Copyright
©The Author(s) 2019.
World J Gastroenterol. Jan 28, 2019; 25(4): 433-446
Published online Jan 28, 2019. doi: 10.3748/wjg.v25.i4.433
Published online Jan 28, 2019. doi: 10.3748/wjg.v25.i4.433
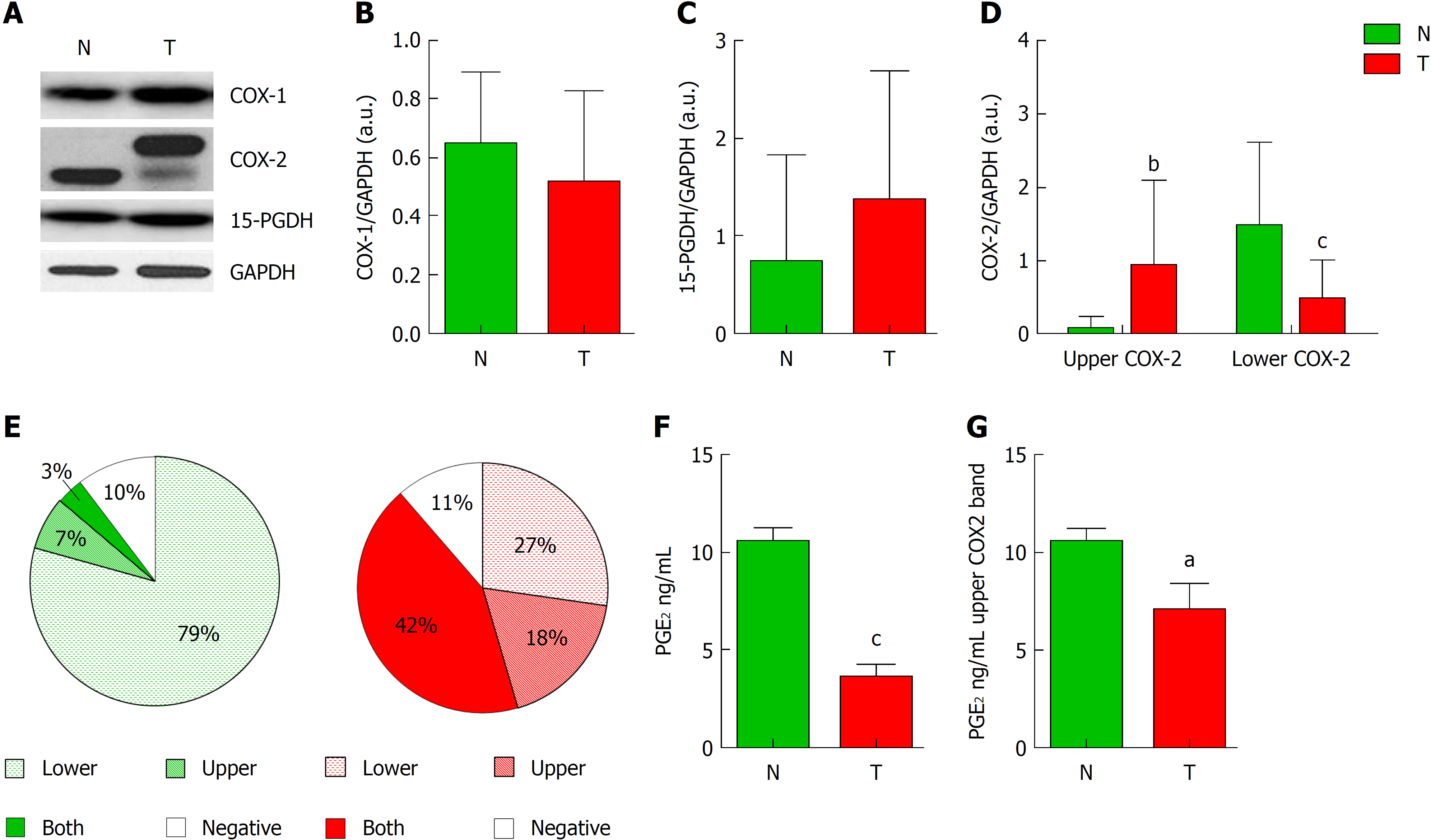
Figure 1 Analysis of cyclooxygenase-2 protein levels and prostaglandin E2 measurement.
