Published online Jul 1, 2004. doi: 10.3748/wjg.v10.i13.1975
Revised: October 5, 2003
Accepted: October 12, 2003
Published online: July 1, 2004
AIM: To analyze loss of heterozygosity (LOH) and homozygous deletion on p53 gene (exon2-3, 4 and 11), chromosome 10q22-10q23 and 22q11.2-22q12.1 in human hepatocellular carcinoma (HCC).
METHODS: PCR and PCR-based microsatellite polymorphism analysis techniques were used.
RESULTS: LOH was observed at D10S579 (10q22-10q23) in 4 of 20 tumors (20%), at D22S421 (22q11.2-22q12.1) in 3 of 20 (15%), at TP53.A (p53 gene exon 2-3) in 4 of 20 (20%), at TP53.B (p53 gene exon 4) in 6 of 20 (30%), and at TP53.G (p53 gene exon 11) in 0 of 20 (0%). Homozygous deletion was detected at 10q22-10q23 (8/20; 40%), 22q11.2-22q12.1 (8/20; 40%), p53 gene exon 2-3 (0/20;0%), p53 gene exon 4 (6/20; 30%), and p53 gene exon 11 (2/20; 10%).
CONCLUSION: There might be unidentified tumor suppressor genes on chromosome 10q22-10q23 and 22q11.2-22q12.1 that contribute to the pathogenesis and development of HCC.
-
Citation: Zhu GN, Zuo L, Zhou Q, Zhang SM, Zhu HQ, Gui SY, Wang Y. Loss of heterozygosity on chromosome 10q22-10q23 and 22q11.2-22q12.1 and
p53 gene in primary hepatocellular carcinoma. World J Gastroenterol 2004; 10(13): 1975-1978 - URL: https://www.wjgnet.com/1007-9327/full/v10/i13/1975.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i13.1975
Hepatocellular carcinoma (HCC) is a primary liver malignancy with high mortality. It is among the most common malignancies worldwide, especially in Asia, Africa and Southern Europe[1]. It has been generally accepted that HCC is highly associated with chronic hepatitis B virus (HBV) or hepatitis C virus (HCV) infection or alcohol intake which induces cirrhosis[2]. High intake of aflatoxin B found in many kinds of food is also reported to be a risk factor for HCC[3,4]. Like other solid tumors, It has been proposed that hepatocarcinogenesis and metastasis of HCC is a multi-step process requiring the accumulation of genetic alterations, but the precise molecular pathogenesis is far from clear.
Loss of heterozygosity (LOH) analysis has become an effective way to identify informative loci and candidate tumor suppressor genes (TSGs). Molecular chromosomal studies of tumors by using polymerase chain reaction (PCR) -based polymorphic markers can detect small loci of anomalies that may harbor TSGs. Search for novel TSGs is based largely on the identification of common regions of deletion on chromosomes. LOH has been found in many types of tumors, including HCC. LOH in HCC has been detected on chromosomal arms 1p, 2q, 4p, 4q, 5q, 6q, 8p, 8q, 9p, 9q, 11p, 13q, 16p, 16q and 17p[5-11]. However, deletion of 10q22-10q23, and 22q11.2-22q12.1 and p53 gene exon 2-3 and 11 in HCC has not been investigated.
In the present study, we detected LOH and homozygous deletion on chromosome 10q, and chromosome 22q near the NF2 gene locus, and p53 gene locus in 20 cases of HCC.
Surgical specimens of HCC were collected from the First Affiliated Hospital of Anhui Medical University and the Affiliated Hospital of Bengbu Medical College. The patients were born and grew in different places of Anhui Province, China. Both tumor and corresponding non-tumor liver tissues were immediately put into liquid nitrogen after separation and then stored at -80 °C until DNA extraction. Diagnosis of HCC was confirmed by pathological examination.
Genomic DNA was extracted from tissues with the standard proteinase K-phenol/choloroform method. To each of the samples, 500 μL of DNA extraction buffer containing 200 mmol/L NaCl, 10 g/L sodium dodecyl sulfate, 2 mmol/L EDTA, 0.1 mol/L Tris-HCl was added during the process of homogenization. After 0.2 mg/mL proteinase K was added, the sample was shaken for 12 h at 37 °C. After phenol-chloroform extraction, DNA was precipitated with cold ethanol overnight at -20 °C. After centrifugation, the pellet was dried and resuspended in 50 μL TE buffer (Tris-EDTA buffer). DNA was stored at -20 °C until polymerase chain reaction (PCR) amplification was performed.
