Copyright
©The Author(s) 2021.
World J Gastroenterol. Apr 28, 2021; 27(16): 1785-1804
Published online Apr 28, 2021. doi: 10.3748/wjg.v27.i16.1785
Published online Apr 28, 2021. doi: 10.3748/wjg.v27.i16.1785
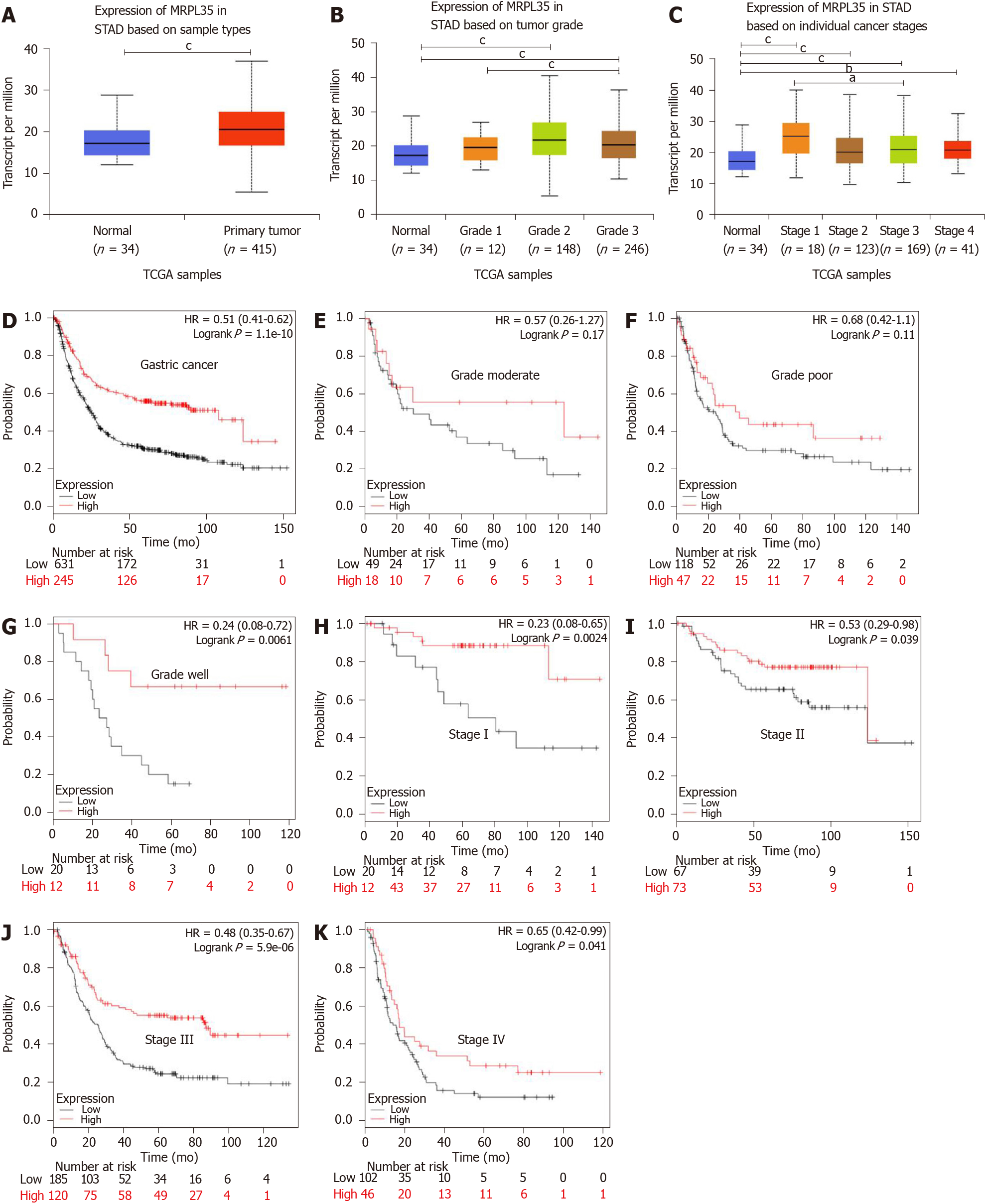
Figure 1 MRPL35 is highly up-regulated in gastric carcinoma.