A: Representative image of western blot analysis of cyclooxygenase-1 (COX-1), cyclooxygenase-2 (COX-2) and 15-hydroxyprostaglandin dehydrogenase (15-PGDH) normalized by glyceraldehyde-3-phosphate dehydrogenase (GAPDH) of tumor (T) or non-tumor (N) samples. B and C: Graphs represent normalized COX-1 (B) or 15-PGDH (C) bands in western blot in T vs N tissue. D: Graph represent normalized COX-2 protein levels corresponding to upper (Up) or lower (Down) band in western blot in T vs N tissue. E: Graphs represent the percentage of samples expressing lower, upper, both or none bands in western blot for COX-2. F: Levels of PGE2 in Tumor vs Non-tumor samples. G: Relative levels of prostaglandin E2 only in samples from colorectal cancer patients showing the upper band of COX-2 in western blot. n = 45. Graphs show mean ± SD. aP ≤ 0.05, bP ≤ 0.01 and cP ≤ 0.001 vs the non-tumor condition. COX-1: Cyclooxygenase-1; COX-2: Cyclooxygenase-2; 15-PGDH: 15-hydroxyprostaglandin dehydrogenase; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; T: Tumor; N: Non-tumor; PGE2: Prostaglandin E2.
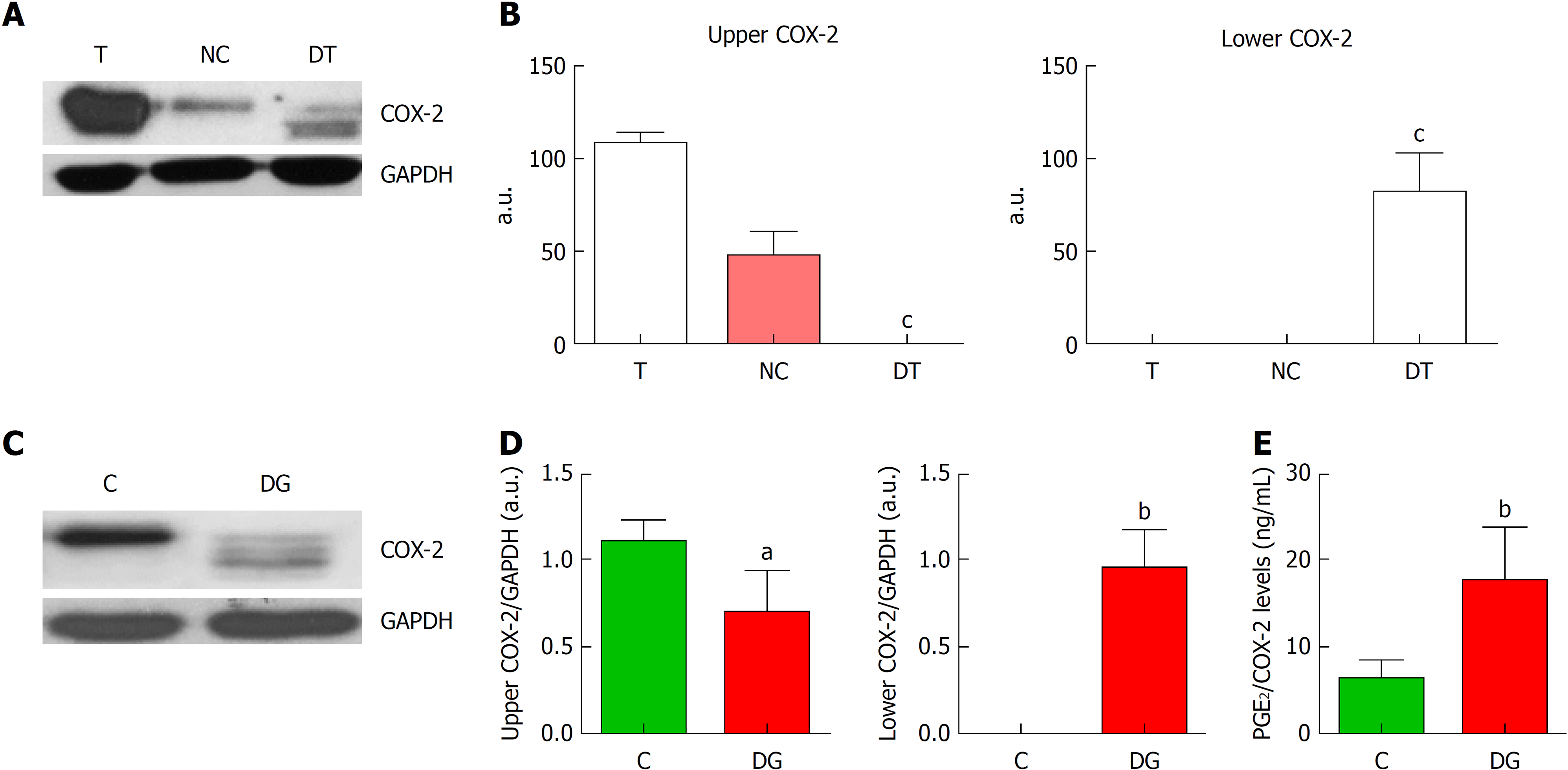
Figure 2 Deglycosylation assays of cyclooxygenase-2.
