| der (1) t (1;8) (p32-36.1;q24.1),der (2) t (2;4) (p12;q12-21),del (3) (p13),del (6) (q25),der (7) t (2;7) (q23;q11.2),-8,i | |||
| (8q), der (9) t (9;10) (p13;q22), der (7;10) (7qter→7p22::13q13→13q11:: 10p11.2→10qter), | |||
| der (10) t (8;10) (q24;p13), +der (10) | |||
| t (10;15) (q11.2;q22),der (11) t (4;11) (q27;q23),der (11) t (1;11) (p13;q13),der (13) t (11;13) (q13;p11.2), | |||
| der (13) t (13;17) (p11.2;p12),der (14) (14qter→14q11.2::3p11→3p26::17q11.2→17q23::3p21→3p23::17q21 | |||
| →17qter), der (17) t (8;17) (q24;q11.2), der (17) t (17;19) (p11.2;q13.1), der (17) t (17;19), +del (18) (q21), | |||
| +der (18) t (18;19) (q11.2;q13.1), -19,der (19) t (19;20) (p13.1;p12), | |||
| +der (20) t (18;20) (q11.2;q11.2), +der (20) (8qter→8q24::20p11.2→20q11.2::2q33→2qter), | |||
| +der (20) t (7;20) (q32;q11.2), der (21) t (4;21) (q12;p11.2),-22 | |||
| MKN28 | Moderate | LN | 47, X, -X, del (1) (p22), +der (1) t (X;1) (q22-24;q32), del (2) (q13), del (2) (q13), +der (2) t (2;18) (p23;q21.3), |
| der (3) del (3) (p13p25) del (3) (q21q25), +der (3) del (3) (p12) del (3) (q21q26.2), -4, -5, -5, -7, | |||
| der (7) t (7;8) (q36;q22), -8, der (8) t (8;10) (p23;q11.2), -10, -13, der (14) t (7;14) (q11.2;q11.2), | |||
| der (14) t (1;14) (q42;q24), +15, +der (16) t (13;16) (q12;q23-24), | |||
| +der (19) t (11;19) (q13;q13.3), der (20) (20pter→20q13.1::8q13→8q24::7q32→7qter), | |||
| +der (20) t (18;20) (q21.1;q11.2), +21, +21 | |||
| MKN74 | Moderate | LT | 39, X, +der (X) (15qter→15q11.2::Xp11.2→Xq28::8q24.1→8qter), |
| +der (X) t (X;7) (q13;q32), -Y, der (1) (9qter→9q32::1p34→1q32::18q21→18qter), +2, -3, | |||
| der (4) t (4;6) (p12;p21.3), -5, der (7) (7pter→7q32::8q22→8q24.1::9q22→9qter), der (7) t (7;8) (q32;q22), der (8) t | |||
| (8;10) (p23;q22), der (9) t (9;11) (q22;q13), -9, -10, +del (11) (q13), -12, -13, | |||
| der (14) t (9;14) (q13;p11.2), -14, -15, der (16) t (9;16) (q13;q24), +der (16) t (9;16) (q13;q24) del (9) (q22), | |||
| der (18) t (2;18) (?;q23), der (20) t (13;20) (q32;q11.1), -20, -21, -21, der (22) t (10;22) (q24;p11.2), | |||
| +der (22) t (13;22) (q14;p11.2) | |||
| MKN1 | Adenosqu amous carcinoma | LN | 56, X, +inv (X) (p11.2p22.1), -Y, +der (1) t (1;8) (p36.1;q24.1), +der (1) t (1;8) (p36.1;q24.1), |
| der (3) t (3;5) (p11;p12), der (3) t (3;20) (q27;q13.1), | |||
| +der (3) t (3;7) (p26;q32.1), -5, -5, -6, +7, +7, +7, +7, del (8) (q24.1), +del (8) (q24.1), | |||
| +del (8) (p11.2), +del (8) (p11.2), -9, -9, +ins (11;17) (q13;q11.2q25), -12, +del (13) (q14q22), | |||
| der (15) t (4;15) (q12;p11.2), -15, +del (16) (q12.1), +del (16) (p11.2), +16, der (17) t (4;17) (p15.2;p11.2), | |||
| der (17) t (4;17) (p15.2;p11.2), del (17) (q11.2), +der (18) t (3;18) (q21;q11.2), del (18) (q11.2), | |||
| del (19) (p13.1), der (19) t (19;21) (q13.1;q11.2), +der (19) t (19;21) (q13.1;q11.2), +del (20) (q11.2), | |||
| +der (20) t (17;20) (q23;q11.2), der (22) t (11;22) (p13;p11.2) |
| Case number. | Age/Sex | Origin1 | Histologic grade2 | G-banding and SKY |
| 1 | 52/M | AS | Poor | 89, XX, der (1) t (1;13) (q44;q14), t (1;2) (p36.3;p11.1), +der (1) t (1;2), +der (1) t (1;8) (p22;q24), |
