Published online Sep 15, 2004. doi: 10.3748/wjg.v10.i18.2719
Revised: January 10, 2004
Accepted: February 18, 2004
Published online: September 15, 2004
AIM: To study the frameshift mutations of the retinoblastoma protein-interacting zinc finger gene RIZ in gastric cancer with microsatellite instability, and to identify two coding polyadenosine tracts of RIZ.
METHODS: Frameshift mutations at (A)8 and (A)9 tracts of RIZ were detected in 70 human gastric cancer (HGC) specimens by DHPLC and DNA sequencing. Microsatellite instability (MSI) status was assessed by two mononucleotide markers, BAT26 and BAT25, by means of denaturing high-performance liquid chromatography (DHPLC).
RESULTS: In 70 HGC samples, 8 (11.4%) were found positive for instabilities at BAT26 and BAT25. In 7 of the 8 cases with instabilities at both BAT26 and BAT25 (MSI-H), 1 was unstable at BAT26 but stable at BAT25. Frameshift mutations were identified in 4 (57.1%) of the 7 samples with MSI-H in the (A)9 tract of RIZ without mutations in the (A)8 tract. In contrast, frameshift mutations were found in neither of the polyadenosine tracts in 63 samples of MSI-L or MSI stable tumors. Pro704 LOH detection in 4 cases with frameshift mutations did not find LOH in these cases.
CONCLUSION: Frameshift mutations of RIZ may play an important role in gastric cancers with MSI.
-
Citation: Pan KF, Lu YY, Liu WG, Zhang L, You WC. Detection of frameshift mutations of
RIZ in gastric cancers with microsatellite instability. World J Gastroenterol 2004; 10(18): 2719-2722 - URL: https://www.wjgnet.com/1007-9327/full/v10/i18/2719.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i18.2719
Two major pathways of genomic instabilities have been recently recognized, namely the chromosomal instability pathway (CIN) and the microsatellite instability (MSI) pathway (MIN), the former is characterized by the loss of heterozygosity (LOH) whereas the latter by MSI[1]. Some genes contain repetitive regions in their coding sequences that are often targets of MSI. Gastrointestinal tumors with DNA mismatch repair (MMR) defects often present MSI and harbor frameshift mutations in coding mononucleotide repeats of cancer-related genes. MSI(+)tumors arise from the defects in the MMR system through the mechanism presumably involving frameshift mutations of the microsatellite repeats within the coding regions of the affected target genes, whose function loss is believed to contribute to tumorigenesis[2-5].
The retinoblastoma protein-interacting zinc finger gene RIZ is a candidate tumor suppressor gene locus on 1p36, a region that commonly harbors alterations in many types of human cancers[6]. The interaction of RIZ with the retinoblastoma protein (Rb) suggests its involvement in the control of proliferation by an alternative mechanism. RIZ gene encodes two protein products, RIZ1 and RIZ2, which differ for a motif present in the N-terminal domain, defined as the PR domain that was previously identified as a common motif in several transcription factors. RIZ1 contains a PR domain indicative of tumor suppressor function, whereas RIZ2 does not contain this motif[7,8]. RIZ1 has the capacity to induce cell cycle arrest at G2/M phase and cell apoptosis, and suppresses tumorigenicity in nude mice[9-11]. A role for RIZ has been recently proposed in cell cycle arrest and cell apoptosis through a transcriptional repression mechanism[7,12].
In MSI(+) tumors, RIZ was found to be affected by frequent frameshift mutations of one or two coding poly(A) tract, an (A)8 tract at the coding nucleotide sequence 4273-4280 and an (A)9 tract at 4462-4471 in exon 8. These mutations generate truncated RIZ1 proteins lacking the COOH-terminal PR-binding motif and are expected to have serious deleterious effects on the PR domain-specific function of RIZ1. RIZ plays an important role in hereditary tumors of the MIN pathway as suggested by the frequent frameshift mutations in HNPCC tumors[11]. The role of RIZ in gastric MSI(+) tumors remains to be investigated. In this study, we used denaturing high-performance liquid chromatography (DHPLC), a highly productive method, to rapidly detect frameshift mutations of (A)8 and (A)9 tracts and LOH of pro704 in gastric cancer specimens and to explore the role of RIZ gene in gastric carcinogenesis.