A: Representative images of western blot of cyclooxygenase-2 (COX-2) protein levels corresponding to tumor tissue (T), negative control (NC) and deglycosylated tumor tissue (DT). B: Graphs correspond to upper or lower COX-2 bands quantifications. n = 4. C: Representative images of western blot of COX-2 protein levels in HT29 cells under basal conditions (C) or deglycosylated (DG). D: Graphs correspond to upper or lower COX-2 bands quantifications. n = 8. E. Relative levels of prostaglandin E2 (PGE2) normalized by total COX-2 band obtained in the corresponding western blot analysis. n = 11. Graphs show mean ± SD. aP ≤ 0.05, bP ≤ 0.01 and cP ≤ 0.001 vs the glycosylated condition. COX-2: Cyclooxygenase-2; T: Tumor; NC: Negative control; DT: Deglycosylated tumor; C: Cells under basal conditions; DG: Deglycosylated; PGE2: Prostaglandin E2.
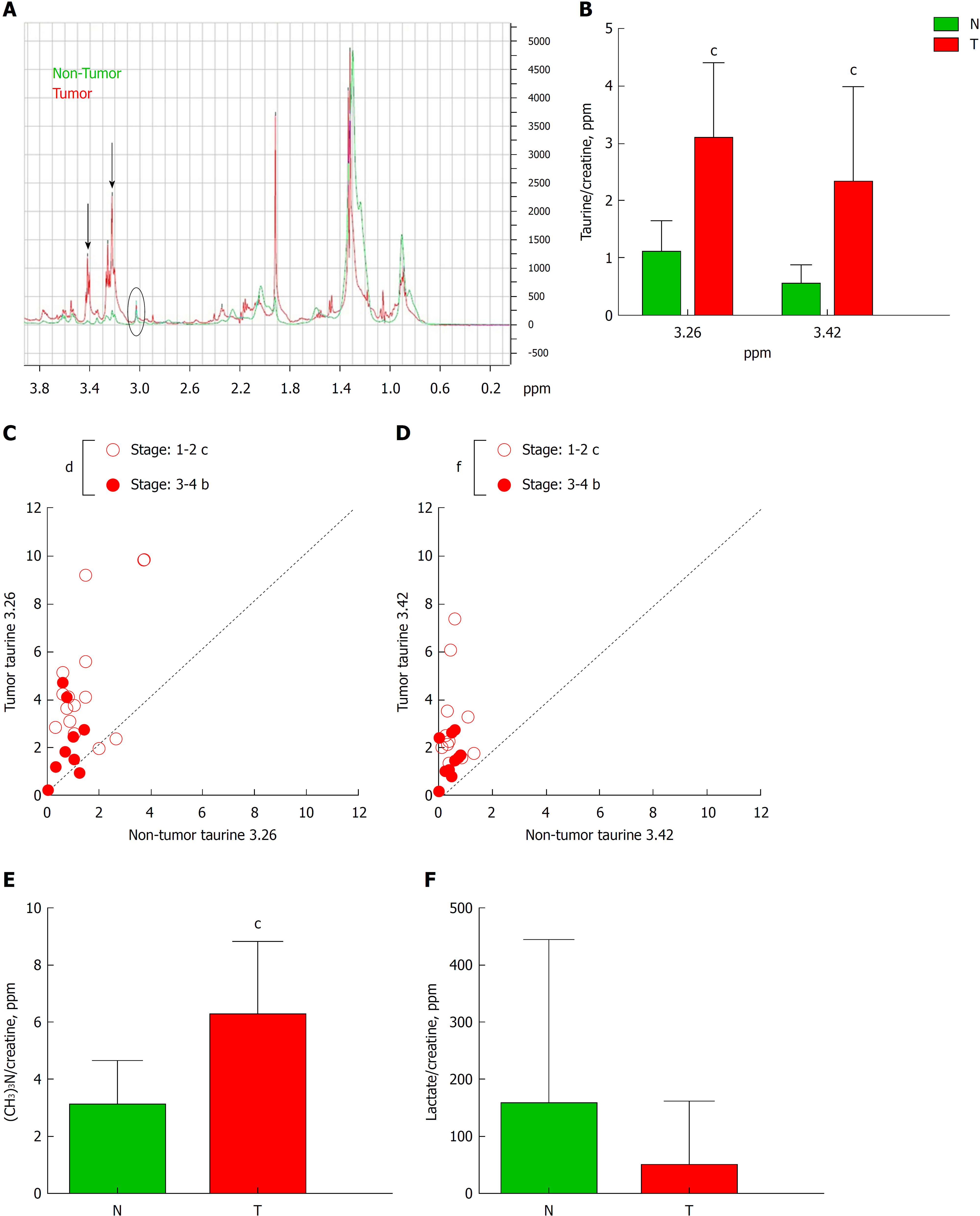
Figure 3 High resolution magic angle spinning analysis of tumor tissue.
A: Schematic diagram of high resolution magic angle spinning spectra of tumor tissue (red line) and non-tumor tissue (blue line). Black arrows indicate two peaks corresponding to taurine δ ppm 3.26 and 3.42. B: Graph represents the quantification of both taurines normalized by creatine of tumor (T) and non-tumor (N) tissue. C and D: Correlation graphs of taurine 3.26 (C) and taurine 3.42 (D) in T vs N subdivided depending on the stage of the Tumor: 1-2 vs 3-4. E and F: Graphs represent the quantification of phosphocholine (E) or lactate (F) in T vs N. n = 29. Graphs