| +der (1) t (1;8) (p22;q24), +der (1) t (1;12) (q32;q13) del (1) (p32), +2, +2, +3, +4, +4, +del (5) (q13), | ||||
| +6, +6, +6, +7, +7, +7, | ||||
| der (8;8) (8pter→8q24.1::8p23→8q24.1::17q23→17qter), +der (8) t (8;11) (q24.1;q13), +del (8) (p11.2), | ||||
| +del (8) (p11.2), +9, +9, +i (9q), del (10) (q24), +der (10) t (8;10) (q24.1;q11.2), | ||||
| +der (10) t (8;10) (q24.1;q11.2), +inv (10) (p15.1q21), +11, +11, +12, +14, i (15q), | ||||
| i (15q), +16, +16, +17, del (18) (q11.2), +der (18) t (10;18) (p11.2;p11.2), | ||||
| +der (18) t (10;18) (p11.2;p11.2), +der (18) t (10;18) (q22;q21), +der (18) t (10;18) (q22;q21), +19, +19, | ||||
| +20, +20, +20, +20, +20, +20, -21, -21, +22 | ||||
| 2 | 72/M | AS | Poor | 47, XY, +X, -3, del (13) (q14), +mar |
| 3 | 63/M | AS | Poor | 54, der (X) t (X;3) (q24;q21), Y, +2, +del (4) (q21), +der (4) (4p15→4q12::17q11.2→17qter), +del (5) (q13), |
| der (6) t (Y;6) (q11.2;p21.1), t (7;16) (q36;q22), der (11) t (9;11) (q13;q13), +del (11) (q13), +11, del (12) | ||||
| (p11.2), der (13) t (8;13) (q13;q14), +15, +16, -21, -21, +22, +22 | ||||
| 4 | 56/M | AS | Signet | 46, X, -Y, +der (1) t (?;1;?) (?::1p13→1q21::?), t (2;20;8) (p11.2;q11.2;q24.1), |
| t (2;20;8) (p11.2;q11.2;q24.1), +2, +2, der (3) (3qter→3p13::7?→7?::11q13→11qter), del (3) (p23) (q25), | ||||
| -4, -4, +del (5) (q13), +del (5) (q11.2), del (6) (q11), | ||||
| +der (6) (6p21.1→6q21::18q11.2→18qter), dic (7;11) (q11.2;p15), del (9) (p11), del (10) (p11.2), | ||||
| del (10) (p11.2), +10, | ||||
| der (11) t (?;11;?) (?::11p13→11q23::?), der (11) (2pter→2p11.2::11q12→11p11.2::7q11.2→7q32:: | ||||
| 17q23→17qter), | ||||
| der (11;20) (20pter→20q11.2::11p13→11q23::11q13→11q14::7q31→7q36::13q14→13qter), | ||||
| del (12) (q13), del (12) (q22), der (13) t (X;13) (p11.2;p11.2), der (13) (3pter→3p13::13p13→13q14::18q21 | ||||
| →18qter), -14, -15, del (16) (q22), add (17) (p11.2), add (18) (q21.3), -18, -19, +20, | ||||
| der (21) t (17;21) (q23;q22), der (21) (7pter→7p11.2::21p13→21q22.1::hsr::21q22.1→21qter), -22 | ||||
| 5 | 55/M | AS | Signet | 50, add (X) (p22.3), Y, +8, +9, +16, +18 |
| 6 | 37/M | AS | Poor | 69, der (X) (Xpter→Xq22::18q11.2→18q21::4q31.1→4qter), |
| Signet | der (X) (Xpter→Xq22::18q11.2→18q21::4q31.1→4qter), -Y, +der (1) t (1;12) (q25;q13), | |||
| +del (1) (p22), del (2) (q23), del (2) (q23), +der (2) t (1;2) (q23;q21), +der (2) t (1;2) (q23;q21), | ||||
| del (3) (p21), +del (3) (p13), +del (3) (q11.2), +5, +5, der (6) t (6;18) (q12;q11.2), -6, +7, +7, +der (8) | ||||
| (5qter→5q31::8p11.2→8q24.1::13q14→13qter), | ||||
| +der (8) (5qter→5q31::8p11.2→8q24.1::13q14→13qter), +der (10) t (4;10) (q12;q22), | ||||
| +der (10) t (4;10) (q12;q22), +der (11) t (7;11) (q22;q23), | ||||
| +der (11) t (11;17) (q25;q11.2), +12, der (13) t (6;13) (p21.1;p11.2), -13, der (14) t (14;17) (p11.2;q11.2), | ||||
| der (14) t (14;17) (p11.2;q11.2), +15, +15, +16, +16, der (17) t (X;17) (q22;q11.2), | ||||
| der (17) t (X;17) (q22;q11.2), +18, +18, +20, +21, +der (21) t (X;21) (q22;p11.2) | ||||
