Published online Mar 1, 2004. doi: 10.3748/wjg.v10.i5.660
Revised: November 12, 2003
Accepted: November 19, 2003
Published online: March 1, 2004
AIM: To prepare thymidine kinase gene (TK gene) nanoparticles and to investigate the expression of TK gene.
METHODS: Poly(D,L-lactic-co-glycolic acid) (PLGA), a biodegradable and biocompatible polymer, was used to prepare recombinant plasmid PEGFP-AFP nanoparticles by a double-emulsion evaporation technique. Characteristics of the nanoparticles were investigated in this study, including morphology, entrapment efficiency, and tissue distribution. The expression of TK gene was also investigated by MTT assay, by which the viable cells were determined after the addition of ganciclovir (GCV). The enhanced green fluorescent protein (EGFP) expression in human hepatocellular carcinoma SMMC-7721 cells and normal parenchymal Chang liver cells were assessed by flow cytometry.
RESULTS: The prepared plasmid-nanoparticles had regular spherical surface and narrow particle size span with a mean diameter of 72 ± 12 nm. The mean entrapment efficiency was 91.25%. A total of 80.14% DNA was found to be localized in the livers after 1-h injection with 32P-DNA-PLGA nanoparticles in mouse caudal vein. The expression of DNA encapsulated in nanoparticles was much higher than that in naked DNA, and human hepatocellular carcinoma SMMC-7721 cells were more sensitive to GCV than human normal parenchymal Chang liver cells.
CONCLUSION: The enhanced transfection efficiency and stronger ability to protect plasmid DNA from being degraded by nucleases are due to nanoparticles encapsulation.
- Citation: He Q, Liu J, Sun X, Zhang ZR. Preparation and characteristics of DNA-nanoparticles targeting to hepatocarcinoma cells. World J Gastroenterol 2004; 10(5): 660-663
- URL: https://www.wjgnet.com/1007-9327/full/v10/i5/660.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i5.660
In recent years, the gene delivery system has attracted much attention[1-3]. However, safe and efficient gene delivery remains a crucial barrier to successful gene therapy. Viral and retroviral vectors have been the most efficient and commonly used delivery modalities for in vivo gene transfer, but viral vector may provoke mutagenesis and carcinogenesis. Repeated administration of a viral vector induces an immune response which abolishes the transgene expression[4-7]. The non-viral delivery system has the potential to be non-immunogenic and stable in vivo[8-11]. Encapsulation of DNA in biodegradable polymer potentially offers a way to protect DNA from degradation and to control DNA release[12,13], and many examples of DNA incorporated in synthetic polymers have been developed in the micron scale. Recently, some studies have showen that intracellular biodistribution of particles with diameter less than 100 nm can be achieved[14].
Among all the present gene therapeutic protocols, combination of the administration of GCV with transfecting thymidine kinase gene of Herpes simplex virus (HSV-TK) into tumor cells is rather practical and potential in intra-tumoral gene therapy. The TK genes in the tumor cells can induce the metabolism of untoxicant prodrug GCV into cytotoxic parent drug, which can cause the suicide of cells. This protocol presents good potential in intra-tumoral gene therapy[15,16]. However, common TK genes (naked genes) do not have the abilities to target to specific organs and tissues, which can be harmful to the normal cells and tissues. In addition, they are easily degraded by nucleases in vivo.
To solve the problems mentioned above, a recombinant plasmid PEGFP -TKAFP was constructed, which can be specifically expressed in hepatocellular carcinoma cells. Furthermore, the plasmid was encapsulated in a biodegradable and biocompatible PLGA polymer to protect plasmid DNA from being digested by nucleases. The following characteristics of the nanoparticles were investigated, including in vitro anti-nuclease ability, tissue distribution in mice and the gene expression in hepatocellular carcinoma cells and normal parenchymal cells in vitro.