show mean ± SD. bP ≤ 0.01, cP ≤ 0.001 of T vs N and dP ≤ 0.05, fP ≤ 0.001 between stages in tumors. T: Tumor; N: Non-tumor.
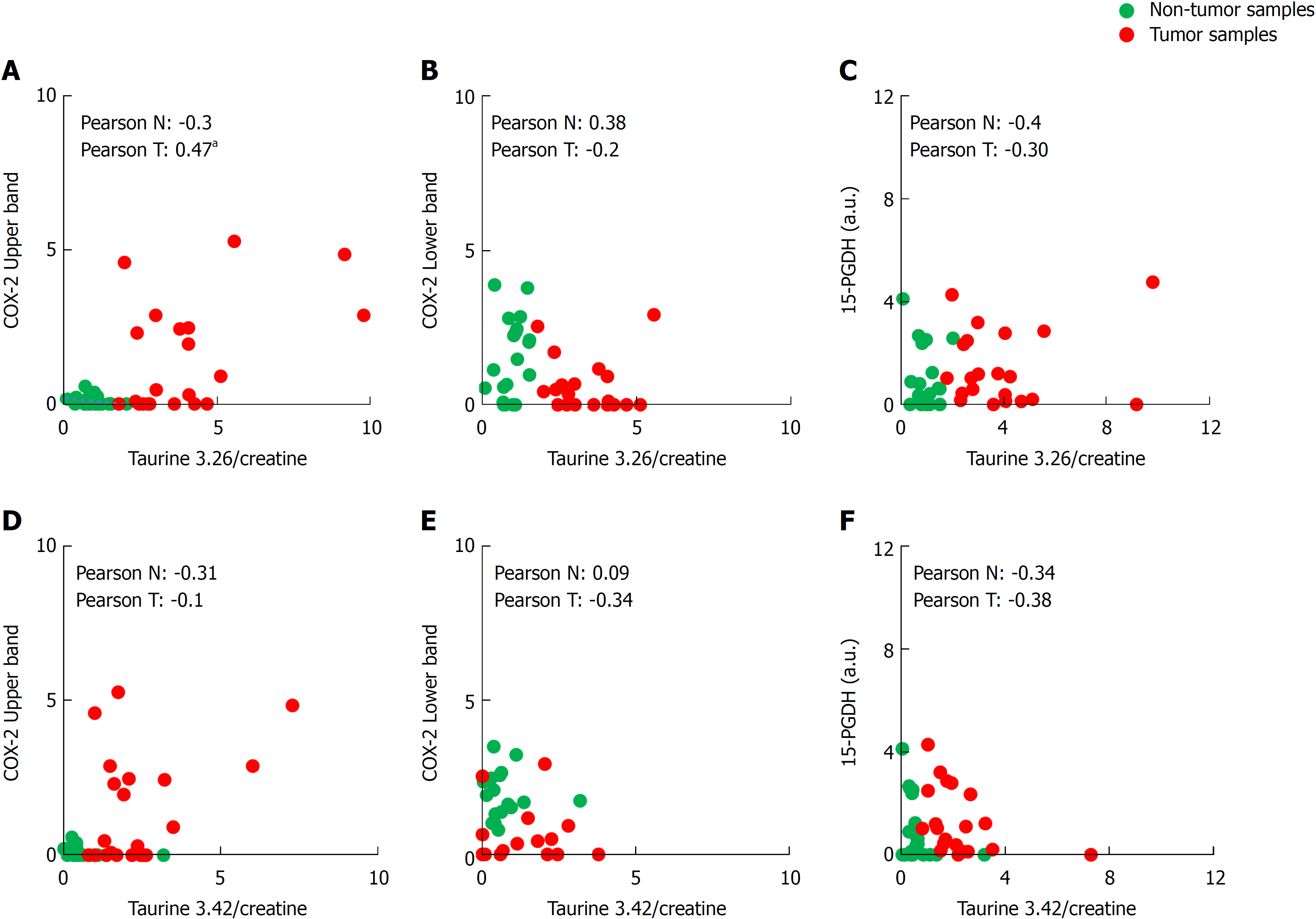
Figure 4 Biostatistical analysis of the correlation between the presence of cyclooxygenase-2 bands in western blot and both taurine peaks.
A-F: Statistical analysis was performed searching the correlation coefficient between the presence of upper or lower cyclooxygenase-2 band or 15-hydroxyprostaglandin dehydrogenase in western blot and the taurine peaks (3.26 and 3.42 δ ppm) levels of 29 Tumor/Non-tumor pairs of samples. The Pearson’s correlation coefficient is shown in each figure for non-tumor or tumor sample. aP ≤ 0.05. COX-2: Cyclooxygenase-2; 15-PGDH: 15-hydroxyprostaglandin dehydrogenase.