A-C: The expression of MRPL35 in stomach adenocarcinoma based on sample type (A), tumor grade (B), and individual cancer stage (C). The results were based on data from the public database UALCAN (www.ualcan.path.uab.edu); D: Kaplan-Meier overall survival chart. The results were based on data from the public database KMplot (www.kmplot.com); E-G: Overall survival of patients with high and low expression of MRPL35 in different pathological grades of gastric carcinoma (GC) (E, moderate; F, poor; G, well); H-K: Overall survival of patients with high and low expression of MRPL35 in different pathological stages of GC (H, stage I; I, stage II; J, stage III; K, stage IV). The results were based on data from the public database KMplot (www.kmplot.com). aP < 0.05, bP < 0.01, cP < 0.001. STAD: Stomach adenocarcinoma; HR: Hazard ratio.
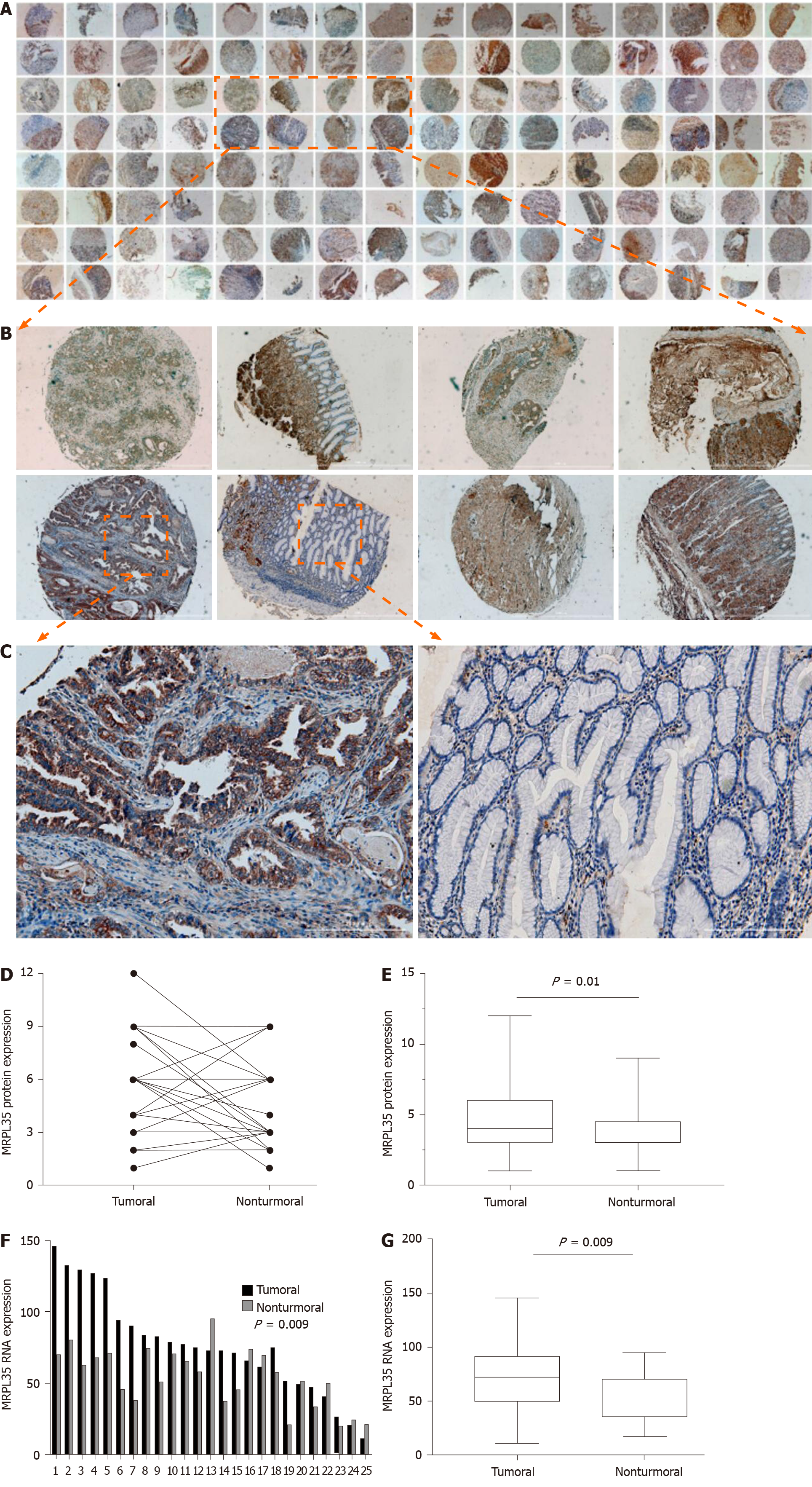
Figure 2 The expression of MRPL35 is up-regulated in gastric carcinoma tissues.