Poly(D,L-lactic-co-glycolic acid) (PLGA; lactic-glycolic acid ratio: 75:25, Mr = 30000, batch number: 020112) was purchased from Chengdu Institute of Organic Chemistry, Chinese Academy of Science. Recombinant plasmid PEGFP-TKAFP was a gift from Dr. Liu Ji (Sichuan University). Human hepatocellular carcinoma SMMC-7721 cells[17] and normal parenchymal Chang liver cells[18] were provided by Shanghai Institute of Cell Biology, Chinese Academy of Science. DNase I was purchased from Chengdu Huamei Biochemicals Cooperation (Sichuan Province, China). Kunming mice, weighted 18-22 g, were provided by Experimental Animal Center of Sichuan University.
Nanoparticles preparation A double-emulsion evaporation technique[19] was used to prepare the nanoparticles. Briefly, plasmid DNA (200 µg) in 100 µL Tris-EDTA (TE) buffer was emulsified in 1 mL methylene chloride solution containing 100 mg of PLGA using a probe sonicator for 5 s. Polyvinyl alcohol (2 mL) was added to the primary emulsion and sonicated for another 5 s to form a double emulsion. The emulsion was added into the same concentration of polyvinyl alcohol and agitated by a magnetic stirrer for 3 h at room temperature to remove methylene chloride.
Particle size and morphology analysis The PLGA nanoparticles were sized by laser diffractometry using a Malvern 2000 laser sizer. The morphology was observed by the scanning electron microscope (JEM-100SX, Akishima, Japan). The samples were placed on to special copper grids and then stained with 20 mL/L phosphato-tungstic acid prior to visualization.
Entrapment ratio analysis The entrapment ratio was determined by measuring the total amount of added DNA and that of DNA being not encapsulated. In detail, colloid solution of DNA-PLGA nanoparticles was centrifuged at 45000 g for 1 h. Then, the concentration of DNA in the supernatant was assessed by fluorescence spectrophotometry after stained with ethidium bromide. The exciting and emission wavelengths were 546 nm and 590 nm, respectively. The entrapment rate (ER) was calculated as follows: ER (%) = DNA added-DNA in the supernatant/DNAadded × 100%
Protection from DNase The PLGA nanoparticles were incubated with DNase I (0.1 unit) at 37 °C in a shaking water bath. The nanoparticles were collected by centrifugation after 4, 8, and 16 h incubation, and then chloroform was added to solubilize the nanoparticles. An equal volume of PBS solution was added, and the mixture was rotated end-over-end to facilitate the extraction of DNA from the organic phase into the aqueous phase. The samples were then centrifuged at 15000 g for 15 min. The resulted supernatant was transferred to another tube and DNA was precipitated with the addition of isopropanol. Precipitate was obtained after centrifugation at 5000 r/min for 15 min. Then, the resulted pellet was rinsed with 700 mL/L ethanol and resuspended in sterile TE buffer. The purified DNA was analyzed by gel electrophoresis.
Tissue distribution One hundred Kunming mice weighed 18-22 g were randomly divided into 10 test groups and 10 control groups with 5 in each group. The nanoparticles of 32P-DNA-PLGA at a dose of 10 µL/g was intravenously administered to each mouse in test groups, and 32P-DNA at the same dose was intravenously administered in control groups. At predetermined intervals, mice were sacrificed for blood collection. Then, heart, livers, spleen, lungs, and kidneys were removed from mice. The radioactivity of each organ was measured by a liquid scintillation analyzer.
The cpmt was the total value of cpm in each organ (cpmi) at a certain time point. The ratio of cpmi/cpmt× 100% represented the relative content of DNA in viscera and blood.
MFI assay Human hepatocellular carcinoma SMMC-7721 cells and normal parenchymal Chang liver cells (5 × 105) were cultured in the DMEM medium containing 100 mL/L fetal bovine serum (FBS) in 12-well plates. The cells were transfected with plasmid DNA or nanoparticles containing DNA, and maintained at 37 °C in an incubator at a 50 mL/L CO2 humidified atmosphere. After incubation for 12 h, the medium was removed and replaced with fresh DMEM containing 100 mL/L FBS for further 48 h incubation. The mean fluorescence intensity (MFI) of the cells was measured by flow cytometry.
