Copyright
©The Author(s) 2021.
World J Gastroenterol. Feb 28, 2021; 27(8): 708-724
Published online Feb 28, 2021. doi: 10.3748/wjg.v27.i8.708
Published online Feb 28, 2021. doi: 10.3748/wjg.v27.i8.708
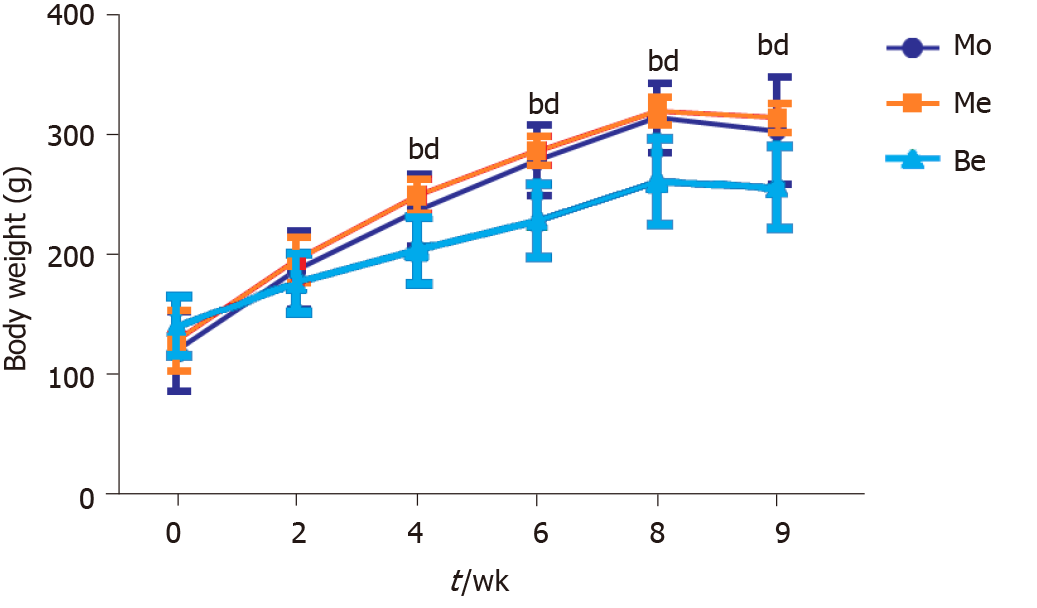
Figure 1 Comparison of body weight.
bP < 0.01 vs the Mo group; dP < 0.01 vs the Me group. Mo: Model group; Me: Metformin group; Be: Berberine group.
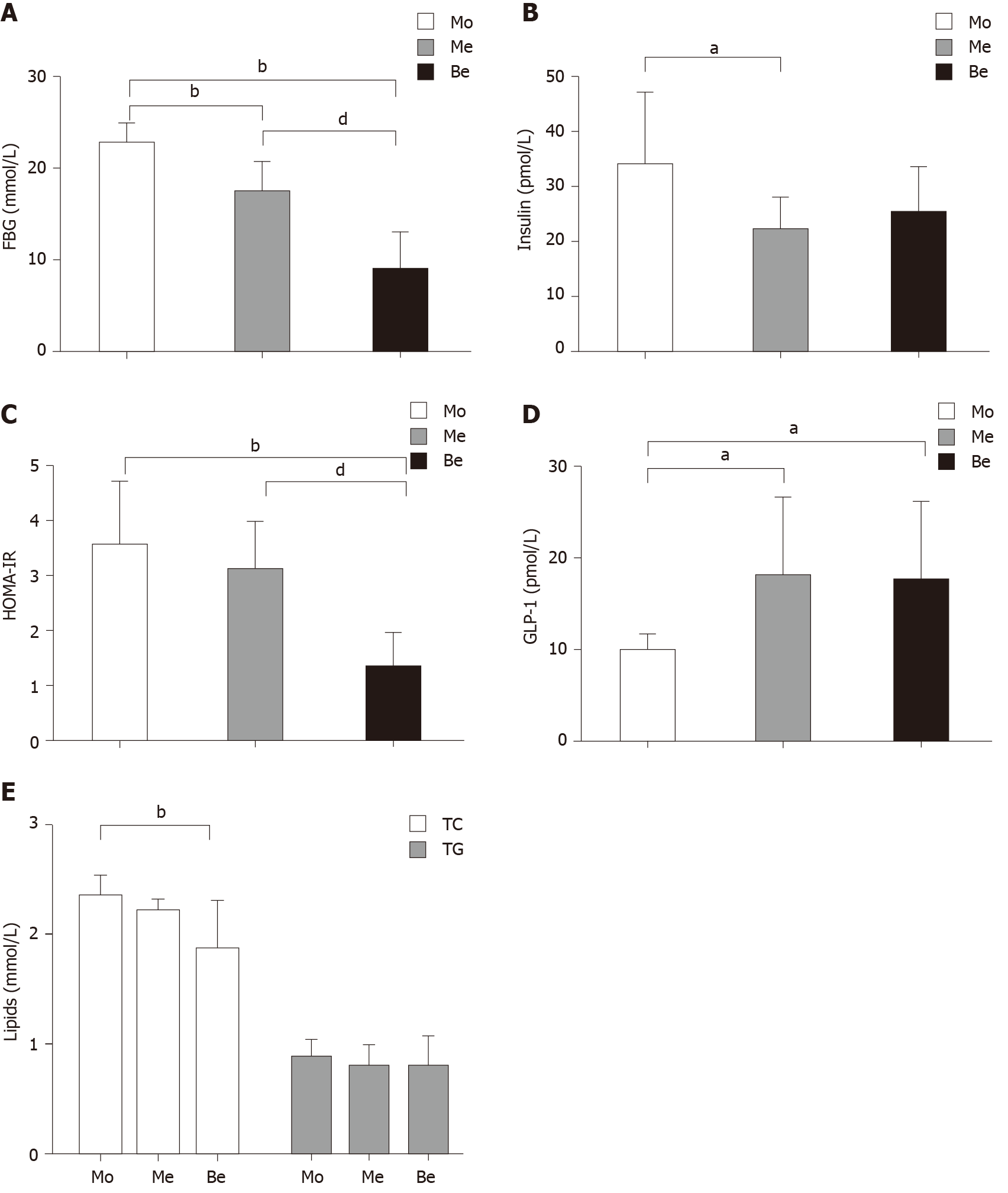
Figure 2 Body weight and metabolic parameter levels after treatment.