| 7 | 73/F | AS | Poor | 67, X, del (X) (q26), der (1) (1qter→1p22::11?→11?::8?→8?::22q11.2→22qter), der (1) (1?→1?::15? |
| Signet | →15?::1?→1qter), der (1) (9pter→9p13::1p12→1q32::9q22→9qter), -2, -5, | |||
| +der (7) t (7;9) (q32;q22), +der (7) t (7;9) (q32;q22), +der (7) t (7;20) (q22;q11.2), | ||||
| +der (7) t (7;20) (q22;q11.2), t (8;14) (q13;p13), +der (8) t (8;14), t (6;8) (p11.2;p21), +8, -9, | ||||
| +der (10) t (2;10) (p13;q24), +11, -12, +der (14) (20qter→20q13.1:: | ||||
| 14p11.2→14q24::11q13→11qter), | ||||
| +der (14) t (3;14) (p21;q13), +14, +15, +16, +17, +18, +18, +18, +18, | ||||
| +der (18) t (18;20) (q11.2;q11.2), +19, +20, +21, +21, +22, +der (22) t (1;22) (p22;p11.2) |
Primary tumors associated with carcinomatous peritonitis were histopathologically diagnosed as poorly differentiated adenocarcinoma in three patients, signet-ring cell carcinoma in two, and coexisting poorly differentiated adenocarcinoma and signet-ring cell carcinoma in two. These diagnoses indicated that all these tumors were diffuse type GC according to Lauren’s classification. Ascitic fluid was aspirated with a syringe and the addition of heparin from the peritoneal cavities of seven patients with carcinomatous peritonitis. Informed consent in accordance with the institutional guidelines was obtained, for using the diagnostic material for research purposes.
G-banding studies were performed as described previously[18]. Briefly, ascitic fluid was diluted in 10 mL of RPMI 1640 medium supplemented with 10% fetal calf serum at a final concentration of 1106 cells/mL. The cells were cultured at 37 °C for 24-48 h in humidified air with 50 mL/L CO2, exposed to colcemid (0.05 μg/mL) for 60 min, processed in 0.075 mol/L potassium chloride for 20 min, and fixed with methanol/glacial acetate (3:1). Chromosomes were stained with a Giemsa solution pretreated with trypsin, and karyotyped according to the International System for Human Cytogenetic Nomenclature (ISCN 1995)[19]. The remaining chromosome pellets were stored at -20 °C for SKY and FISH analyses.
Chromosomes prepared on a slide glass were denatured and hybridized with a cocktail probe mixture for 2 d at 37 °C. The SKY probe mixture and hybridization reagents were purchased from Applied Spectral Imaging Inc. (Carlsbad, CA, USA) and signal detection was performed according to the manufacturer’s protocol. Chromosomes were counterstained with 4, 6-diaminido-2-phenylindole dihydrochloride (DAPI) combined with an anti-fade solution (Vectaschield; Vector Laboratories, Burlingame, CA, USA). Images were acquired by means of an SD200 Spectracube (Applied Spectral Imaging) mounted on an Olympus BX50-RF (Olympus, Tokyo, Japan) using a custom-designed optical filter (SKY-1; Chroma Technology, Brattleboro, VT). With another special optical filter, the inverted DAPI images were captured in conjunction with spectral classifications as QFH band patterns for the identification of chromosomal breakpoints[20,21]. For each case, 10-20 metaphase spreads were analyzed, and karyotypes were described according to the ISCN 1995.