Cytotoxicity assay Cells were cultured in the same way as the MFI assay. After incubation for 12 h, the medium was removed, replaced with DMEM containing 100 mL/L FBS, and incubated with 0.1, 1, or 10 µg/mL GCV. The cytotoxicity of GCV was detected by MTT assay.
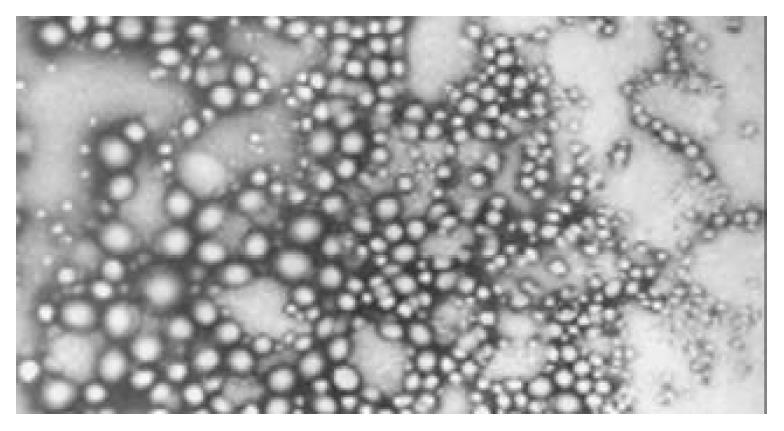
The resulted plasmid-nanoparticles had regular spherical surface (Figure 1) and a narrow size distribution with a mean diameter of 72 ± 12 nm.
The mean entrapment efficiency was 91.25%, which was rather high in the nanoparticles preparation with PLGA as a carrier.
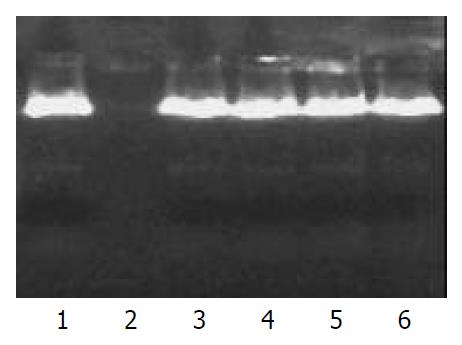
Plasmid DNA encapsulated in nanoparticles remained intact in the presence of DNase I for up to 16 h incubation. On the other hand, control plasmid DNA was completely digested within 1 h incubation with the equal amount of DNase I. This result demonstrated that PLGA nanoparticles could protect encapsulated plasmid DNA from nuclease digestion (Figure 2).
The tissue distribution of DNA-PLGA nanoparticles was investigated by the technique of gamma scintigraphy. The results showed that 1 h after injection with 32P-DNA-PLGA nanoparticles in mouse caudal vein, the ratio of radioactivity in livers against total radioactivity was more than 80%, which was 1.5-fold of that after injection with 32P-DNA alone (Table 1 and Table 2).
| Time | Heart | Liver | Spleen | Lung | Kidney | Blood |
| 5 min | 3.14 ± 0.60 | 36.67 ± 2.65 | 3.56 ± 1.58 | 16.32 ± 0.79 | 23.52 ± 1.48 | 16.79 ± 1.62 |
| 15 min | 3.15 ± 0.97 | 41.26 ± 2.89 | 5.25 ± 0.86 | 14.16 ± 0.99 | 20.61 ± 1.62 | 15.57 ± 1.58 |
| 30 min | 6.94 ± 1.57 | 52.59 ± 4.02 | 10.46 ± 0.75 | 7.95 ± 0.58 | 13.10 ± 0.85 | 2.95 ± 0.65 |
| 1 h | 6.11 ± 1.95 | 54.62 ± 3.12 | 14.85 ± 1.23 | 7.82 ± 0.67 | 13.50 ± 0.94 | 3.10 ± 0.78 |
| 2 h | 4.56 ± 1.01 | 51.34 ± 2.88 | 14.93 ± 0.68 | 9.65 ± 1.41 | 15.03 ± 1.34 | 4.49 ± 0.28 |
| 6 h | 5.12 ± 0.79 | 48.66 ± 1.87 | 13.15 ± 0.84 | 9.66 ± 1.27 | 18.26 ± 1.58 | 5.15 ± 1.67 |