A: Comparison of the fasting FBG levels. bP < 0.01 vs the model (Mo) group; dP < 0.01 vs the metformin (Me) group; B: Comparison of the insulin levels. aP < 0.05 vs the Mo group; C: Comparison of the homeostatic model assessment-insulin resistance levels. bP < 0.01 vs the Mo group; dP < 0.05 vs the Me group; D: Comparison of the glucagon-like peptide-1 levels. aP < 0.05 vs the Mo group; E: Comparison of the lipid levels. bP < 0.01 vs the Mo group. FBG: Fasting blood glucose; HOMA-IR: Homeostatic model assessment-insulin resistance; GLP-1: Glucagon-like peptide-1; TC: Total cholesterol; TG: Triglycerides; Mo: Model group; Me: Metformin group; Be: Berberine group.
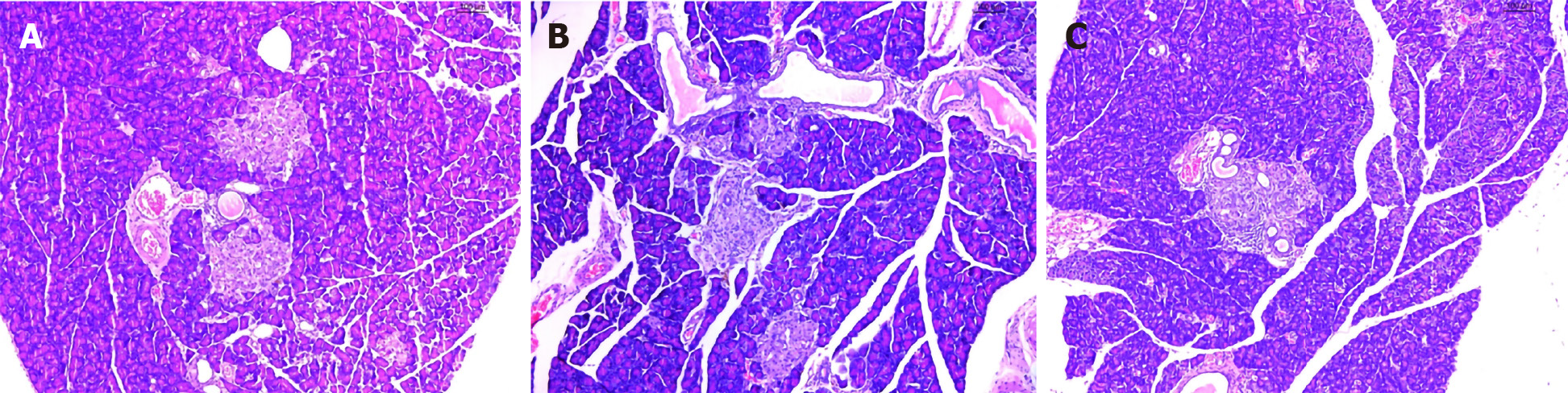
Figure 3 Histopathological images of the pancreas.
Hematoxylin and eosin staining (original magnification × 100). A: Model group; B: Metformin group; C: Berberine group.
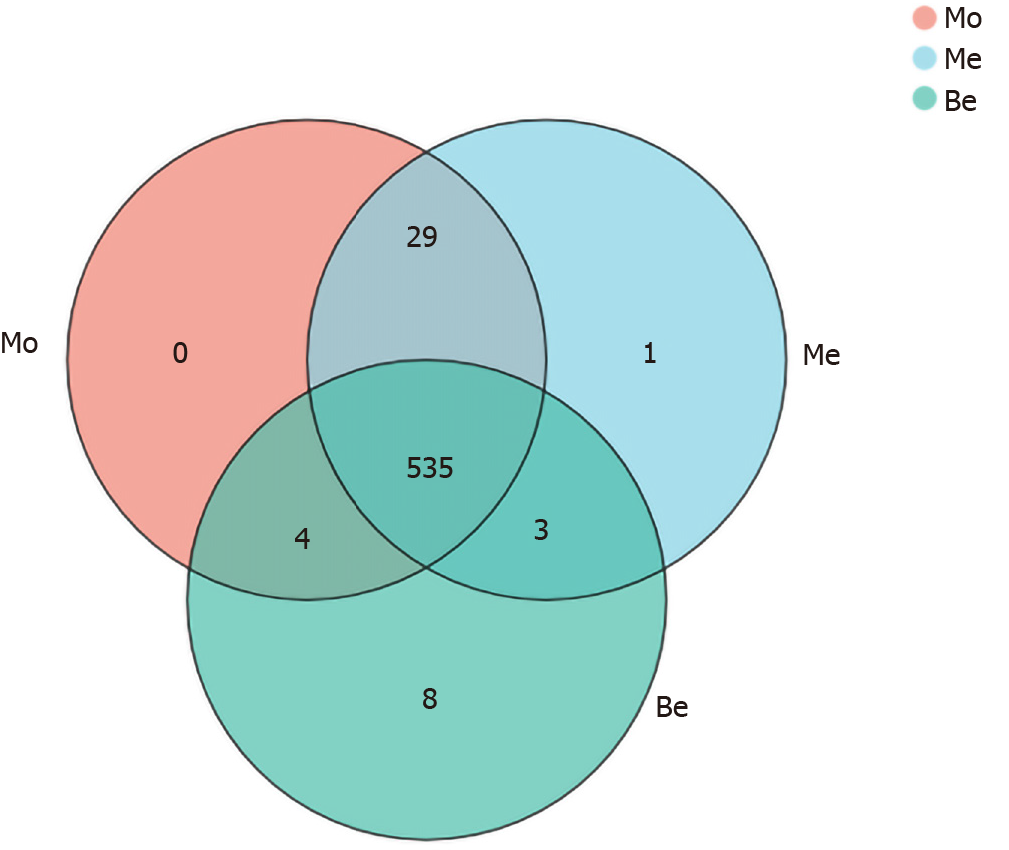
Figure 4 Comparison of the operational taxonomic units.