PCR amplification primer pairs for p53 gene, 10q22-10q23 and 22q11.2-22q12.1 are as follows (Table 1).
| Markers name | Forward | Reverse | Annealing (T °C) | Size (bp) |
| TP53.A1/TP53.A2 (p53 gene exon 2-3) | TGGATCCTCTTGCAGCAGCC | AACCCTTGTCCTTACCAGAA | 54 | 270 |
| TP53.B1/TP53.B2 (p53 gene exon 4) | ATCTACAGTCCCCCTTGCCGGC | AACTGACCGTGCAAGTCA | 57 | 296 |
| TP53.G1/TP53.G2 (p53 gene exon 11) | TCTCCTACAGCCACCTGAAG | CTGACGCACACCTATTGCAA | 58 | 122 |
| D10S579 | CCGATCAATGAGGAGTGCC | ATACACCCAGCCAATGCTGC | 60 | 260 |
| D22S421 | CTGCTGCCCCTAACATATCAC | GGCCAGGAGTGTCTGAATTTTA | 65 | 163 |
| CDK41 | GGAGGTCGGTACCAGAGTG | CATGTAGACCAGGACAGG | 60 | 364 |
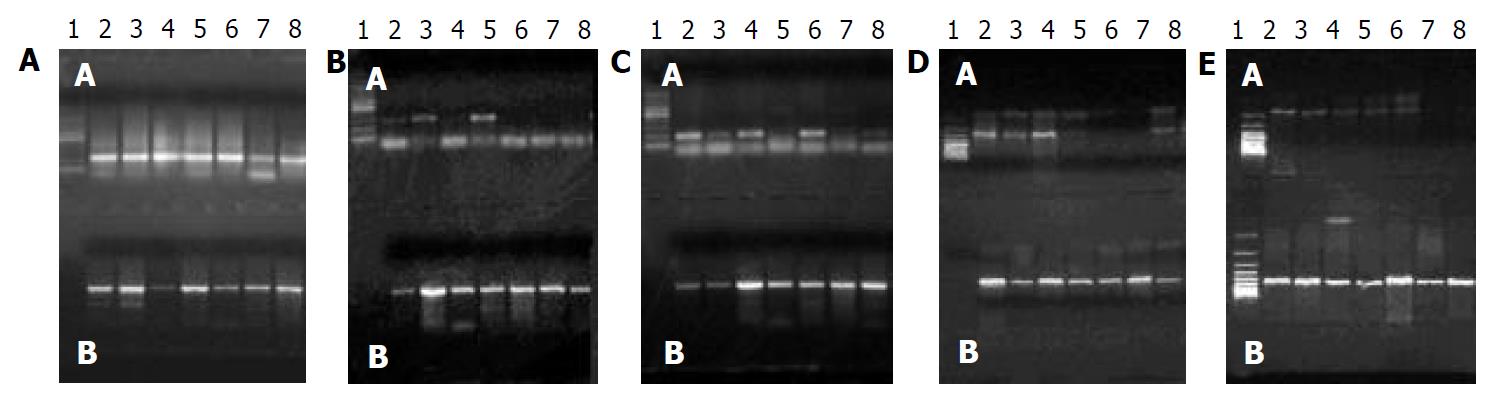
PCR product (12 μL) was mixed with 3 μL 950 g/L deionized formamide and 3 μL DNA loading buffer containing 2.5 g/L xylene cyanol FF, 2.5 g/L bromophenol blue, and 300 g/L glycerin. The mixture was denatured at 95 °C for 5 min, put onto ice for 10 min, loaded onto 80 g/L denaturing polyacrylamide gel containing 3.3 mol/L urea and then electrophoresed at 100 V for 2 h. The gel was silver-stained[13]. LOH was determined by visual evaluation, which compared the allele bands from tumors and the corresponding non-tumor tissues. The complete loss of one polymorphic allele from those seen in the paired control DNA was scored as allelic loss by three independent observers. PCR reactions were performed twice to confirm LOH.