| 12 h | 5.48 ± 0.99 | 44.52 ± 2.02 | 11.32 ± 1.33 | 11.36 ± 1.34 | 21.83 ± 2.54 | 5.49 ± 0.58 |
| 24 h | 3.93 ± 0.65 | 43.31 ± 1.67 | 11.48 ± 1.11 | 10.12 ± 0.64 | 25.01 ± 3.12 | 6.16 ± 0.94 |
| 48 h | 3.43 ± 0.84 | 36.75 ± 1.65 | 11.56 ± 0.82 | 13.36 ± 1.60 | 28.52 ± 2.15 | 6.38 ± 0.83 |
| 72 h | 4.52 ± 0.77 | 34.48 ± 1.85 | 12.03 ± 1.73 | 13.88 ± 1.63 | 28.31 ± 3.01 | 6.78 ± 0.91 |
| Time | Heart | Liver | Spleen | Lung | Kidney | Blood |
| 5 min | 2.22 ± 0.51 | 57.32 ± 2.36 | 2.96 ± 0.69 | 8.29 ± 1.33 | 16.58 ± 2.02 | 12.63 ± 1.24 |
| 15 min | 3.52 ± 0.64 | 69.69 ± 3.32 | 3.19 ± 0.58 | 5.77 ± 0.55 | 13.87 ± 0.99 | 3.96 ± 0.36 |
| 30 min | 2.38 ± 0.67 | 73.37 ± 3.62 | 7.55 ± 1.03 | 3.64 ± 0.41 | 11.28 ± 1.28 | 1.78 ± 0.65 |
| 1 h | 2.08 ± 0.54 | 80.14 ± 4.56 | 3.37 ± 0.67 | 3.08 ± 0.62 | 9.34 ± 0.68 | 1.99 ± 0.54 |
| 2 h | 2.59 ± 0.28 | 78.45 ± 4.02 | 3.73 ± 0.76 | 5.35 ± 0.58 | 8.36 ± 0.94 | 1.47 ± 0.23 |
| 6 h | 3.18 ± 0.60 | 75.03 ± 3.69 | 7.38 ± 1.03 | 3.74 ± 0.39 | 9.61 ± 0.96 | 1.06 ± 0.24 |
| 12 h | 2.21 ± 0.51 | 69.34 ± 3.96 | 6.17 ± 1.24 | 7.88 ± 0.64 | 10.17 ± 1.04 | 4.22 ± 0.32 |
| 24 h | 1.94 ± 0.32 | 65.59 ± 3.25 | 6.46 ± 1.04 | 7.95 ± 0.57 | 12.10 ± 1.23 | 5.96 ± 0.59 |
| 48 h | 2.21 ± 0.29 | 61.18 ± 2.69 | 7.01 ± 0.48 | 9.27 ± 0.71 | 12.87 ± 0.86 | 6.46 ± 0.86 |
| 72 h | 2.51 ± 0.37 | 60.56 ± 2.98 | 6.45 ± 0.69 | 9.98 ± 0.65 | 13.54 ± 1.11 | 6.96 ± 0.75 |
The EGFP expression in human hepatocellular carcinoma SMMC-7721 cells and normal parenchymal Chang liver cells were assessed by flow cytometry. The expression of TK gene was also investigated by MTT assay, by which the viable cells were quantitated after the addition of parent drug GCV. The results showed that the EGFP and TK expression in human hepatocellular carcinoma SMMC-7721 cells was much higher than that in human normal parenchymal Chang liver cells (P < 0.05) (Table 3). It also showed that plasmid DNA encapsulated in nanoparticles could enhance the expression of TK or EGFP gene compared with naked plasmid DNA (P < 0.05) (Table 4).