Mo: Model group; Me: Metformin group; Be: Berberine group.
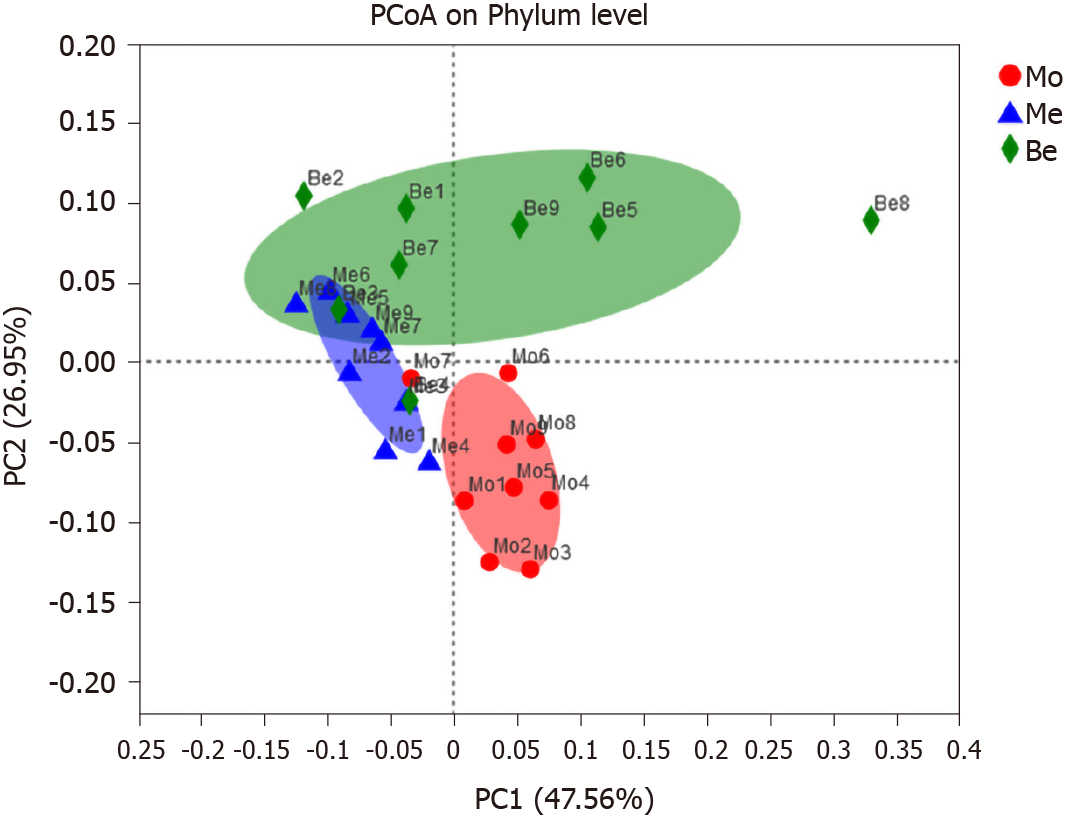
Figure 5 Comparison of gut microbiota structure using principal coordinates analysis.
PCoA: Principal coordinates analysis; PC1: The first principal component; PC2: The second principal component; Mo: Model group; Me: Metformin group; Be: Berberine group.
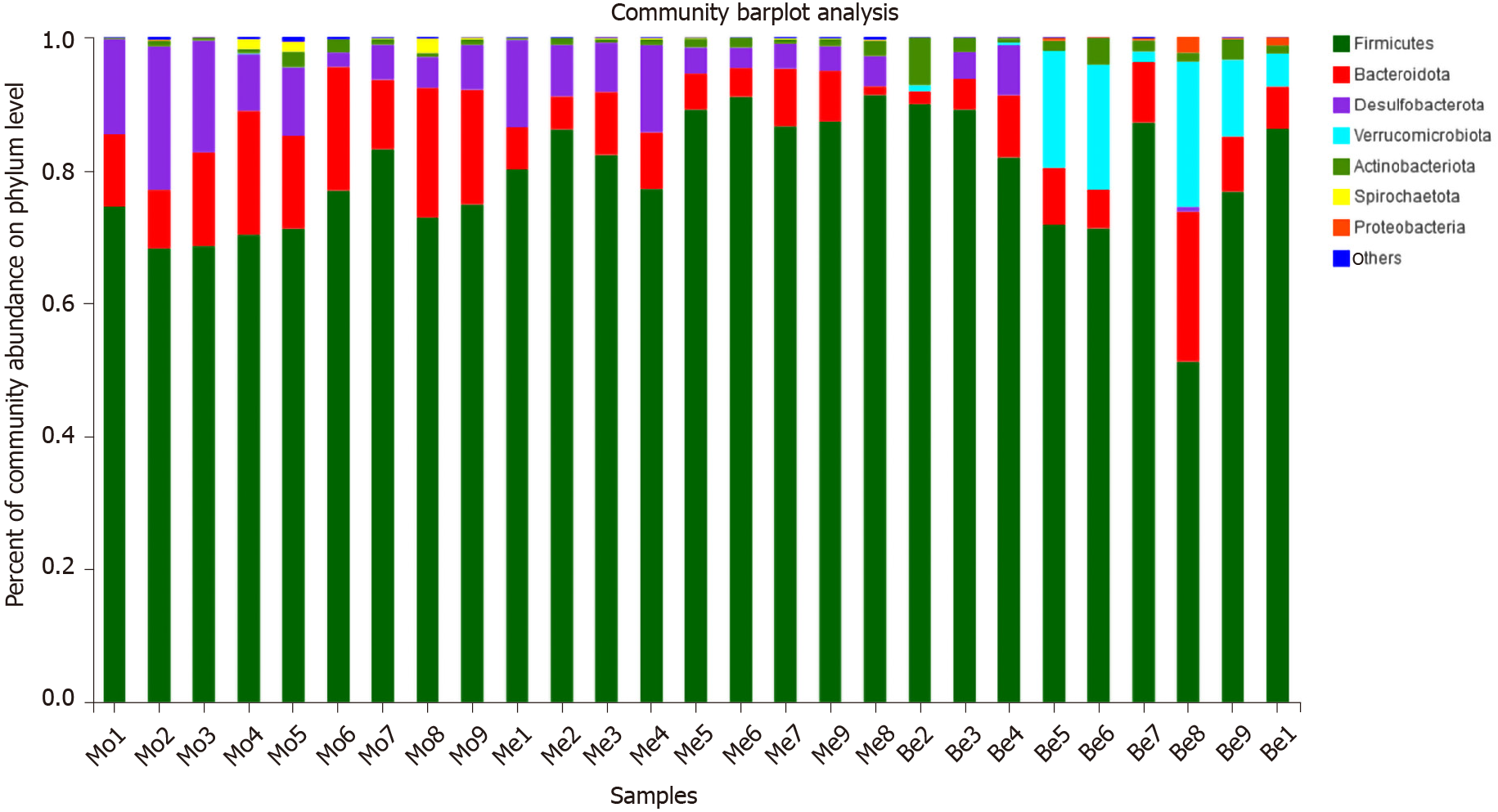
Figure 6
Comparison of the composition abundance in gut bacterial phylum.