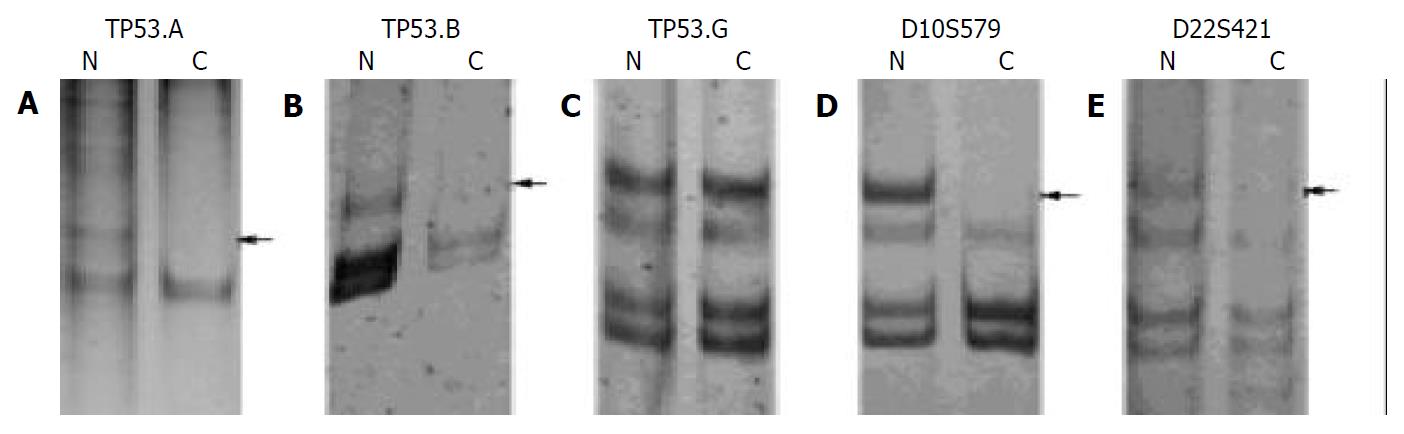
HCC tumor and corresponding non-tumor liver tissues of 20 patients were studied for LOH on 10q22-10q23 (D10S579), 22q11.2-22q12.1 (D22S421), and 17p13.1 by five microsatellite markers, and the rate of LOH was 20% (4/20), 15% (3/20), 50% (10/20), respectively (Table 2). Homozygous deletion was observed in 8 of 20 cases (40%) for the marker D10S579, 8 of 20 cases (40%) for D22S421, 6 of 20 cases (30%) for TP53.B, 2 of 20 cases (10%) for TP53.G, and in 0 of 20 cases (0%) for the marker TP53.A.
| Case No. | Age (yr) | Sex | HBsAg | TP53.A | TP53.B | TP53.G | D10S579 | D22S421 |
| 1 | 49 | M | + | * | * | O | * | O |
| 2 | 55 | M | + | O | △ | O | O | △ |
| 3 | 39 | M | + | O | △ | O | △ | △ |
| 4 | 55 | F | + | * | O | O | O | * |
| 5 | 72 | F | + | O | * | △ | △ | O |
| 6 | 40 | M | + | O | O | O | O | O |
| 7 | 34 | F | + | O | * | O | △ | △ |
| 8 | 56 | M | + | O | O | O | O | O |
| 9 | 27 | F | - | O | * | O | △ | O |
| 10 | 50 | M | + | O | O | O | O | △ |
| 11 | 48 | M | + | O | △ | O | △ | △ |
| 12 | 52 | M | + | O | O | O | △ | O |
| 13 | 63 | F | + | O | △ | △ | △ | △ |
| 14 | 60 | M | + | O | △ | O | * | △ |
| 15 | 65 | M | + | O | O | O | * | O |
| 16 | 34 | M | + | O | * | O | O | O |
| 17 | 64 | M | + | * | * | O | * | * |
| 18 | 52 | F | + | O | O | O | △ | O |
| 19 | 32 | F | + | O | △ | O | O | △ |
| 20 | 38 | M | + | * | O | O | O | * |
| LOH rate (%) | 20 | 30 | 0 | 20 | 15 | |||
| Homozygous deletion rate (%) | 0 | 30 | 10 | 40 | 40 |
Results of 20 g/L agarose gel electrophoresis are shown in Figure 1. LOH in tumor and corresponding non-tumor liver tissues are shown in Figure 2.
HCC is one of most malignant tumors. The mechanism of hepato-carcinogenesis is a multi-factor and multi-step process requiring the accumulation of genetic alterations, including chromosomal aberration, oncogene activation, inactivation of TSGs and abnormality of growth factors and growth factor receptors. Of these factors, inactivation of TSGs is a very important factor.