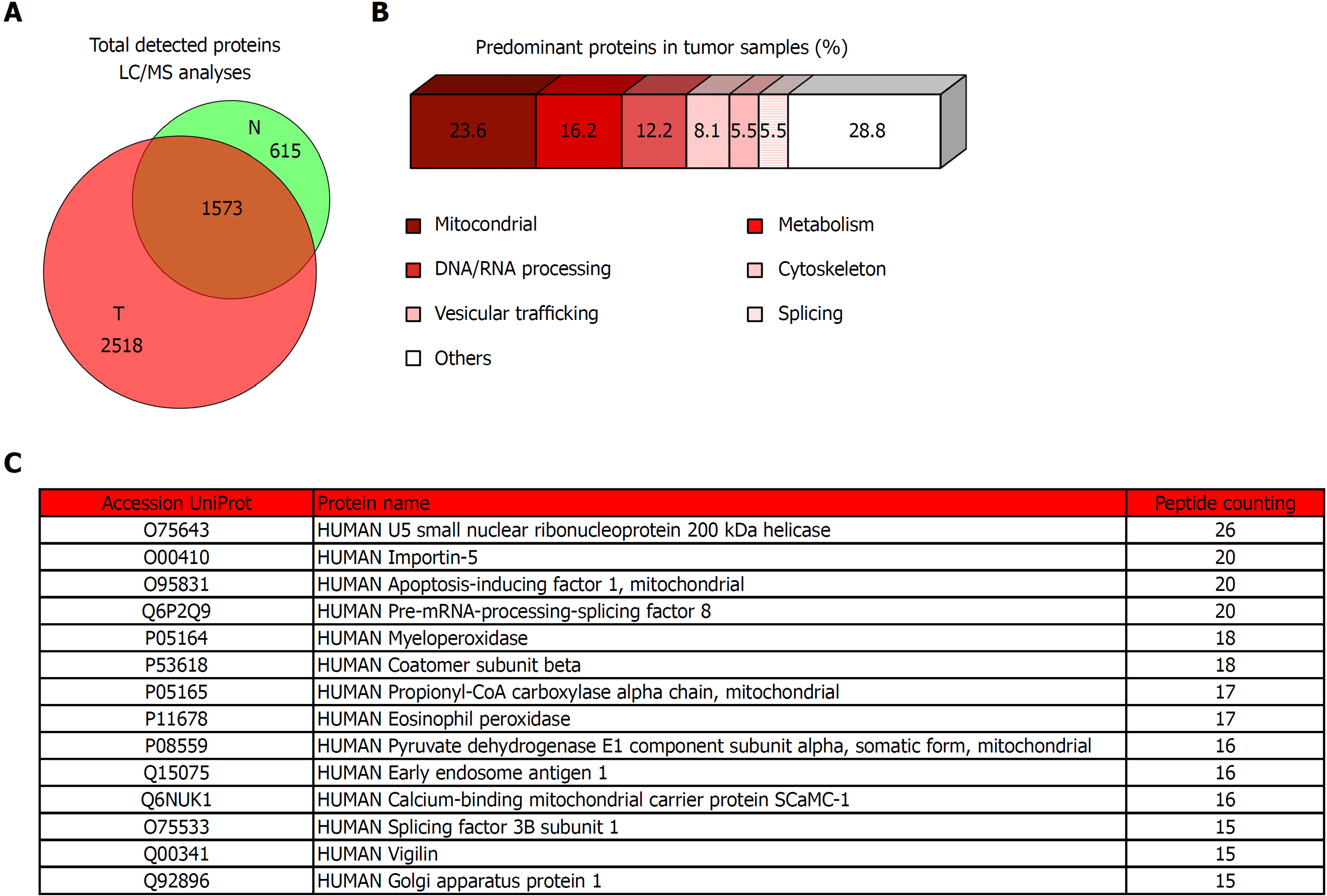
Figure 5 Proteomic analysis of tumor tissue of colorectal cancer patients and the corresponding non-tumor adjacent tissue.
A: Venn diagram with specific and overlapped proteins. 2188 and 4091 proteins were identified (FDR < 1%) in non-tumor and tumor tissue, respectively. Among them, 1573 were identified in both tissues, and 615 and 2518 were unique in non-tumor and tumor tissue, respectively. B: Enrichment analysis of specific proteins identified in tumor tissue by mass spectrometry performed with proteins that were identified with 5 or more peptides (271 most abundant proteins). Most of these proteins had mitochondrial and metabolic function, or were involved in processes related to nucleic acids processing, cytoskeleton, vesicular trafficking, or splicing. C: Specific tumor tissue proteins identified with 15 or more peptides in liquid chromatography-mass spectrometry analyses. T: Tumor; N: Non-tumor; LC/MS: Liquid chromatography-mass spectrometry.
- Citation: Prieto P, Jaén RI, Calle D, Gómez-Serrano M, Núñez E, Fernández-Velasco M, Martín-Sanz P, Alonso S, Vázquez J, Cerdán S, Peinado MÁ, Boscá L. Interplay between post-translational cyclooxygenase-2 modifications and the metabolic and proteomic profile in a colorectal cancer cohort. World J Gastroenterol 2019; 25(4): 433-446
- URL: https://www.wjgnet.com/1007-9327/full/v25/i4/433.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i4.433