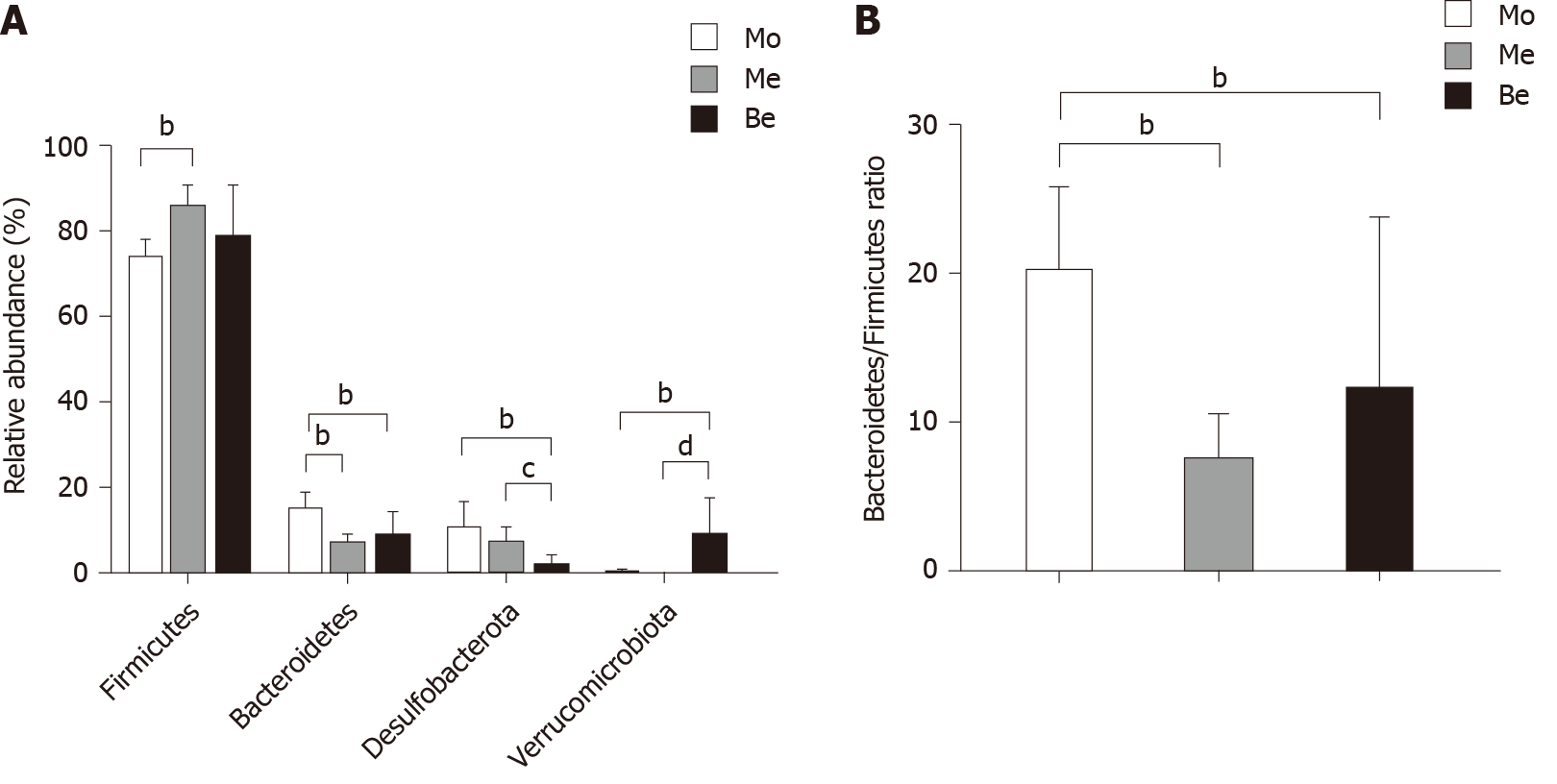
Figure 7 Relative abundance of gut bacterial phylum and the Bacteroidetes/Firmicutes ratio.
A: Comparison of the relative abundance of gut bacterial phylum; bP < 0.01 vs the model group; cP < 0.05, dP < 0.01 vs the metformin group; B: Comparison of the Bacteroidetes/Firmicutes ratio; bP < 0.01 vs the model group. Mo: Model group; Me: Metformin group; Be: Berberine group.