Two cell lines, SNU16 and KMK2, were studied by means of DC-FISH using an oncogene and whole chromosome painting (WCP) probes. DC-FISH was performed as described previously[18]. The oncogene probe consisted of an I2 yeast artificial chromosome (YAC) clone containing MYC on 8q24.1 and a CPP29 cosmid containing CCND1 on 11q13.3[22,23]. Human-specific DNA sequences were amplified from YAC by means of Alu-polymerase chain reaction[24], DNA probes were labeled by biotin-16-dUTP and digoxigenin-16-dUTP (Boehringer Mannheim Biochemicals, Mannheim, Germany). Metaphase spreads were counterstained with 0.04 μg/mL of DAPI, mounted in an anti-fade solution (Vectaschield), and observed with a BX40-RF fluorescence microscope (Olympus). Images were captured with a CCD camera (SenSys0400-G1; Photometrics Ltd, Tucson, AZ, USA).
All established cell lines and patients with GC showed clonal karyotypic abnormalities. The modal chromosome number varied from 39 to 89. SKY detected chromosomal deletions, translocations (mainly unbalanced), inversions, insertions, dicentrics, duplication, ring chromosomes, isochromosomes, double minute chromosomes (DMs), and homogeneously staining regions (HSRs). Complex rearrangements involving more than three chromosomes were frequently encountered (Figure 1). Polysomy of chromosomes no. 16 (8 cases), no.20 (8), no. 11 (7), and no. 9, no. 15, and no. 22 (5 each), and monosomy of chromosomes no. 5 (5 cases), and no. 4, no. 6, no. 13, no. 14, no. 15, no. 21, and no. 22 (4 each) were frequently observed. Structural rearrangements most frequently involved chromosome no.8 (65 breaks), followed by chromosomes no. 11 (53), no. 1 (49), no. 7 (46), no. 13 (37), no. 3 (36), no. 17 (33), and no. 20 (29) (Figure 2). Chromosomal segments and bands were identified by combining SKY with DAPI banding (Figure 1). Representative karyotypes are listed in Tables 1 and 2.
A total of 630 chromosomal breaks that occurred in 177 breakpoints were identified by means of SKY analysis as shown in Figure 2 with several breakpoints highlighted: 8q24.1 (30 breaks), 11q13 (29), 13q14 (16), 20q11.2 (14), 7q32 (13), 17q11.2 (13), 18q21 (12), 17q23 (9), 18q11.2 (9), 1p22 (8), 1q32 (8), 7q22 (8), 8p11.2 (8), 8q22 (8), 9q22 (8), 10q22 (8), 20q13 (8), Xq22 (8), 7q11.2 (7), 11q23 (7), 22q11.2 (7), 3p13 (6), 3p21 (6), 3q21 (6), 4q12 (6), 9q13 (6), 13q12 (6), and 14p11.2 (6).
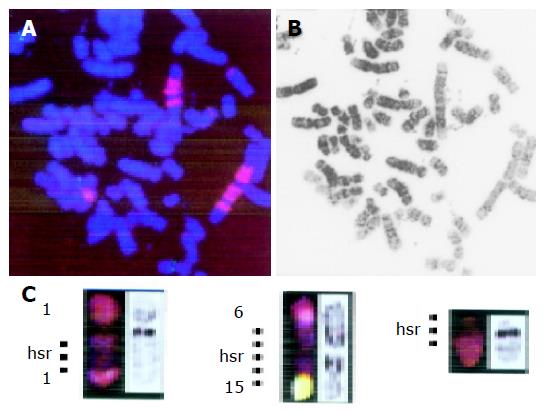
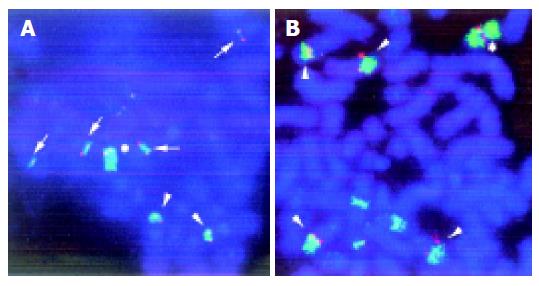
DC-FISH using YAC I2 and applied to two cell lines (SNU16 and KMK2) demonstrated that the chromosomal locus of the MYC gene (8q24.1) was involved in various types of chromosomal rearrangements including translocation, HSRs, and frequent insertion, thus resulting in gain and amplification of the gene (Figure 3). FISH using the MYC probe showed a periodic staining pattern, although we initially defined HSRs as abnormalities of chromosome no. 2 based on its SKY staining property as shown in Figure 3.