Recombinant plasmid PEGFP-AFP was constructed, which could be specifically expressed in hepatocellular carcinoma cells because of alpha-fetoprotein-albumin (AFP-alb) promoter[20-27]. Meanwhile, EGFP as the reporter gene of plasmid DNA, can be assessed by confocal laser scanning microscopy and flow cytometry[28-32]. A polymer, PLGA, was selected due to its biocompatible and biodegradable properties, which was already approved for in vivo applications[33-39]. Non-toxicity of the carrier may permit repeated administration of the nanoparticles to compensate for transient transgene expression.
Our results showed that plasmid DNA could be encapsulated in PLGA nanoparticles without compromising its structural and functional integrity. Additionally, PLGA nanoparticles could protect plasmids from nuclease degradation, and therefore offer an effective approach for gene delivery in vivo. However, the relatively low transfection efficiency was obtained in comparison with viral vector, which still remains to be a problem.
The DNA nanoparticles probably permeate the cells through endocytotic mechanism due to their small size and negative charged surface[40]. The encapsulation of plasmid DNA in cationic liposomes offers another choice to be protected from DNases. However, cationic liposomes may be toxic to cells due to an excess of positive charge[41,42], and can be easily influenced by the substances in plasma. In recent years, nanoparticles attract more and more attention because of many advantages, including high stability at room temperature, favorable safety, the ability to deliver plasmid DNA at a controllable rate, and easy adaptability. Unlike most viral vector, there is no limit on the size of plasmids encapsulated into the nanoparticles[43,44].
Edited by Chao JCJ Proofread by Xu FM
| 1. | Wiethoff CM, Middaugh CR. Barriers to nonviral gene delivery. J Pharm Sci. 2003;92:203-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 377] [Cited by in RCA: 348] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 2. | Guo SY, Gu QL, Zhu ZG, Hong HQ, Lin YZ. TK gene combined with mIL-2 and mGM-CSF genes in treatment of gastric cancer. World J Gastroenterol. 2003;9:233-237. [PubMed] |
| 3. | Mhashilkar A, Chada S, Roth JA, Ramesh R. Gene therapy. Therapeutic approaches and implications. Biotechnol Adv. 2001;19:279-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 4. | Liu F, Huang L. Development of non-viral vectors for systemic gene delivery. J Control Release. 2002;78:259-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 196] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 5. | Corsi K, Chellat F, Yahia L, Fernandes JC. Mesenchymal stem cells, MG63 and HEK293 transfection using chitosan-DNA nanoparticles. Biomaterials. 2003;24:1255-1264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 289] [Cited by in RCA: 260] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 6. | Brown MD, Schätzlein AG, Uchegbu IF. Gene delivery with synthetic (non viral) carriers. Int J Pharm. 2001;229:1-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 279] [Cited by in RCA: 250] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 7. | Kircheis R, Wightman L, Wagner E. Design and gene delivery activity of modified polyethylenimines. Adv Drug Deliv Rev. 2001;53:341-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 541] [Cited by in RCA: 508] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 8. | Hashida M, Nishikawa M, Yamashita F, Takakura Y. Cell-specific delivery of genes with glycosylated carriers. Adv Drug Deliv Rev. 2001;52:187-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 142] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 9. | Kamiya H, Tsuchiya H, Yamazaki J, Harashima H. Intracellular trafficking and transgene expression of viral and non-viral gene vectors. Adv Drug Deliv Rev. 2001;52:153-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 71] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 10. | Zhdanov RI, Podobed OV, Vlassov VV. Cationic lipid-DNA complexes-lipoplexes-for gene transfer and therapy. Bioelectrochemistry. 2002;58:53-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 123] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 11. | Oku N, Yamazaki Y, Matsuura M, Sugiyama M, Hasegawa M, Nango M. A novel non-viral gene transfer system, polycation liposomes. Adv Drug Deliv Rev. 2001;52:209-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 42] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 12. | Crommelin DJ, Storm G, Jiskoot W, Stenekes R, Mastrobattista E, Hennink WE. Nanotechnological approaches for the delivery of macromolecules. J Control Release. 