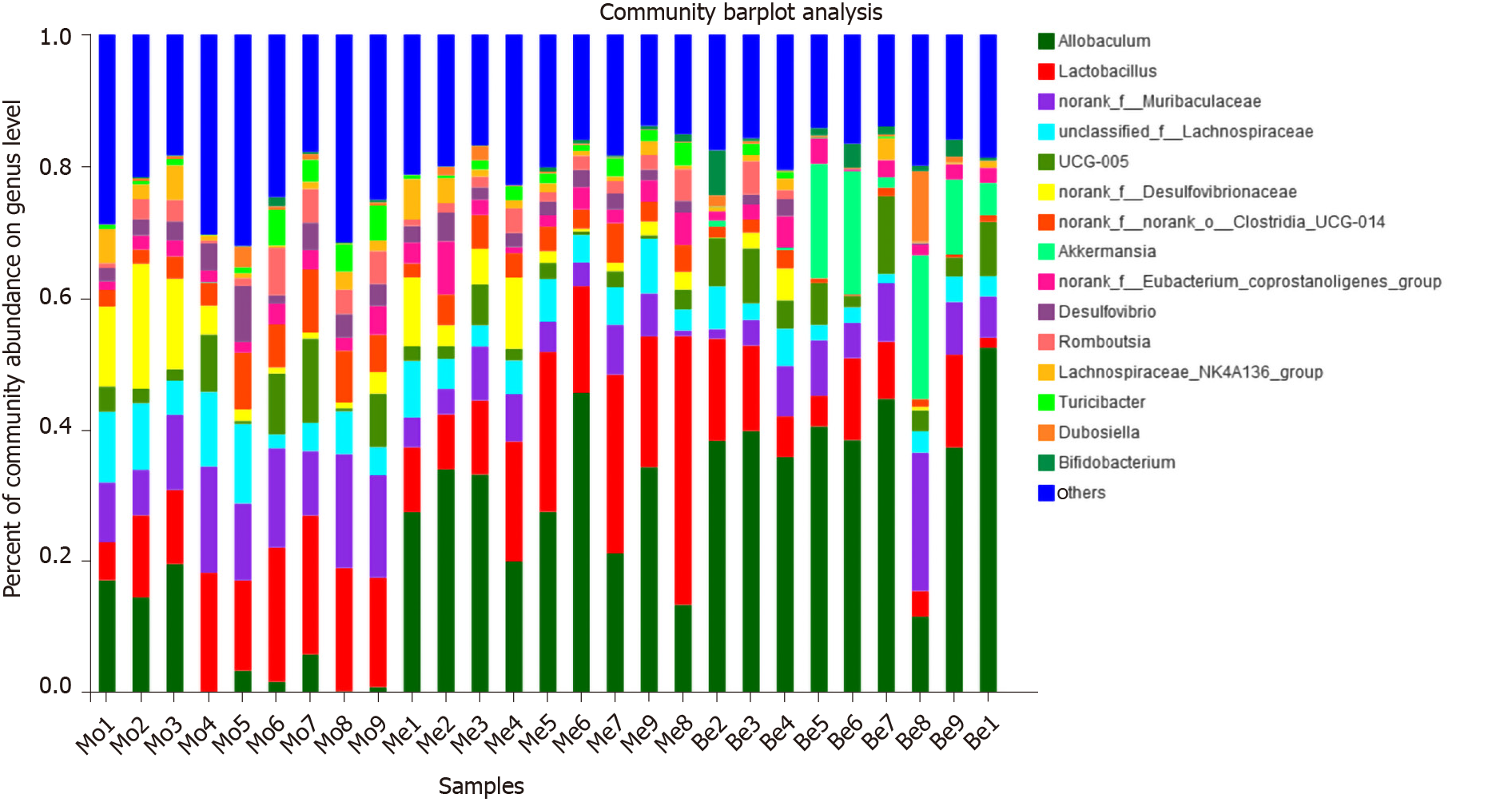
Figure 8
Comparison of the composition abundance in gut bacterial genera.
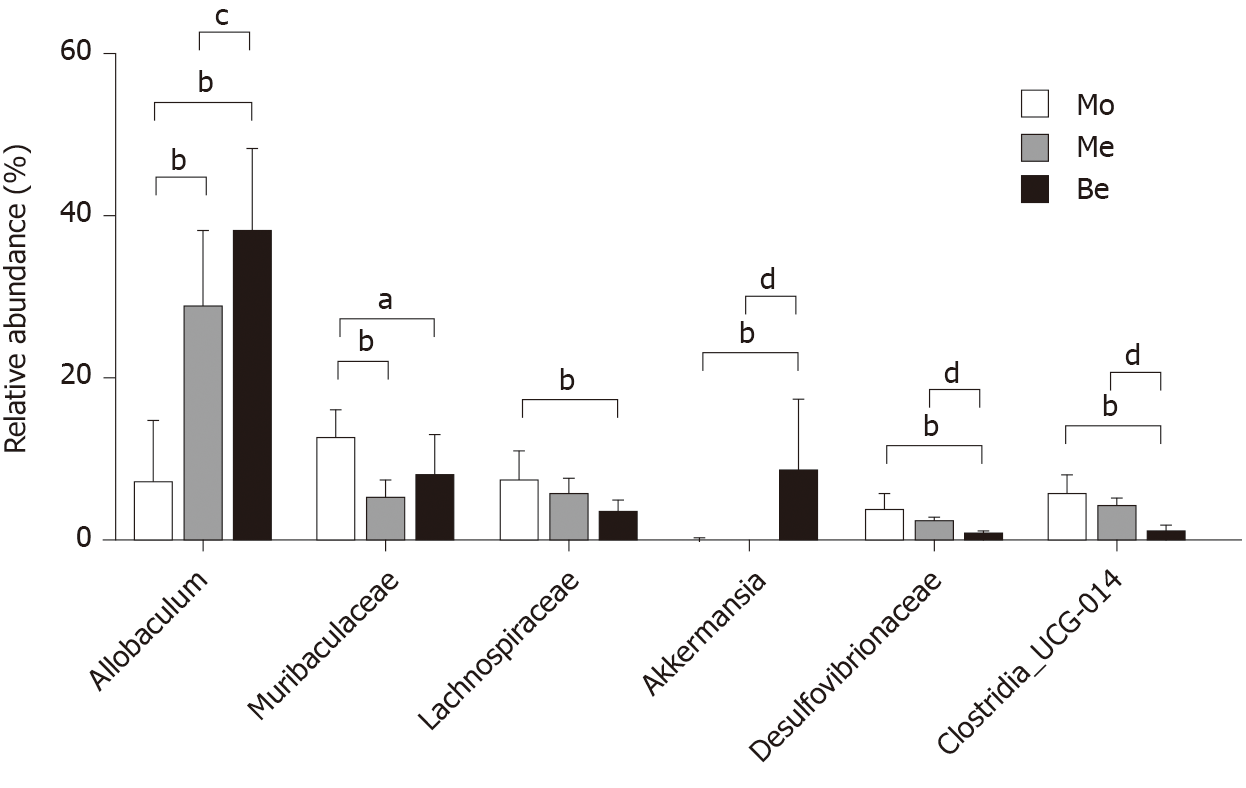
Figure 9 Comparison of the relative abundance in gut bacterial genera.