In KMK2, DC FISH with WCP 11 and cosmid CPP29 demonstrated that the chromosomal locus of the CCND1 gene (11q13) was involved in translocations, resulting in increased copy number of the gene (Figure 4).
A total of 242 chromosomal rearrangements including 190 translocations were identified. The four combinations of chromosomal bands repeatedly involved in translocations comprised 8q24.1 and 13q14 (SNU5, KMK2, and patient no. 6), 8q24.1 and 11q13 (SNU16, KMK2, and patient no. 1), 11q13 and 17q11.2 (KMK2 and MKN1), and 18q11.2 and 20q11.2 (GT3TKB and patient no. 7) as shown in Figure 5. Table 3 shows common translocation partners, some of which were previously undetectable because of the structurally complicated chromosomal rearrangements. The same der(20)t(18;20)(q11.2;q11.2) was found in two cases (GT3TKB and patient no. 7; Figure 3), and chromosome deletions of del(7)(q32), del(10)(q22), del(11)(q13), and del(18)(q11.2) were detected in two cases each.
| Translocation | Number of | Candidate | Partner breakpoints involved in chromsomal translocations or deletions1 | |
| breakpoint | breakpoints | genes | Common | Single |
| 8q24.1 | 30 | MYC | 11q13 [3], 13q14 [4] | 1p22, 1p32-36.1, 1p34, 1p36.1, 1p36.3, 1q42, 2p11.2, 3p21.3, 7q22, |
| 7q32, 7q36, 8p23, 9p13, 9q22, 10p13, 10q11.2, 17q11.2, 17q23, | ||||
| 18q21, 20p11.2, 20q11.2, 20q13.1, Xq28, deletion | ||||
| 11q13 | 26 | FGFR4 | 8q24.1 [3], 8q24.3 [3], | 1p13, 2p21, 7p15, 7q11.1, 7q22, 7q36, 7?, 9q13, 9q22, 11q23, 12q22, |
| 13p11.2, 14q24, 17q25, 19q13.3 | ||||
| FGF3 | 3q24-25 [2], 17q11.2 [2], | |||
| CCND1 | deletion [2] | |||
| 20q11.2 | 14 | Unknown | 18q11.2 [2] | 2p11.2, 2q33, 7p11.2, 7q22, 7q32, 8p11.2, 8q24, 11p13, 13q12, |
| 17q23, 18q21.1, deletion | ||||
| 1p32-36.3 | 12 | RUNX3 | 8q24 [4] | 2p11.1, 2q21, 9q32, 16q22, 17q21, 20q13.1, Xp11.4, deletion |
| 13q14 | 12 | RB | 8q24.1 [3] | 1q44, 3p13, 4q12, 7q36, 8q13, 13q22, 18q21, 22p11.2, deletion |
| 17q11.2 | 12 | ERBB2 | 11q13 [2] | 3p26, 4q12, 8p23, 8q24, 11q25, 12p11.2, 14p11.2, 16q22, Xq22, deletion |
| 7q32 | 11 | MET | deletion [2] | 3p26, 5q13, 8q22, 8q24, 9q22, 12q21, 17q23, 20q11.2, Xq13 |
| 18q21 | 11 | BCL2 | None | 1q32, 2p23, 3p21.3, 4q31.1, 8q24.1, 10q22, 13q14, 14q24, 20q11.2, deletion |
| 17q23 | 10 | Unknown | 11q25 [2] | 3p21, 4q33, 5?, 7q32, 8q24.1, 13q32, 20q11.2, 21q22 |
| 10q22 | 9 | KSAM | deletion [2] | 4q12, 6p25, 8p23, 9p13, 15q15, 15q22, 18q21 |
| 18q11.2 | 9 | Unknown | 20q11.2 [2],deletion [2] | 3q21, 6q12, 6q21, 19q13.1, Xq22 |
SKY analysis proved to be capable of identifying 630 breaks and 242 rearrangements of chromosomes in 10 established cell lines and seven cancerous ascitic fluid samples of GC, as well as of identifying recurrent breakpoints. Chromosomal rearrangements frequently involved well-known oncogene and tumor suppressor gene loci that may be associated with gastric carcinogenesis, such as 8q24.1 (MYC), 11q13 (HST1/FGF4, INT2/FGF3, and CCND1), 7q32 (MET), 13q14 (RB), 17q11.2 (adjacent to the ERBB2 locus), 18q21 (DCC and BCL2), 7q22 (MET), 3p21 (CTNNB1), and 16q22 (CDH1). The other breakpoints that were detected more than eight times were 20q11.2 (14 breaks), 17q23 (9), 18q11.2 (9), 1p22 (8), 1q32 (8), 8p11.2 (8), 8q22 (8), 9q22 (8), 10q22 (8), 20q13 (8), and Xq22 (8). However, it is not clear which breakpoints represent key genes during the earlier stages of tumorigenesis in this lineage, because when the entire tumor was examined in our study, it was already highly progressed and in the case of cell lines, they were further selected for liver culture conditions.