A-C: Immunohistochemistry was performed on 64 pairs of gastric carcinoma (GC) tissues and matched adjacent tissues. Magnification: 4 × (A), 40 × (B), 200 × (C); D and E: The relative expression of MRPL35 in GC tissues and matched adjacent tissues; F and G: The relative expression of MRPL35 mRNA in GC tissues and matched adjacent tissues (n = 25).
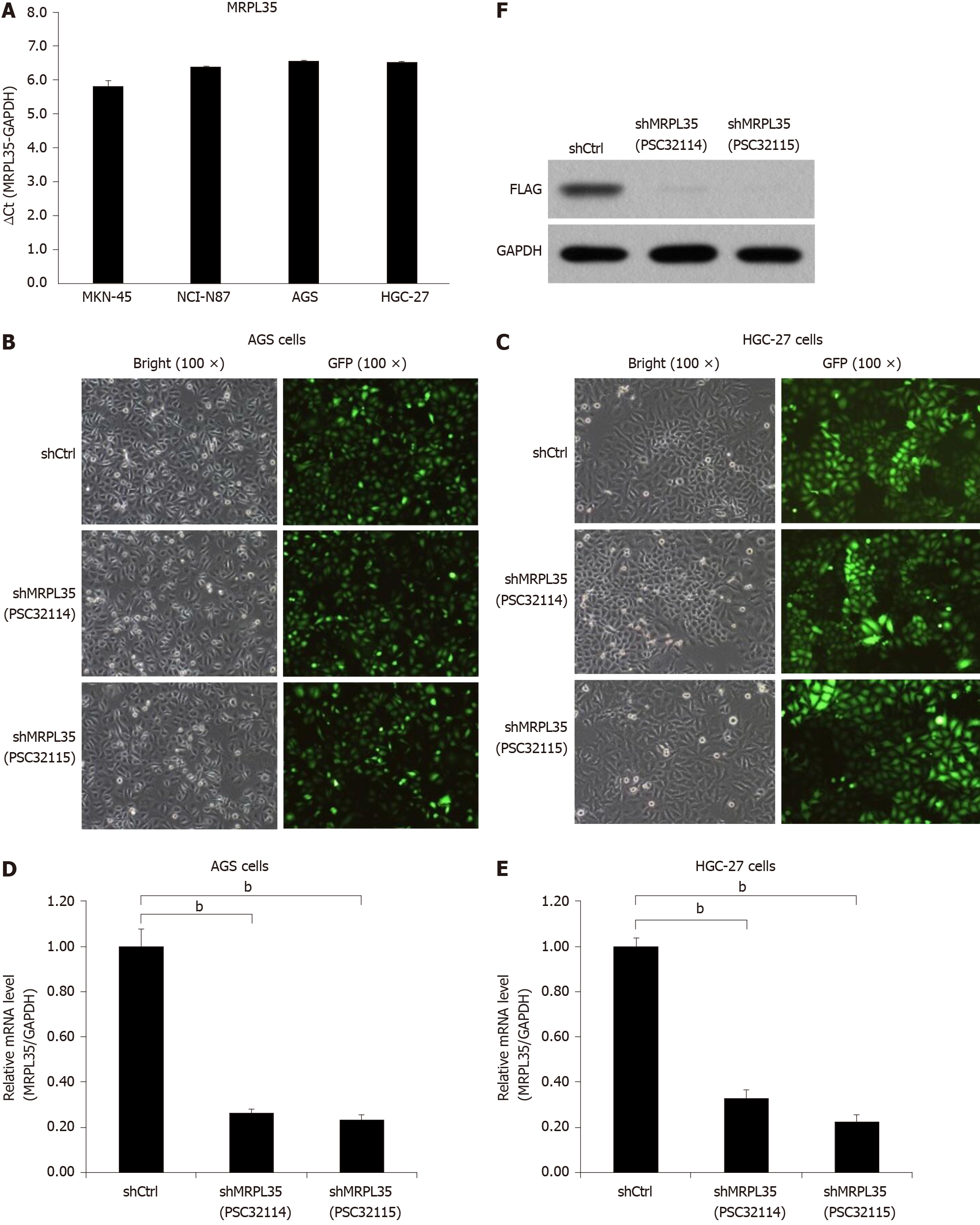
Figure 3 The expression of MRPL35 in gastric carcinoma cells.