aP < 0.05, bP < 0.01 vs the model group; cP < 0.05, dP < 0.01 vs the metformin group. Mo: Model group; Me: Metformin group; Be: Berberine group.
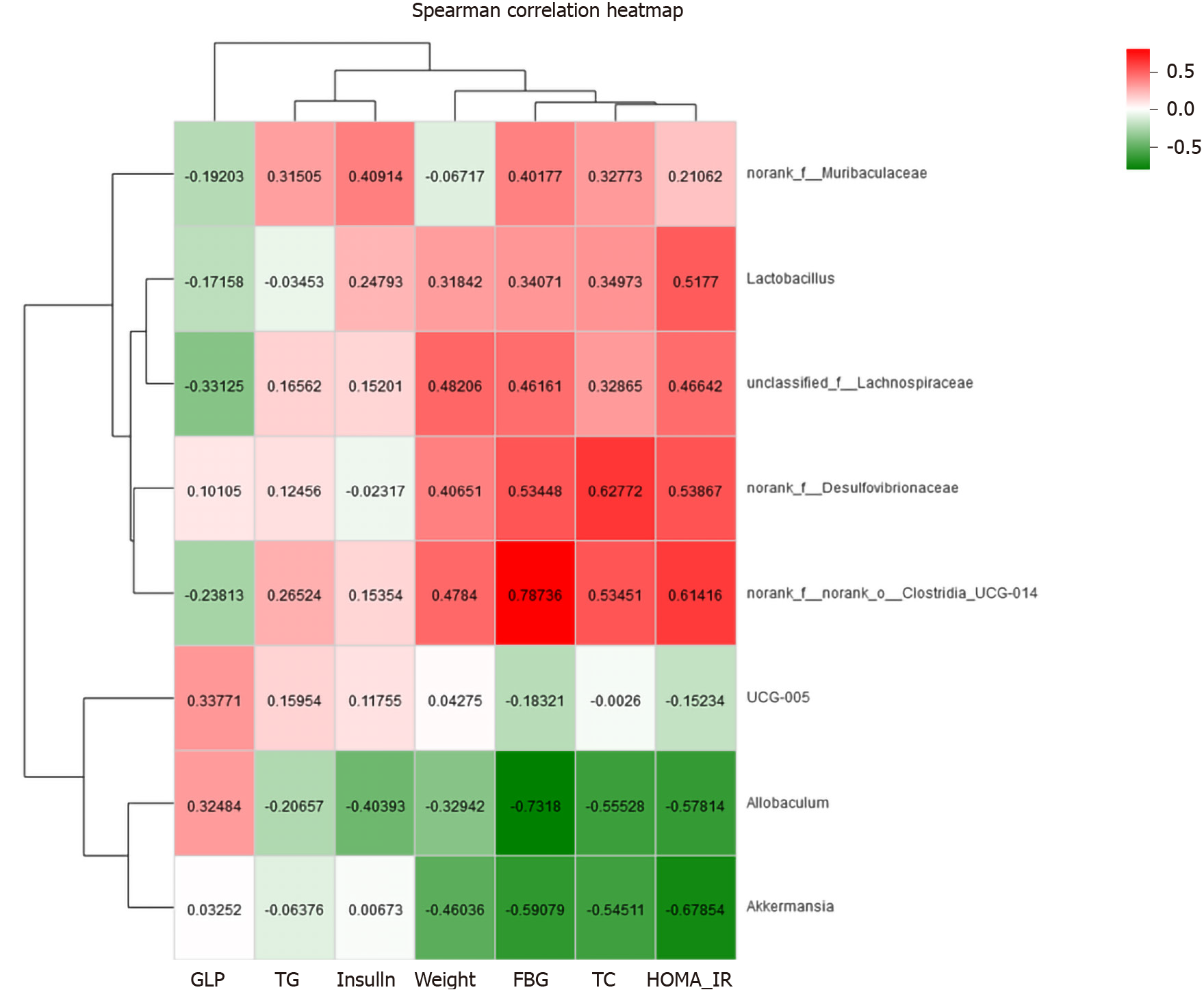
Figure 10 Correlation analysis between type 2 diabetes mellitus-related biochemical factors and gut microbiota at the genus level.
The colours ranged from green (negative correlation) to red (positive correlation). Statistical significance was based on a P value of 0.05 (Spearman’s correlation coefficient). r values are indicated. GLP: Glucagon-like peptide-1; TG: Triglycerides; FBG: Fasting blood glucose; TC: Total cholesterol; HOMA-IR: Homeostatic model assessment-insulin resistance.