2003;87:81-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 62] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 13. | Guang Liu W, De Yao K. Chitosan and its derivatives--a promising non-viral vector for gene transfection. J Control Release. 2002;83:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 133] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 14. | Hirosue S, Müller BG, Mulligan RC, Langer R. Plasmid DNA encapsulation and release from solvent diffusion nanospheres. J Control Release. 2001;70:231-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 45] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 15. | Singh S, Cunningham C, Buchanan A, Jolly DJ, Nemunaitis J. Toxicity assessment of intratumoral injection of the herpes simplex type I thymidine kinase gene delivered by retrovirus in patients with refractory cancer. Mol Ther. 2001;4:157-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 16. | Vlachaki MT, Chhikara M, Aguilar L, Zhu X, Chiu KJ, Woo S, Teh BS, Thompson TC, Butler EB, Aguilar-Cordova E. Enhanced therapeutic effect of multiple injections of HSV-TK + GCV gene therapy in combination with ionizing radiation in a mouse mammary tumor model. Int J Radiat Oncol Biol Phys. 2001;51:1008-1017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 17. | Dong RC, Zhou RH, Lu FD, Tao WZ. Primary study on the es-tablishment of Human hepatocellular carcinoma cell line SMMC-7721 and its biological characteristics. Dier Junyi Daxue Xuebao. 1980;1:5-9. |
| 18. | CHANG RS. Continuous subcultivation of epithelial-like cells from normal human tissues. Proc Soc Exp Biol Med. 1954;87:440-443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 195] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 19. | Prabha S, Zhou WZ, Panyam J, Labhasetwar V. Size-dependency of nanoparticle-mediated gene transfection: studies with fractionated nanoparticles. Int J Pharm. 2002;244:105-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 441] [Cited by in RCA: 399] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 20. | Sa Cunha A, Bonte E, Dubois S, Chrétien Y, Eraiser T, Degott C, Bréchot C, Tran PL. Inhibition of rat hepatocellular carcinoma tumor growth after multiple infusions of recombinant Ad.AFPtk followed by ganciclovir treatment. J Hepatol. 2002;37:222-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 21. | Li MS, Li PF, He SP, Du GG, Li G. The promoting molecular mechanism of alpha-fetoprotein on the growth of human hepatoma Bel7402 cell line. World J Gastroenterol. 2002;8:469-475. [PubMed] |
| 22. | Cao G, Kuriyama S, Gao J, Nakatani T, Chen Q, Yoshiji H, Zhao L, Kojima H, Dong Y, Fukui H. Gene therapy for hepatocellular carcinoma based on tumour-selective suicide gene expression using the alpha-fetoprotein (AFP) enhancer and a housekeeping gene promoter. Eur J Cancer. 2001;37:140-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 23. | Kim J, Lee B, Kim JS, Yun CO, Kim JH, Lee YJ, Joo CH, Lee H. Antitumoral effects of recombinant adenovirus YKL-1001, conditionally replicating in alpha-fetoprotein-producing human liver cancer cells. Cancer Lett. 2002;180:23-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 24. | Ishikawa H, Nakata K, Mawatari F, Ueki T, Tsuruta S, Ido A, Nakao K, Kato Y, Ishii N, Eguchi K. Retrovirus-mediated gene therapy for hepatocellular carcinoma with reversely oriented therapeutic gene expression regulated by alpha-fetoprotein enhancer/promoter. Biochem Biophys Res Commun. 2001;287:1034-1040. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 25. | Takahashi M, Sato T, Sagawa T, Lu Y, Sato Y, Iyama S, Yamada Y, Fukaura J, Takahashi S, Miyanishi K. E1B-55K-deleted adenovirus expressing E1A-13S by AFP-enhancer/promoter is capable of highly specific replication in AFP-producing hepatocellular carcinoma and eradication of established tumor. Mol Ther. 2002;5:627-634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 52] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 26. | Ye X, Liang M, Meng X, Ren X, Chen H, Li ZY, Ni S, Lieber A, Hu F. Insulation from viral transcriptional regulatory elements enables improvement to hepatoma-specific gene expression from adenovirus vectors. Biochem Biophys Res Commun. 2003;307:759-764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 27. | Lu SY, Sui YF, Li ZS, Pan CE, Ye J, Wang WY. Construction of a regulable gene therapy vector targeting for hepatocellular carcinoma. World J Gastroenterol. 2003;9:688-691. [PubMed] |