A: Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) detection of MRPL35 in gastric carcinoma cell lines (AGS, NCI-N87, MKN-45 and HGC-27); B and C: shCtrl (negative control virus) and shMRPL35 (lentiviral particles of MRPL35) (PSC32114 or PSC32115) infected AGS cells (B) and HGC-27 cells (C) at 72 h with bright and fluorescent micrographs (100 × magnification); D and E: qRT-PCR was used to analyze the expression of MRPL35 in AGS cells (D) and HGC-27 cells (E) infected with shCtrl and shMRPL35; F: Western blot was used to detect the expression of MRPL35 in AGS cells transfected with shCtrl and shMRPL35, and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as an internal control. n = 3. bP < 0.01. shCtrl: Negative control virus; shMRPL35: Lentiviral particles of MRPL35; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; GFP: Green fluorescent protein.
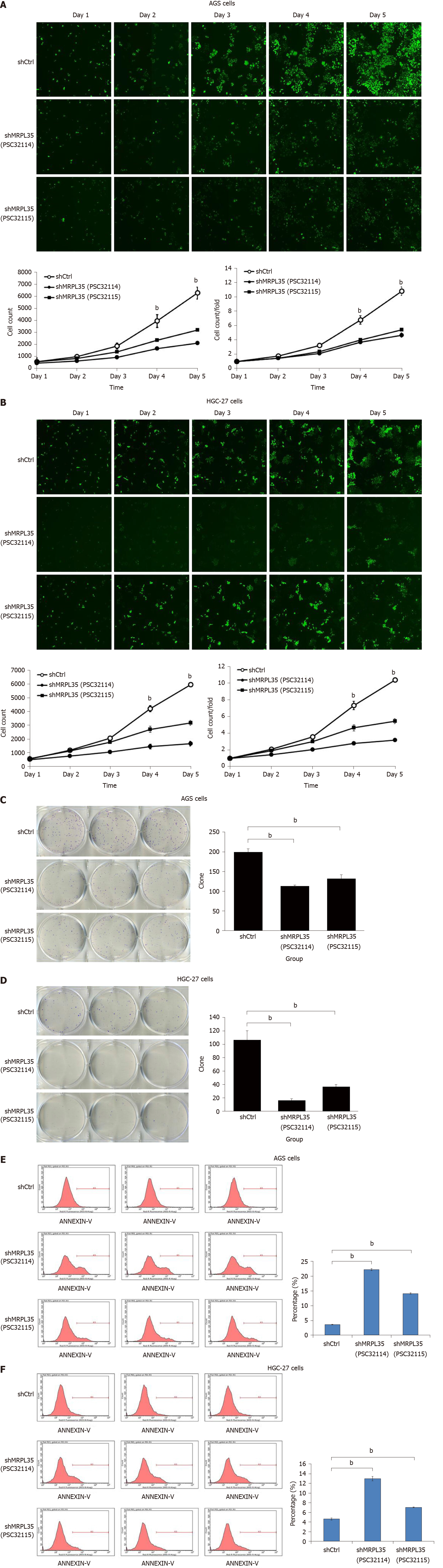
Figure 4 Knockdown of MRPL35 inhibits gastric carcinoma cell proliferation and monoclonal formation and promotes gastric carcinoma cell apoptosis.
A and B: The Celigo cell count assay was used to analyze the proliferation of AGS cells (A) and HGC-27 cells (B) transfected with shCtrl (negative control virus) and shMRPL35 (lentiviral particles of MRPL35); C and D: Colony formation assay of shCtrl and shMRPL35 infected AGS cells (C) and HGC-27 cells (D); E and F: Flow cytometry analysis of apoptosis of shCtrl and shMRPL35 infected AGS cells (E) and HGC-27 cells (F). n = 3. bP < 0.01. shCtrl: Negative control virus; shMRPL35: Lentiviral particles of MRPL35.