Two distinct breakpoints, which were not noted as unique occurrences by G-banding, 8q24.1 (30 breaks, 4.8% of 630 breaks) and 11q13 (29, 4.6%), were identified by a combination of SKY and DAPI banding analysis. The currently most likely candidate at 8q24 is the MYC gene, because it was frequently amplified and overexpressed in advanced GC, particularly in patients with carcinomatous peritonitis or distant metastasis[25,26]. The DC-FISH analysis used in our study showed that the MYC locus was involved in amplification resulting from the HSR as well as in multiple chromosomal translocations, insertions, and duplications in KMK2 (Figures 3 and 4). However, in SNU16, the MYC locus was amplified in HSRs but not on DMs, suggesting that other genes, for example the K-sam gene, may be involved in DMs (Figure 3)[27].
The second common breakpoint of 11q13 contains a variety of genes associated with cell proliferation and differentiation such as HST1/FGF4, INT2/FGF3, CCND1, SEA, MYEOV, and SPA1[28,29]. It has been demonstrated that HST1/FGF4 is co-amplified with INT2/FGF3 in human GC, although amplification of FGF4 and FGF3 does not correlate with mRNA overexpression[30], while high-level amplification of 11q13 has been frequently detected in CGH analysis[12,13]. All these genes are assigned to band 11q13.3. Band 11q13 may thus span a region approximately 25 Mb in size as estimated from both the physical length of 11q (86 Mb) and the number and size of the bands, which are divided into five sub-bands including three light bands (q13.1, q13.3, and q13.5). Since it is difficult to distinguish these subregions in a karyotype of 400 bands, 11q13.1 and 11q13.5 also warrant molecular dissection for the identification of target genes.
Combining SKY and DAPI banding analysis led to the identification of recurrent co-localization of the chromosomal bands involved in translocations: 8q24.1 and 13q14 (3 cases), 8q24.1 and 11q13 (3), 11q13 and 17q11.2 (2), and 18q11.2 and 20q11.2 (2). These translocations used to be frequently undetectable because of the complicated rearrangements of chromosomes. In most cases, each co-localized breakpoint contains chromosomal segments to which oncogenes or tumor suppressor genes have been assigned.
In conclusion, SKY analysis identified the frequently occurring breakpoints 8q24.1, 11q13, 20q11.1-13.1, and 13q14 and the recurrent translocations 8q24.1 and 13q14, 8q24.1 and 11q13, 11q13 and 17q11.2, and 18q11.2 and 20q11.2. SKY thus proved to be extremely useful for a comprehensive analysis of chromosomal translocations in GC and derived cell lines. The chromosomal breakpoints defined in our study may well contain critical genes which are involved in multistage carcinogenesis of GC and thus can serve as landmarks for crucial regions that warrant molecular dissection.
We are grateful to Professor Carlo M. Croce (Kimmel Cancer Center, Thomas Jefferson University, Philadelphia, PA) for providing DNA probes I2 and P72 (MYC) and to Dr. Masao Seto (Aichi Cancer Center Research Institute, Nagoya) for providing CPP29 (CCND1). We also wish to thank Professor Takeshi Okanoue (Department of Gastroenterology and Hepatology, Kyoto Prefectural University of Medicine, Kyoto) for critical reading of the manuscript.