Gastric cancer samples and matched adjacent normal gastric tissues were obtained from 41 male and 29 female patients during surgical resection of the tumors with informed consent from the patients at Beijing Institute for Cancer Research, Beijing Cancer Hospital. The fresh samples were collected at the time of surgery and frozen at -80 °C. The sections from each specimen were examined by a pathologist. There were 39 intestinal-type tumors and 31 diffuse-type tumors. High-molecular weight genomic DNA was extracted by standard proteinase K digestion and phenol/chloroform extraction[13].
Primers used for (A)8 tract and (A)9 tract in the RIZ gene amplification were as follows: RIZ A8-F 5’-GAGCTCAGCAAAATGTCGTC-3’, RIZ A8-R 5’-CAAGTCGGCCTTCTGCTTTG-3’; RIZ A9-F 5’-TCTCACATCTGCCCTTACTG-3’, RIZ A9-R 5’-GTGATGAGTGTCCACCTTTC-3’. The RIZ Pro704 deletion polymorphism was assayed by PCR followed by DHPLC. The PCR primers were: RP145 5’-CCCAAGATAAACTAACTCCT-3’, RP105 5’-ACTCCATGCTGGTGAGTC-3’.
The samples used for mutation screening and sequencing were amplified in 25 μL reaction solution containing 50 ng genomic DNA, 0.4 μmol/L sense and antisense primers for each tract, 200 μmol/L dNTPs (Perkin-Elmer, Foster City, CA, USA), 0.2 μL Taq polymerase (Ampli Taq Gold: Perkin-Elmer), and 2.0 mmol/L MgCl2. After an initial activation of the enzyme by denaturation at 95 °C for 9 min, PCR amplification was performed for 35 cycles in the following sequence: at 94 °C for 30 s, at optimized annealing temperature for 45 s, and at 72 °C for 45 s, with a final extension at 72 °C for 10 min. The annealing temperature for various primer sets was: 58 °C for RIZ A8 tract, 60 °C for RIZ A9 tract, and 55 °C for pro704.
For examining the heteroduplex content, 50-100 ng of the PCR products were subjected to DHPLC (WAVETM system, Transgenomic, USA) under partial denaturation condition. The mobile phase consisted of a mixture of 0.1 mol/L triethylamine acetate (TEAA, pH7.0) with or without 25% acetonitrile. The flow rate used in this study was 0.9 mL/min. The column temperatures for the PCR products were 57 °C for RIZ (A)8 tract and 56 °C for RIZ (A)9 tract. The PCR products were heated to 95 °C for 3 min followed by cooling to 25 °C over 45 min. Homozygous mutant DNA must be combined with the wild type at the ratio of approximately 1:1 prior to hybridization.
The pro704 PCR products were directly used without a denaturation and reannealing process under non-denaturing conditions on the WAVETM system. The gradient of buffer B from 1 to 7 min was 49%-55%. The column temperature was 50 °C and flow rate was 0.75 mL/min.
The PCR product was treated with exonuclease and shrimp alkaline phosphatase based on the protocol provided by the United States Biochemical and sequenced by the Mayo Clinic DNA sequencing facility. Sequencing reactions were performed in the GeneAmp PCR System 9600 with fluorescent terminations, and the products were analyzed on an ABI 377 sequencer (Perkin-Elmer, Foster City, CA, USA). All sequence alterations were confirmed by bidirectional sequencing of the PCR products generated by at least two independent reactions.
MSI analysis by DHPLC was performed as described previously[14]. Briefly, the PCR products were examined by DHPLC under fully denaturing conditions. The flow rate used in this study was 0.9 mL/min, with the column temperature of 80 °C. The gradient of buffer B from 0.1 to 7.1 min was 30%-51%.