| 28. | Kantakamalakul W, Jaroenpool J, Pattanapanyasat K. A novel enhanced green fluorescent protein (EGFP)-K562 flow cytometric method for measuring natural killer (NK) cell cytotoxic activity. J Immunol Methods. 2003;272:189-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 37] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 29. | Bi JX, Wirth M, Beer C, Kim EJ, Gu MB, Zeng AP. Dynamic characterization of recombinant Chinese hamster ovary cells containing an inducible c-fos promoter GFP expression system as a biomarker. J Biotechnol. 2002;93:231-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 30. | Henry SC, Schmader K, Brown TT, Miller SE, Howell DN, Daley GG, Hamilton JD. Enhanced green fluorescent protein as a marker for localizing murine cytomegalovirus in acute and latent infection. J Virol Methods. 2000;89:61-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 50] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 31. | Jakobs S, Subramaniam V, Schönle A, Jovin TM, Hell SW. EFGP and DsRed expressing cultures of Escherichia coli imaged by confocal, two-photon and fluorescence lifetime microscopy. FEBS Lett. 2000;479:131-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 120] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 32. | Hanson P, Mathews V, Marrus SH, Graubert TA. Enhanced green fluorescent protein targeted to the Sca-1 (Ly-6A) locus in transgenic mice results in efficient marking of hematopoietic stem cells in vivo. Exp Hematol. 2003;31:159-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 33. | Yamaguchi Y, Takenaga M, Kitagawa A, Ogawa Y, Mizushima Y, Igarashi R. Insulin-loaded biodegradable PLGA microcapsules: initial burst release controlled by hydrophilic additives. J Control Release. 2002;81:235-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 110] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 34. | Pérez-Rodriguez C, Montano N, Gonzalez K, Griebenow K. Stabilization of alpha-chymotrypsin at the CH2Cl2/water interface and upon water-in-oil-in-water encapsulation in PLGA microspheres. J Control Release. 2003;89:71-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 59] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 35. | Dorta MJ, Santoveña A, Llabrés M, Fariña JB. Potential applications of PLGA film-implants in modulating in vitro drugs release. Int J Pharm. 2002;248:149-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 37] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 36. | Lu L, Yaszemski MJ, Mikos AG. Retinal pigment epithelium engineering using synthetic biodegradable polymers. Biomaterials. 2001;22:3345-3355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 75] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 37. | Jain RA, Rhodes CT, Railkar AM, Malick AW, Shah NH. Controlled release of drugs from injectable in situ formed biodegradable PLGA microspheres: effect of various formulation variables. Eur J Pharm Biopharm. 2000;50:257-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 80] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 38. | Panyam J, Labhasetwar V. Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Adv Drug Deliv Rev. 2003;55:329-347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2387] [Cited by in RCA: 2035] [Article Influence: 92.5] [Reference Citation Analysis (0)] |
| 39. | Vila A, Sánchez A, Tobío M, Calvo P, Alonso MJ. Design of biodegradable particles for protein delivery. J Control Release. 2002;78:15-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 436] [Cited by in RCA: 364] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 40. | Garcia-Chaumont C, Seksek O, Grzybowska J, Borowski E, Bolard J. Delivery systems for antisense oligonucleotides. Pharmacol Ther. 2000;87:255-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 73] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 41. | Han So RI, Kim SW. Water-soluble lipopolymer for gene delivery. Bioconjug Chem. 2001;12:337-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 165] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 42. | Pouton CW, Seymour LW. Key issues in non-viral gene delivery. Adv Drug Deliv Rev. 2001;46:187-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 237] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 43. | del Barrio GG, Novo FJ, Irache JM. Loading of plasmid DNA into PLGA microparticles using TROMS (Total Recirculation One-Machine System): evaluation of its integrity and controlled release properties. J Control Release. 2003;86:123-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 44. | Davis ME. Non-viral gene delivery systems. Curr Opin Biotechnol. 2002;13:128-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 327] [Cited by in RCA: 301] [Article Influence: 13.1] [Reference Citation Analysis (0)] |