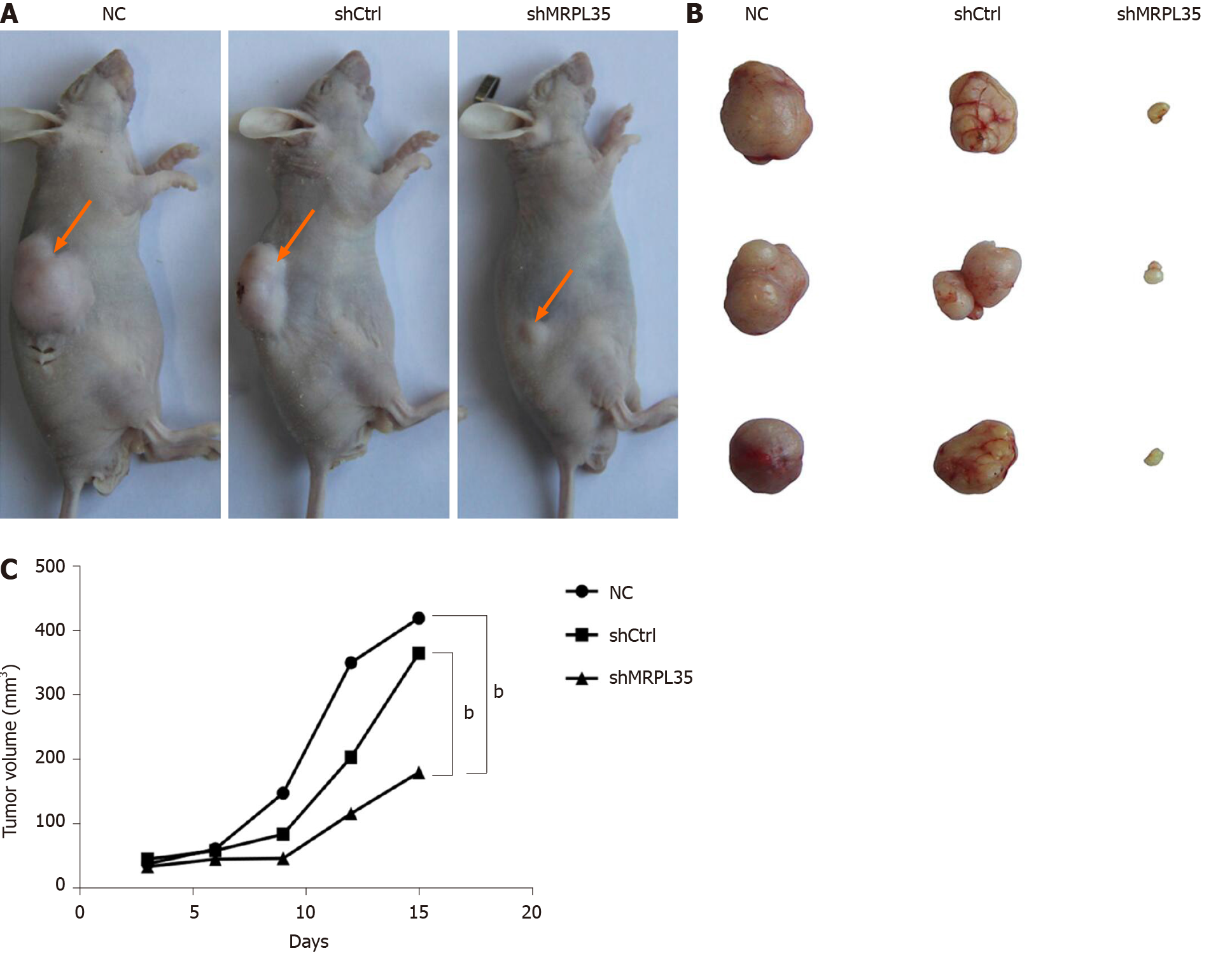
Figure 5 Knockdown of MRPL35 inhibits tumor formation in nude mice.
A: Nude mouse gastric carcinoma xenograft model; B: Subcutaneously transplanted tumor in nude mice; C: Tumor growth curve of transplanted tumor model. n = 30. bP < 0.01. NC: Negative control; shCtrl: Negative control virus; shMRPL35: Lentiviral particles of MRPL35.