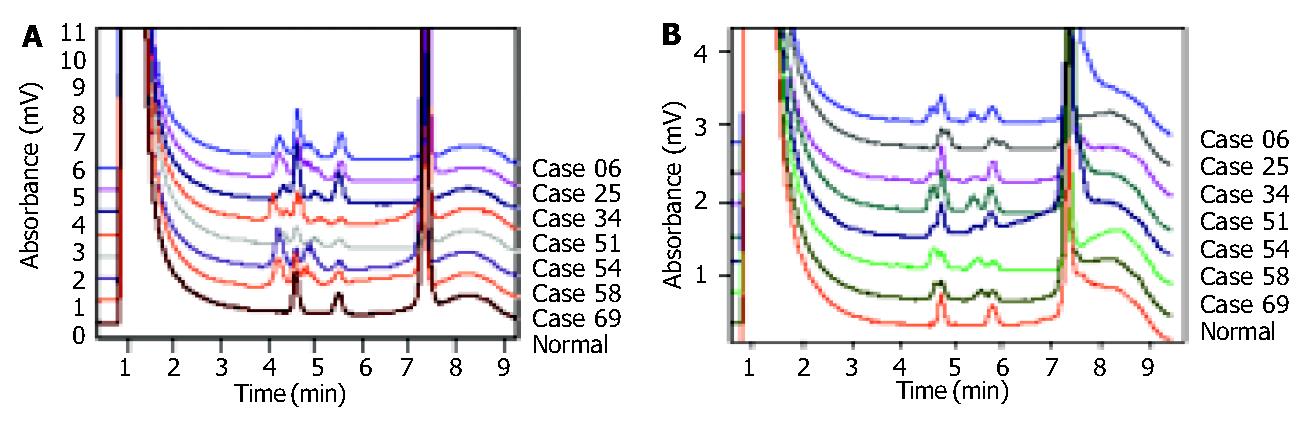
A total of 70 HGC samples and their matched normal tissues were analyzed for MSI status by two mononucleotide markers, BAT26 and BAT25, by means of DHPLC. In 70 HGC samples, 8 (11.4%) were found to contain sequence variation at BAT26 and BAT25 and 7 of them were shown to be unstable at both BAT26 and BAT25 (Figure 1), classified as MSI-H. One was unstable at BAT26 but stable at BAT25.
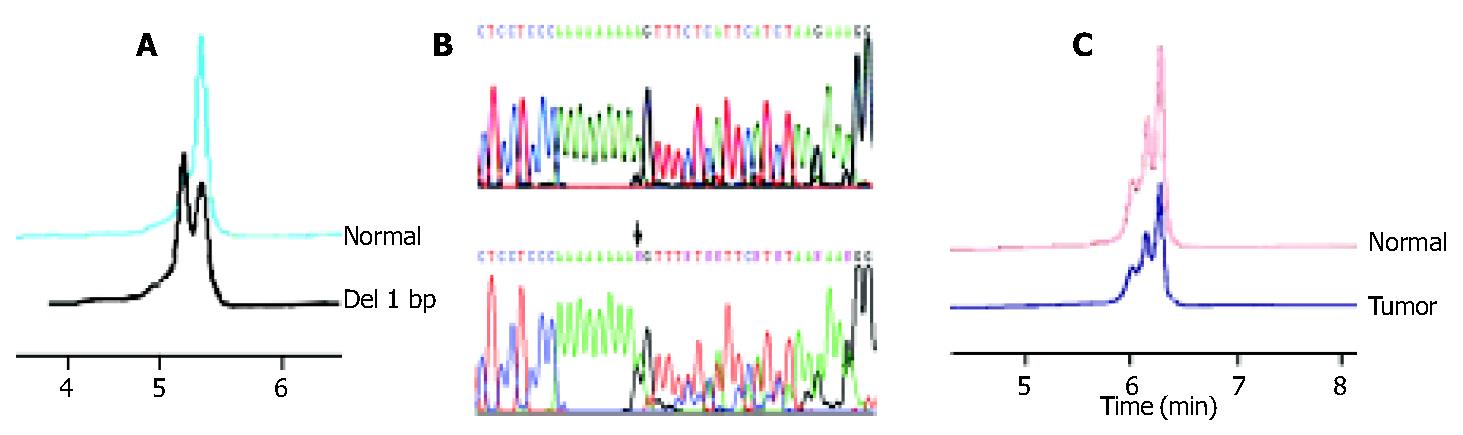
Frameshift mutations of RIZ (A)9 and (A)8 tract were identified. HCT-116 cell line had been studied previously and frameshift mutations were identified in the polyadenosine tracts[11], which served as the positive control in this study. In 7 samples with MSI-H, 4 (57.1%) were found to have mutations in the (A)9 tract of RIZ by DHPLC and DNA sequencing, including 3 intestinal-type tumors and 1 diffuse-type tumor. All of them had a 1-bp deletion (Figure 2A, Figure 2B). No mutations were detected in the (A)8 tract. No mutations in the (A)9 or (A)8 tract were found in the 63 MSI-L or MSI stable samples.