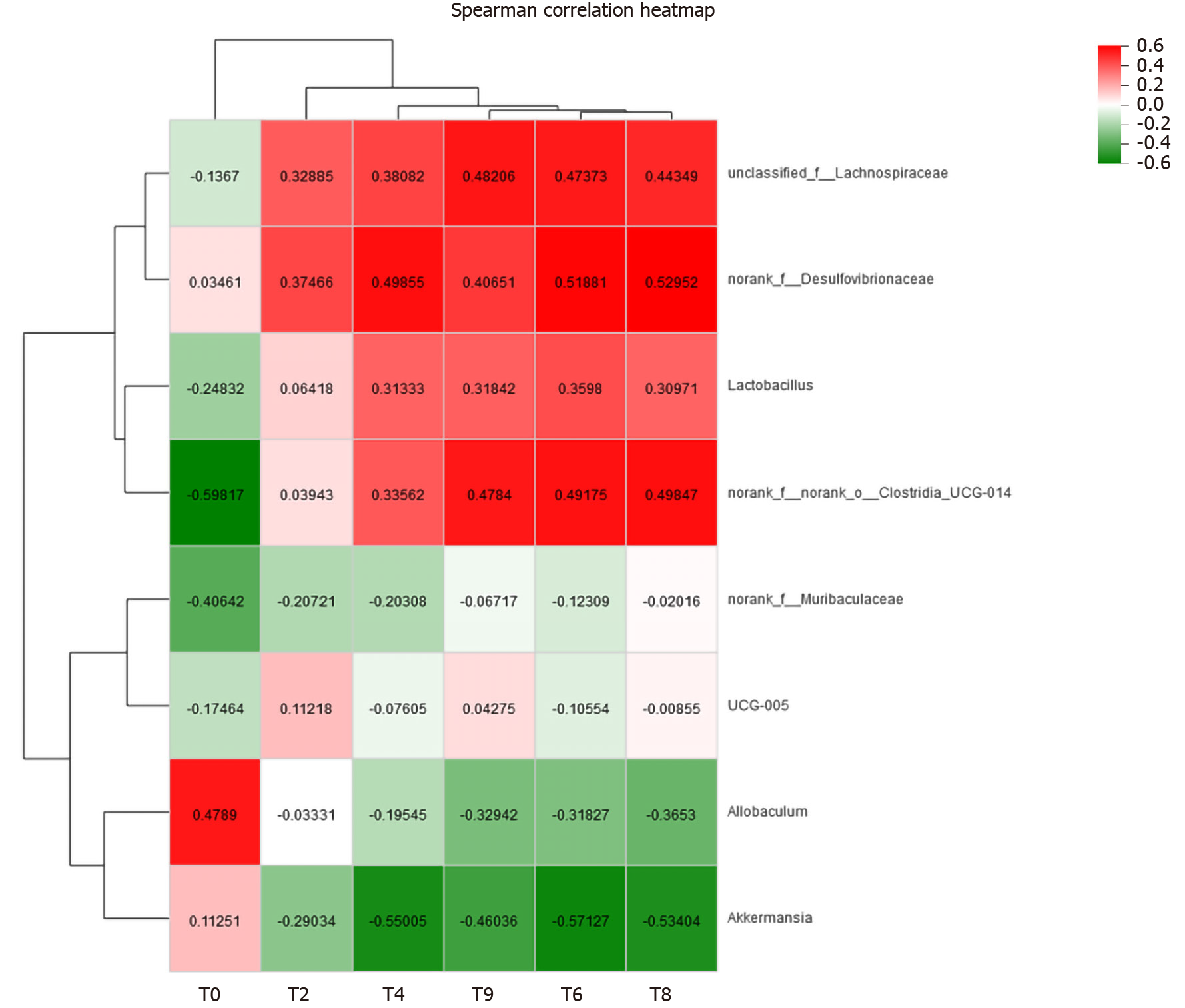
Figure 11 Correlation analysis between type 2 diabetes mellitus-related body weight and gut microbiota at the genus level.
The colours range from green (negative correlation) to red (positive correlation). Statistical significance was based on a P value of 0.05 (Spearman’s correlation coefficient). r values are indicated.
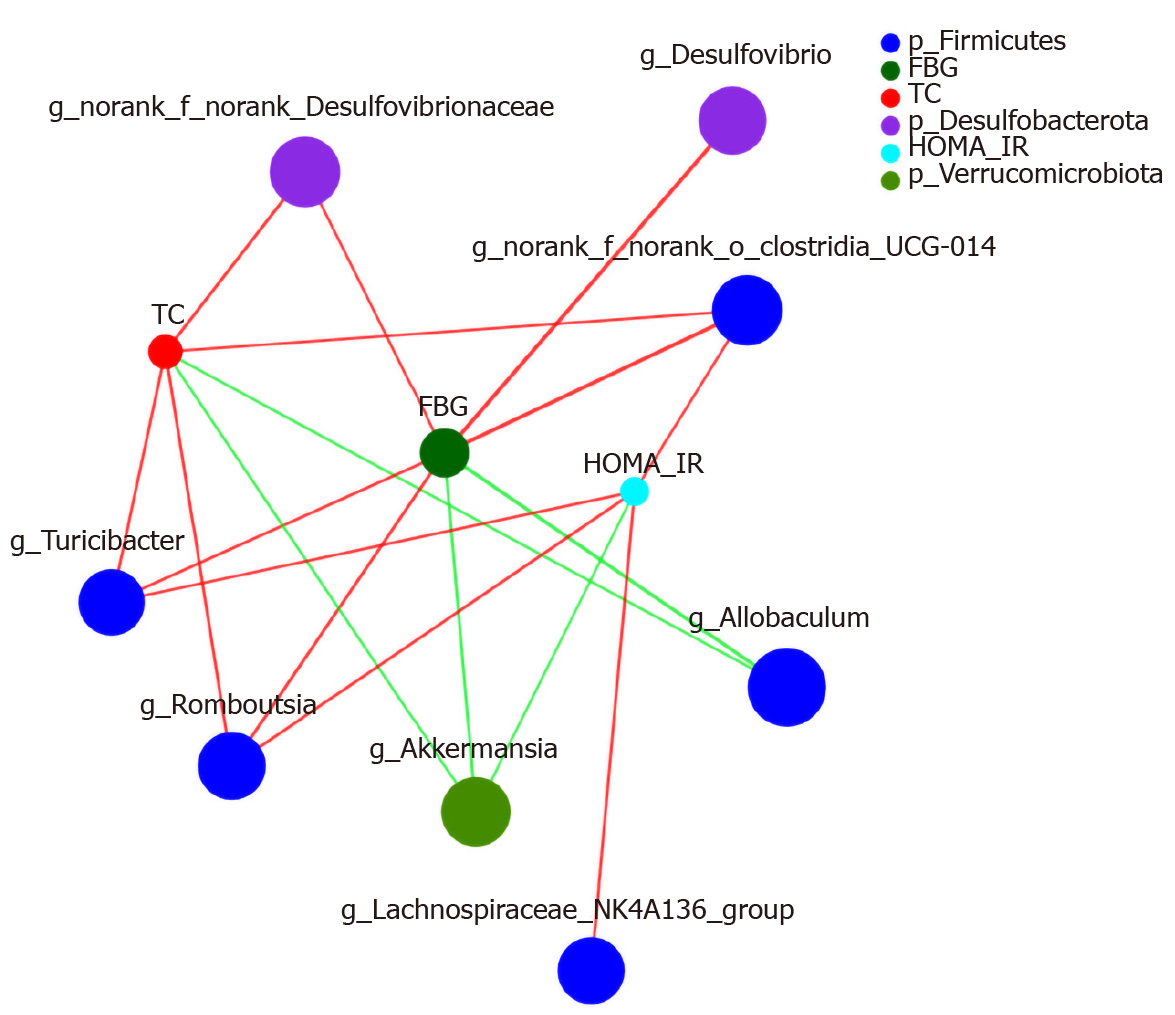
Figure 12 Construct of a species and type 2 diabetes mellitus biochemical factors-related network at the genus level.