Science Editor Guo SY Language Editor Elsevier HK
| 1. | Kameda T, Yasui W, Yoshida K, Tsujino T, Nakayama H, Ito M, Ito H, Tahara E. Expression of ERBB2 in human gastric carcinomas: relationship between p185ERBB2 expression and the gene amplification. Cancer Res. 1990;50:8002-8009. [PubMed] |
| 2. | Kuniyasu H, Yasui W, Kitadai Y, Yokozaki H, Ito H, Tahara E. Frequent amplification of the c-met gene in scirrhous type stomach cancer. Biochem Biophys Res Commun. 1992;189:227-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 241] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 3. | Grady WM. Genetics of gastric cancer. Molecular Genetics of Cancer. San Diego: Bios Scientific Publishers 2001; 115-148. |
| 4. | Becker KF, Atkinson MJ, Reich U, Becker I, Nekarda H, Siewert JR, Höfler H. E-cadherin gene mutations provide clues to diffuse type gastric carcinomas. Cancer Res. 1994;54:3845-3852. [PubMed] |
| 5. | Li QL, Ito K, Sakakura C, Fukamachi H, Inoue Ki, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 789] [Cited by in RCA: 835] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 6. | Ochi H, Douglass HO, Sandberg AA. Cytogenetic studies in primary gastric cancer. Cancer Genet Cytogenet. 1986;22:295-307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 78] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 7. | Misawa S, Horiike S, Taniwaki M, Tsuda S, Okuda T, Kashima K, Abe T, Sugihara H, Noriki S, Fukuda M. Chromosome abnormalities of gastric cancer detected in cancerous effusions. Jpn J Cancer Res. 1990;81:148-152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 8. | Seruca R, Castedo S, Correia C, Gomes P, Carneiro F, Soares P, de Jong B, Sobrinho-Simões M. Cytogenetic findings in eleven gastric carcinomas. Cancer Genet Cytogenet. 1993;68:42-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 40] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 9. | Panani AD, Ferti A, Malliaros S, Raptis S. Cytogenetic study of 11 gastric adenocarcinomas. Cancer Genet Cytogenet. 1995;81:169-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 10. | Kokkola A, Monni O, Puolakkainen P, Larramendy ML, Victorzon M, Nordling S, Haapiainen R, Kivilaakso E, Knuutila S. 17q12-21 amplicon, a novel recurrent genetic change in intestinal type of gastric carcinoma: a comparative genomic hybridization study. Genes Chromosomes Cancer. 1997;20:38-43. [RCA] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 11. | Kokkola A, Monni O, Puolakkainen P, Nordling S, Haapiainen R, Kivilaakso E, Knuutila S. Presence of high-level DNA copy number gains in gastric carcinoma and severely dysplastic adenomas but not in moderately dysplastic adenomas. Cancer Genet Cytogenet. 1998;107:32-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 26] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 12. | Nessling M, Solinas-Toldo S, Wilgenbus KK, Borchard F, Lichter P. Mapping of chromosomal imbalances in gastric adenocarcinoma revealed amplified protooncogenes MYCN, MET, WNT2, and ERBB2. Genes Chromosomes Cancer. 1998;23:307-316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 13. | Sakakura C, Mori T, Sakabe T, Ariyama Y, Shinomiya T, Date K, Hagiwara A, Yamaguchi T, Takahashi T, Nakamura Y. Gains, losses, and amplifications of genomic materials in primary gastric cancers analyzed by comparative genomic hybridization. Genes Chromosomes Cancer. 1999;24:299-305. [RCA] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 14. | Rodriguez E, Rao PH, Ladanyi M, Altorki N, Albino AP, Kelsen DP, Jhanwar SC, Chaganti RS. 11p13-15 is a specific region of chromosomal rearrangement in gastric and esophageal adenocarcinomas. Cancer Res. 1990;50:6410-6416. [PubMed] |