To determine whether RIZ was also affected by chromosomal deletion in MSI(+)tumors, pro704 LOH studies were performed on 4 samples with frameshift mutations for which the matched normal DNAs were available. We observed that both the PCR products of the tumor DNA and matched normal DNA resulted in the same peak chromatogram on DHPLC under the condition used for DNA sizing at 50 °C. LOH was not found at the RIZ locus in these tumors (Figure 2C).
Approximately 10%-15% of gastrointestinal tumors are caused by defective MMR[15], characterized by the presence of tumor MSI (MSI-H) and the absence of protein expression for any of the various genes involved in DNA MMR including hMLH1, hMSH2, hMSH6 or hPMS2[16]. Gastric cancer with an MSI-H phenotype often harbors somatic frameshift mutations in the coding mononucleotide repeats of cancer-related genes. Frameshift mutations in TGFßRII, BAX, IGFIIR and E2F4 genes are often observed in cancers exhibiting a high frequency of MSI[17]. Recently, the new candidate tumor suppressor gene, RIZ, which may be targeted for deletion, was identified. RIZ is a protein with two alternative forms of RIZ1 and RIZ2, which differ for a PR domain present in the N-terminal domain. The PR domain is necessary for the negative regulatory function of RIZ. Frameshift mutations in either (A)8 or (A)9 tract are thought to lead to C-terminal domain loss of the RIZ protein involved in PR binding. In this study, we detected frameshift mutations in RIZ (A)9 tract, whereas no mutation was found in (A)8 tract. All of the frameshift mutations here found in RIZ are assumed to lead to the production of the COOH-terminal domain-truncated protein, which is likely to seriously affect RIZ1 functions.
Frameshift in short mononucleotide tracts is common in gastrointestinal tumors of the microsatellite mutator phenotype (MMP). MSI is considered a hallmark of the mutator phenotype, and determination of MSI is critical for understanding tumor biology. Separation of HGCs based on their mutator phenotypes is an effective first step to allow the distinction of these two different pathways of carcinogenesis. In the present study, we analyzed MSI status by two mononucleotide markers, and detected RIZ mutations in 4 (57.1%) of the 7 MSI-H tumors but in none of the 63 MSI-L or MSI stable gastric cancers, indicating that these mutations are specific for MSI-H tumors that exhibit a tendency to accumulate frameshift mutations in reiterated sequence of the coding regions of cancer-related genes known to facilitate cancer development and progression. These mutations may contribute to cancer progression either by inactivating their tumor suppressor functions or acting as secondary mutator mutations in MMP(+) gastric tumors[18]. Our study has shown that MSI-H gastric cancers accumulate frameshift mutations in the RIZ gene. On the basis of our findings, we suppose that RIZ is a candidate target gene in MSI tumorigenesis.
Two kinds of genetic instability, MIN and CIN, have been documented in colorectal cancers. To determine whether RIZ is also affected by chromosomal deletion in MSI-H cancers, we detected the pro704 LOH in the 4 cases with frameshift mutations but failed to find LOH at the RIZ locus in these tumors, suggesting that RIZ frameshift mutations are common in MSI(+)gastric cancers, whereas LOH is not. More extensive studies on gastric cancers are needed to clarify whether RIZ is affected by two different ways. In MSI(+)tumors (MIN pathway), frameshift mutations in the COOH-terminal interfere with the interactions between the C terminus of the protein and its PR domain. In MSI(-) tumors (CIN pathway), mutations or deletions of the PR domain of RIZ1 may have similar effects.
In this study, we detected MSI status and LOH by DHPLC analysis. DHPLC has been described as a novel technology for the detection of gene mutations in inherited diseases or cancers and for the identification of single nucleotide polymorphisms (SNPs)[19]. The present study demonstrates the efficacy of DHPLC in analysis of the MSI status and LOH, which allows automated examination of MSI and LOH with considerable precision at relatively low cost, without any special labeling procedure.