Allelic loss on chromosome 17p is among the most common genetic abnormalities in many human cancers. p53 gene is thought to be the gene associated with the genesis of these cancer types, including HCC[12]. p53 is activated in response to DNA damage, inducing either cell cycle arrest to permit DNA repair or apoptosis. Loss of p53 function occurs mainly through allelic deletions at chromosome 17p13, where p53 gene is located. In human HCC, LOH at chromosome 17p13 has been reported in 25%-60% of tumors, and the worldwide prevalence of p53 mutation is around 28% with, however, important geographic variations. In this study, LOH was observed at exon 2 and 3 (TP53.A) and exon 4 (TP53.B), of the gene in 20% and 30% of HCC cases, respectively, but not detected at exon 11 (TP53.G). In addition, all but one (19/20) patients were positive with HBsAg. These data also support the idea that LOH at p53 gene and HBV infection are highly associated with the pathogenesis and development of HCC.
LOH on D10S579 has been reported in renal cell carcinoma (RCC)[13]. We investigated 20 HCCs in the present study, and found four cases had LOH and eight cases had homozygous deletion on 10q22-10q23 (D10S579). Our finding suggests that on 10q22-10q23, there might be unidentified TSG(s) that plays an important role in the pathogenesis of hepatocellular carcinoma.
22q11.2-22q12.1 (D22S421) is near the locus of NF2 gene. NF2 (neurofibromatosis 2) gene, which is located on chromosome 22q12.2-22q12.2, is postulated to be a tumor suppressor gene. It encodes for a protein with 595 amino acids, designated as merlin or schwannomin which belongs to a family of cytoskeletal proteins. The majority of NF2 gene mutations are deletions, insertions, and point mutations, all of which lead to a nonfunctional, truncated protein[14].
LOH at the NF2 locus has been observed in many tumors, including schwannoma[15], meningioma[16], malignant mesothelioma[17], gastrointestinal stromal tumor[18], colorectal carcinoma[19]. However, Handel-Fernandez et al[20] found that there was no LOH at NF2 gene in pancreatic adenocarcinoma, but 37% of the cases had deletions which were clustered into two separate areas of chromosome 22 - one proximal and one distal to NF2 gene. In the present study, we detected LOH on 22q11.2-22q12.1 in three of 20 HCCs and homozygous deletion on 22q11.2-22q12.1 in eight of 20 HCCs. Our finding suggests that 22q11.2-22q12.1 likely contains an unidentified tumor suppressor gene that contributes to the pathogenesis and the development of HCC, that the region plays an important role of cis-acting element similar to NF2 gene, or that it acts the part of trans-acting factor similar to other TSGs, such as p53 gene.
In conclusion, we have obtained important new information on LOH and homozygous deletion in chromosome 10q, 22q and 17p, in a subset of HCC. Inactivation of p53 gene and unidentified tumor suppressor gene(s), present in regions of 10q22-10q23 and 22q11.2-22q12.1, may play an important role in the pathogenesis of HCC.
Edited by Xia HHX and Xu FM
| 1. | Simonetti RG, Cammà C, Fiorello F, Politi F, D'Amico G, Pagliaro L. Hepatocellular carcinoma. A worldwide problem and the major risk factors. Dig Dis Sci. 1991;36:962-972. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 256] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 2. | Harris CC. Hepatocellular carcinogenesis: recent advances and speculations. Cancer Cells. 1990;2:146-148. [PubMed] |
| 3. | Yakicier MC, Legoix P, Vaury C, Gressin L, Tubacher E, Capron F, Bayer J, Degott C, Balabaud C, Zucman-Rossi J. Identification of homozygous deletions at chromosome 16q23 in aflatoxin B1 exposed hepatocellular carcinoma. Oncogene. 2001;20:5232-5238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 36] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 4. | Martins C, Kedda MA, Kew MC. Characterization of six tumor suppressor genes and microsatellite instability in hepatocellular carcinoma in southern African blacks. World J Gastroenterol. 1999;5:470-476. [PubMed] |
| 5. | Fujimori M, Tokino T, Hino O, Kitagawa T, Imamura T, Okamoto E, Mitsunobu M, Ishikawa T, Nakagama H, Harada H. Allelotype study of primary hepatocellular carcinoma. Cancer Res. 1991;51:89-93. [PubMed] |