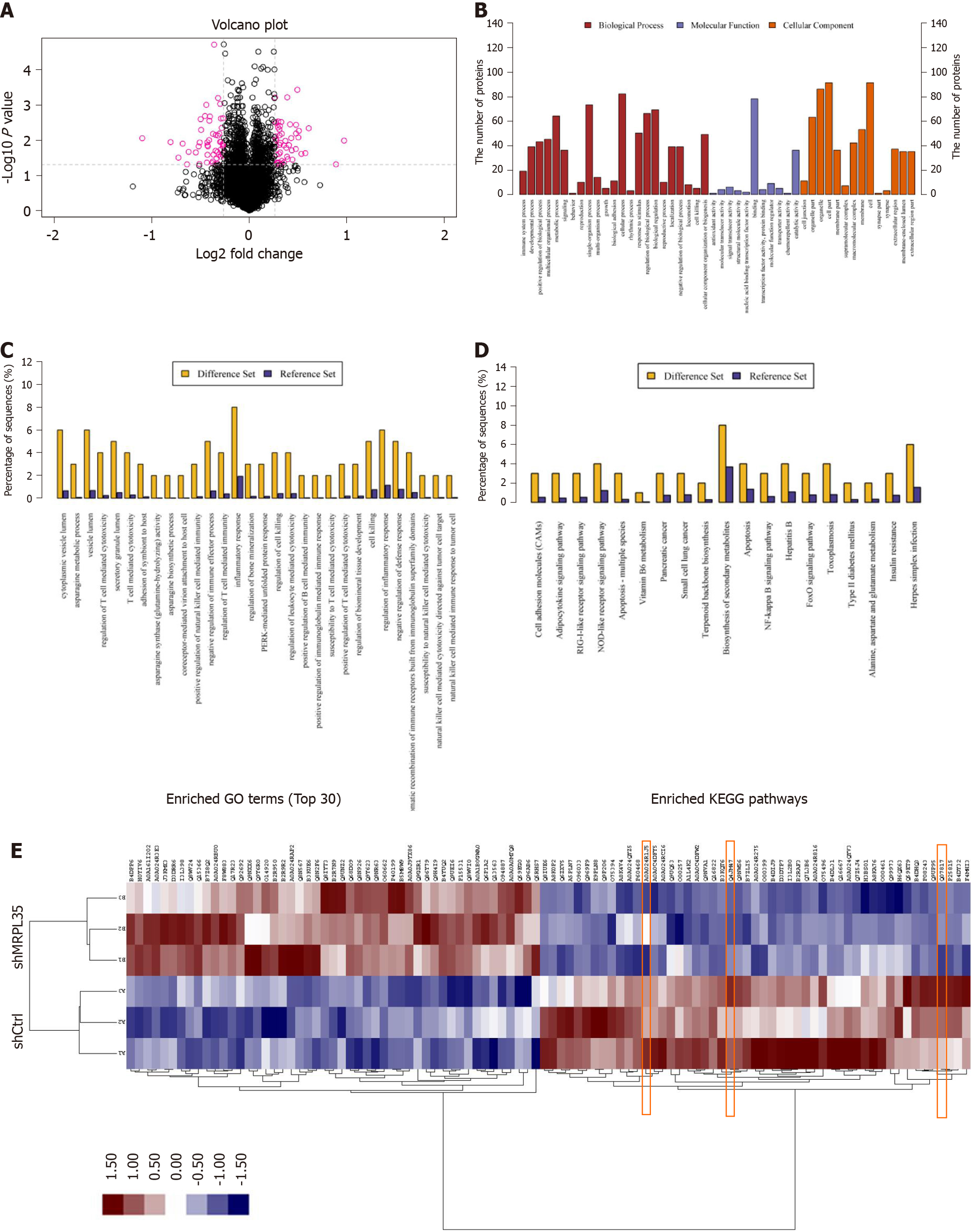
Figure 6 Proteomic and bioinformatic analysis of identified proteins.
A: A total of 5993 proteins were identified by isobaric tags for relative and absolute quantification. Red circle indicates 100 differentially expressed proteins (DEPs); B: The result of Gene Ontology annotation. The identified proteins were categorized into three types: Biological process, molecular function, and cellular component; C: Enriched Gene Ontology annotation statistics of DEPs; D: Enriched KEGG pathway statistics of DEPs; E: Protein cluster analysis of DEPs. The red box part is AGR2, PICK1, and BCL-XL. shCtrl: Negative control virus; shMRPL35: Lentiviral particles of MRPL35.