| 15. | Chun YH, Kil JI, Suh YS, Kim SH, Kim H, Park SH. Characterization of chromosomal aberrations in human gastric carcinoma cell lines using chromosome painting. Cancer Genet Cytogenet. 2000;119:18-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 16. | Stamouli MI, Ferti AD, Panani AD, Raftakis J, Consoli C, Raptis SA, Young BD. Application of multiplex fluorescence in situ hybridization in the cytogenetic analysis of primary gastric carcinoma. Cancer Genet Cytogenet. 2002;135:23-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 17. | Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. an attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31-49. [PubMed] |
| 18. | Taniwaki M, Nishida K, Ueda Y, Misawa S, Nagai M, Tagawa S, Yamagami T, Sugiyama H, Abe M, Fukuhara S. Interphase and metaphase detection of the breakpoint of 14q32 translocations in B-cell malignancies by double-color fluorescence in situ hybridization. Blood. 1995;85:3223-3228. [PubMed] |
| 19. | International Standing Committee on Human Cytogenetic Nomenclature. In: Mittelman F, ed. An International System for Human Cytogenetic Nomenclature. Basel, Switzerland: S. Karger AG; 1995; PMC162727. |
| 20. | Schröck E, du Manoir S, Veldman T, Schoell B, Wienberg J, Ferguson-Smith MA, Ning Y, Ledbetter DH, Bar-Am I, Soenksen D. Multicolor spectral karyotyping of human chromosomes. Science. 1996;273:494-497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1223] [Cited by in RCA: 1046] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 21. | Kakazu N, Taniwaki M, Horiike S, Nishida K, Tatekawa T, Nagai M, Takahashi T, Akaogi T, Inazawa J, Ohki M. Combined spectral karyotyping and DAPI banding analysis of chromosome abnormalities in myelodysplastic syndrome. Genes Chromosomes Cancer. 1999;26:336-345. [DOI] [Full Text] |
| 22. | Veronese ML, Ohta M, Finan J, Nowell PC, Croce CM. Detection of myc translocations in lymphoma cells by fluorescence in situ hybridization with yeast artificial chromosomes. Blood. 1995;85:2132-2138. [PubMed] |
| 23. | Takashima T, Itoh M, Ueda Y, Nishida K, Tamaki T, Misawa S, Abe T, Seto M, Machii T, Taniwaki M. Detection of 14q32.33 translocation and t(11; 14) in interphase nuclei of chronic B-cell leukemia/lymphomas by in situ hybridization. Int J Cancer. 1997;72:31-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 24. | Taniwaki M, Matsuda F, Jauch A, Nishida K, Takashima T, Tagawa S, Sugiyama H, Misawa S, Abe T, Kashima K. Detection of 14q32 translocations in B-cell malignancies by in situ hybridization with yeast artificial chromosome clones containing the human IgH gene locus. Blood. 1994;83:2962-2969. [PubMed] |
| 25. | Ninomiya I, Yonemura Y, Matsumoto H, Sugiyama K, Kamata T, Miwa K, Miyazaki I, Shiku H. Expression of c-myc gene product in gastric carcinoma. Oncology. 1991;48:149-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 26. | Hajdú J, Kozma L, Kiss I, Szentkereszty Z, Szakáll S, Ember I. Is the presence of distant metastasis associated with c-myc amplification in gastric cancer? Acta Chir Hung. 1997;36:119-121. [PubMed] |
| 27. | Hara T, Ooi A, Kobayashi M, Mai M, Yanagihara K, Nakanishi I. Amplification of c-myc, K-sam, and c-met in gastric cancers: detection by fluorescence in situ hybridization. Lab Invest. 1998;78:1143-1153. [PubMed] |
| 28. | Wada Y, Kubota H, Maeda M, Taniwaki M, Hattori M, Imamura S, Iwai K, Minato N. Mitogen-inducible SIPA1 is mapped to the conserved syntenic groups of chromosome 19 in mouse and chromosome 11q13.3 centromeric to BCL1 in human. Genomics. 1997;39:66-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 29. | Katoh M, Katoh M. FLJ10261 gene, located within the CCND1-EMS1 locus on human chromosome 11q13, encodes the eight-transmembrane protein homologous to C12orf3, C11orf25 and FLJ34272 gene products. Int J Oncol. 2003;22:1375-1381. [PubMed] |
| 30. | Yoshida MC, Wada M, Satoh H, Yoshida T, Sakamoto H, Miyagawa K, Yokota J, Koda T, Kakinuma M, Sugimura T. Human HST1 (HSTF1) gene maps to chromosome band 11q13 and coamplifies with the INT2 gene in human cancer. Proc Natl Acad Sci USA. 1988;85:4861-4864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 70] [Article Influence: 1.9] [Reference Citation Analysis (0)] |