Edited by Chen WW Proofread by Zhu LH and Xu FM
| 1. | Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643-649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2881] [Cited by in RCA: 2818] [Article Influence: 104.4] [Reference Citation Analysis (0)] |
| 2. | Parsons R, Myeroff LL, Liu B, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Microsatellite instability and mutations of the transforming growth factor beta type II receptor gene in colorectal cancer. Cancer Res. 1995;55:5548-5550. [PubMed] |
| 3. | Shin KH, Park JG. Microsatellite instability is associated with genetic alteration but not with low levels of expression of the human mismatch repair proteins hMSH2 and hMLH1. Eur J Cancer. 2000;36:925-931. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 4. | Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336-1338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1648] [Cited by in RCA: 1590] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 5. | Rampino N, Yamamoto H, Ionov Y, Li Y, Sawai H, Reed JC, Perucho M. Somatic frameshift mutations in the BAX gene in colon cancers of the microsatellite mutator phenotype. Science. 1997;275:967-969. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 968] [Cited by in RCA: 934] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 6. | Huang S. The retinoblastoma protein-interacting zinc finger gene RIZ in 1p36-linked cancers. Front Biosci. 1999;4:D528-D532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 29] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 7. | Huang S, Shao G, Liu L. The PR domain of the Rb-binding zinc finger protein RIZ1 is a protein binding interface and is related to the SET domain functioning in chromatin-mediated gene expression. J Biol Chem. 1998;273:15933-15939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 125] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 8. | Abbondanza C, Medici N, Nigro V, Rossi V, Gallo L, Piluso G, Belsito A, Roscigno A, Bontempo P, Puca AA. The retinoblastoma-interacting zinc-finger protein RIZ is a downstream effector of estrogen action. Proc Natl Acad Sci USA. 2000;97:3130-3135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 9. | He L, Yu JX, Liu L, Buyse IM, Wang MS, Yang QC, Nakagawara A, Brodeur GM, Shi YE, Huang S. RIZ1, but not the alternative RIZ2 product of the same gene, is underexpressed in breast cancer, and forced RIZ1 expression causes G2-M cell cycle arrest and/or apoptosis. Cancer Res. 1998;58:4238-4244. [PubMed] |
| 10. | Jiang Gl, Liu L, Buyse IM, Simon D, Huang S. Decreased RIZ1 expression but not RIZ2 in hepatoma and suppression of hepatoma tumorigenicity by RIZ1. Int J Cancer. 1999;83:541-546. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 11. | Chadwick RB, Jiang GL, Bennington GA, Yuan B, Johnson CK, Stevens MW, Niemann TH, Peltomaki P, Huang S, de la Chapelle A. Candidate tumor suppressor RIZ is frequently involved in colorectal carcinogenesis. Proc Natl Acad Sci USA. 2000;97:2662-2667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 121] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 12. | Xie M, Shao G, Buyse IM, Huang S. Transcriptional repression mediated by the PR domain zinc finger gene RIZ. J Biol Chem. 1997;272:26360-26366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 39] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 13. | Albrecht S, von Deimling A, Pietsch T, Giangaspero F, Brandner S, Kleihues P, Wiestler OD. Microsatellite analysis of loss of heterozygosity on chromosomes 9q, 11p and 17p in medulloblastomas. Neuropathol Appl Neurobiol. 1994;20:74-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 47] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 14. | Pan KF, Liu W, Lu YY, Zhang L, Li ZP, Lu WL, Thibodeau SN, You WC. High throughput detection of microsatellite instability by denaturing high-performance liquid chromatography. Hum Mutat. 2003;22:388-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 15. | Peltomäki P. Deficient DNA mismatch repair: a common etiologic factor for colon cancer. Hum Mol Genet. 2001;10:735-740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 336] [Cited by in RCA: 318] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 16. | Aaltonen LA, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomäki P, Chadwick RB, Kääriäinen H, Eskelinen M, Järvinen H. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med. 1998;338:1481-1487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 767] [Cited by in RCA: 764] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 17. | Wu MS, Lee CW, Shun CT, Wang HP, Lee WJ, Chang MC, Sheu JC, Lin JT. Distinct clinicopathologic and genetic profiles in sporadic gastric cancer with different mutator phenotypes. Genes Chromosomes Cancer. 2000;27:403-411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 18. | Piao Z, Fang W, Malkhosyan S, Kim H, Horii A, Perucho M, Huang S. Frequent frameshift mutations of RIZ in sporadic gastrointestinal and endometrial carcinomas with microsatellite instability. Cancer Res. 2000;60:4701-4704. [PubMed] |
| 19. | Liu W, Smith DI, Rechtzigel KJ, Thibodeau SN, James CD. Denaturing high performance liquid chromatography (DHPLC) used in the detection of germline and somatic mutations. Nucleic Acids Res. 1998;26:1396-1400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 172] [Article Influence: 6.4] [Reference Citation Analysis (0)] |