The colours range from green (negative correlation) to red (positive correlation). Statistical significance was based on a P value of 0.05 (Networkx). The size of the node represents the abundance of species. The thickness of the line indicates the size of the correlation coefficient. The thicker the line, the higher the correlation between species; the more lines, the closer the relationship between the species and other species. FBG: Fasting blood glucose; HOMA-IR: Homeostatic model assessment-insulin resistance.
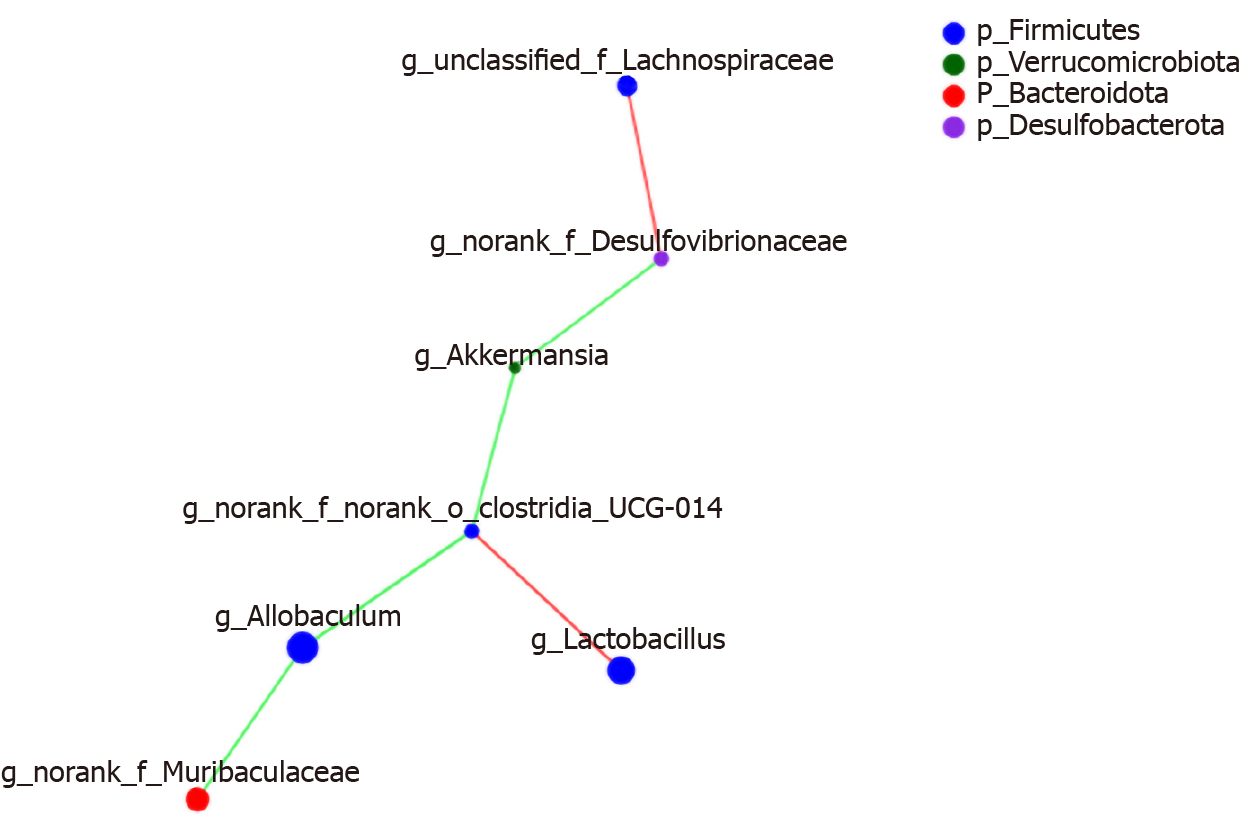
Figure 13 Construct of a species-related network at the genus level.
The colours range from green (negative correlation) to red (positive correlation). Statistical significance was based on a P value of 0.05 (Networkx). The size of the node represents the abundance of species. The thickness of the line indicates the size of the correlation coefficient. The thicker the line, the higher the correlation between species; the more lines, the closer the relationship between the species and other species.
- Citation: Zhao JD, Li Y, Sun M, Yu CJ, Li JY, Wang SH, Yang D, Guo CL, Du X, Zhang WJ, Cheng RD, Diao XC, Fang ZH. Effect of berberine on hyperglycaemia and gut microbiota composition in type 2 diabetic Goto-Kakizaki rats. World J Gastroenterol 2021; 27(8): 708-724
- URL: https://www.wjgnet.com/1007-9327/full/v27/i8/708.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i8.708