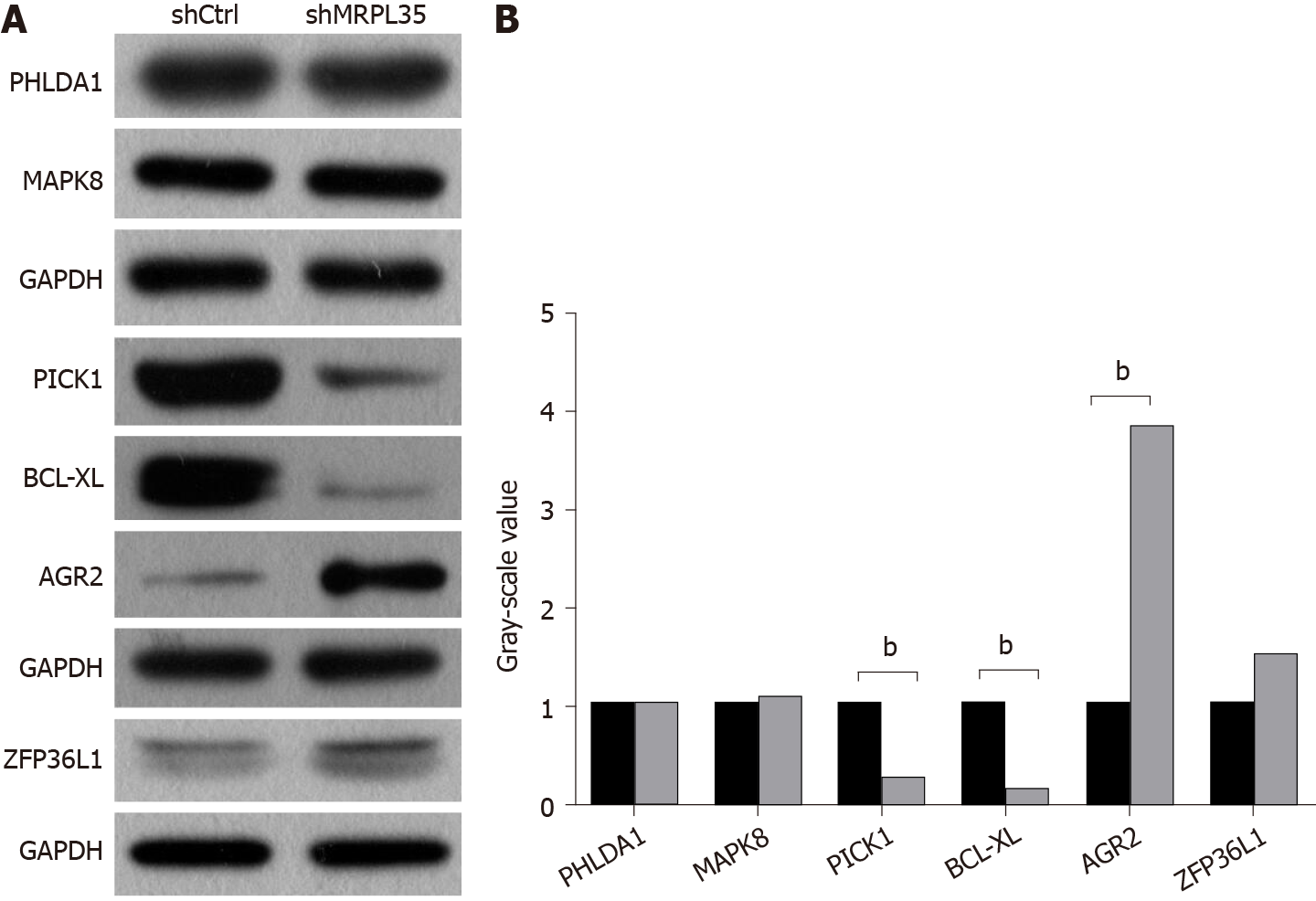
Figure 7 Expression of related proteins in AGS cells after knockdown of MRPL35.
A and B: The expression of PHLDA1, MAPK8, PICK1, BCL-XL, AGR2, and ZFP36L1 proteins was detected by Western blot. GAPDH was used as an internal control. A: Western blot images; B: Gray-scale value. n = 3. bP < 0.01. shCtrl: Negative control virus; shMRPL35: Lentiviral particles of MRPL35.
- Citation: Yuan L, Li JX, Yang Y, Chen Y, Ma TT, Liang S, Bu Y, Yu L, Nan Y. Depletion of MRPL35 inhibits gastric carcinoma cell proliferation by regulating downstream signaling proteins. World J Gastroenterol 2021; 27(16): 1785-1804
- URL: https://www.wjgnet.com/1007-9327/full/v27/i16/1785.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i16.1785