| 6. | Fujimoto Y, Hampton LL, Wirth PJ, Wang NJ, Xie JP, Thorgeirsson SS. Alterations of tumor suppressor genes and allelic losses in human hepatocellular carcinomas in China. Cancer Res. 1994;54:281-285. [PubMed] |
| 7. | Hsu HC, Peng SY, Lai PL, Sheu JC, Chen DS, Lin LI, Slagle BL, Butel JS. Allelotype and loss of heterozygosity of p53 in primary and recurrent hepatocellular carcinomas. A study of 150 patients. Cancer. 1994;73:42-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 8. | Boige V, Laurent-Puig P, Fouchet P, Fléjou JF, Monges G, Bedossa P, Bioulac-Sage P, Capron F, Schmitz A, Olschwang S. Concerted nonsyntenic allelic losses in hyperploid hepatocellular carcinoma as determined by a high-resolution allelotype. Cancer Res. 1997;57:1986-1990. [PubMed] |
| 9. | Nagai H, Pineau P, Tiollais P, Buendia MA, Dejean A. Comprehensive allelotyping of human hepatocellular carcinoma. Oncogene. 1997;14:2927-2933. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 204] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 10. | Shao J, Li Y, Li H, Wu Q, Hou J, Liew C. Deletion of chromosomes 9p and 17 associated with abnormal expression of p53, p16/MTS1 and p15/MTS2 gene protein in hepatocellular carcinomas. Chin Med J (Engl). 2000;113:817-822. [PubMed] |
| 11. | Herath NI, Kew MC, Walsh MD, Young J, Powell LW, Leggett BA, MacDonald GA. Reciprocal relationship between methylation status and loss of heterozygosity at the p14(ARF) locus in Australian and South African hepatocellular carcinomas. J Gastroenterol Hepatol. 2002;17:301-307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 12. | Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5482] [Cited by in RCA: 5459] [Article Influence: 195.0] [Reference Citation Analysis (0)] |
| 13. | Alimov A, Li C, Gizatullin R, Fredriksson V, Sundelin B, Klein G, Zabarovsky E, Bergerheim U. Somatic mutation and homozy-gous deletion of PTEN/MMAC1 gene of 10q23 in renal cell carcinoma. Anticancer Res. 1999;19(5B):3841-3846. |
| 14. | Lasota J, Fetsch JF, Wozniak A, Wasag B, Sciot R, Miettinen M. The neurofibromatosis type 2 gene is mutated in perineurial cell tumors: a molecular genetic study of eight cases. Am J Pathol. 2001;158:1223-1229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 46] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 15. | Mohyuddin A, Neary WJ, Wallace A, Wu CL, Purcell S, Reid H, Ramsden RT, Read A, Black G, Evans DG. Molecular genetic analysis of the NF2 gene in young patients with unilateral vestibular schwannomas. J Med Genet. 2002;39:315-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 47] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 16. | Leuraud P, Marie Y, Robin E, Huguet S, He J, Mokhtari K, Cornu P, Hoang-Xuan K, Sanson M. Frequent loss of 1p32 region but no mutation of the p18 tumor suppressor gene in meningiomas. J Neurooncol. 2000;50:207-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 17. | Pylkkänen L, Sainio M, Ollikainen T, Mattson K, Nordling S, Carpén O, Linnainmaa K, Husgafvel-Pursiainen K. Concurrent LOH at multiple loci in human malignant mesothelioma with preferential loss of NF2 gene region. Oncol Rep. 2002;9:955-959. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 18. | Fukasawa T, Chong JM, Sakurai S, Koshiishi N, Ikeno R, Tanaka A, Matsumoto Y, Hayashi Y, Koike M, Fukayama M. Allelic loss of 14q and 22q,NF2 mutation, and genetic instability occur inde-pendently of c-kit mutation in gastrointestinal stromal tumor. Jpn J Cancer Res. 2000;91:1241-1249. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 54] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 19. | Sugai T, Habano W, Nakamura S, Yoshida T, Uesugi N, Sasou S, Itoh C, Katoh R. Use of crypt isolation to determine loss of heterozygosity of multiple tumor suppressor genes in colorectal carcinoma. Pathol Res Pract. 2000;196:145-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 20. | Handel-Fernandez ME, Nassiri M, Arana M, Perez MM, Fresno M, Nadji M, Vincek V. Mapping of genetic deletions on the long arm of chromosome 22 in human pancreatic adenocarcinomas. Anticancer Res. 2000;20:4451-4456. [PubMed] |