Copyright
©The Author(s) 2015.
World J Gastroenterol. Jun 21, 2015; 21(23): 7208-7217
Published online Jun 21, 2015. doi: 10.3748/wjg.v21.i23.7208
Published online Jun 21, 2015. doi: 10.3748/wjg.v21.i23.7208
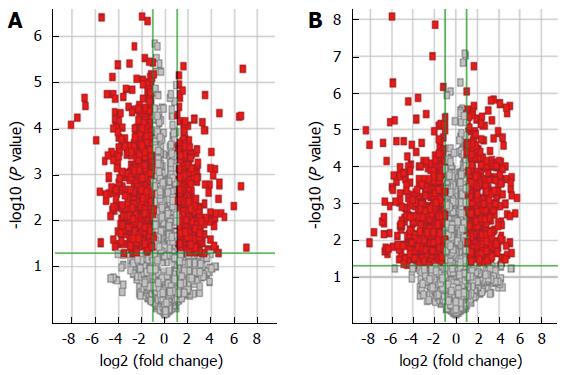
Figure 1 Long noncoding RNAs and mRNA profile comparisons between the hepatocellular carcinoma and adjacent non-tumor tissues.
Volcano plots used to distinguish the different expressed A: Long noncoding RNAs; and B: mRNAs. The vertical green lines correspond to 2.0-fold up- and downregulation, respectively, and the horizontal green lines represent P = 0.05. The red points represent the differentially expressed genes.
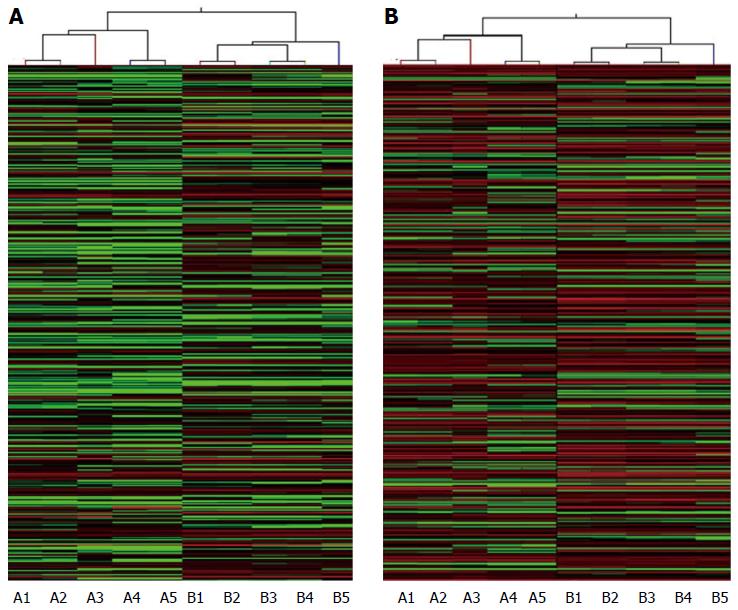
Figure 2 Hierarchical clustering of long noncoding RNAs and mRNAs in hepatocellular carcinoma.
Hierarchical clustering analysis of A: 1772 Long noncoding RNAs; and B: 2508 mRNAs that were differentially expressed between hepatocellular carcinoma samples (A1-A5) and the adjacent non-tumor samples (B1-B5). The red and the green shades indicate the expression above and below the relative expression, respectively, across all samples.
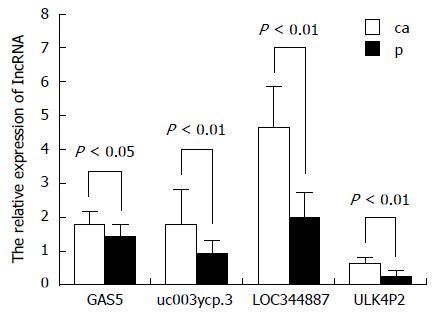
Figure 3 Quantitative real-time reverse-transcription PCR.
Long noncoding (lnc)RNAs GAS5, uc003ycp.3, uc004bdv.3, and ULK4P2 were differentially expressed between hepatitis B virus-related hepatocellular carcinoma tissues (ca) and the paired non-tumor samples (p).
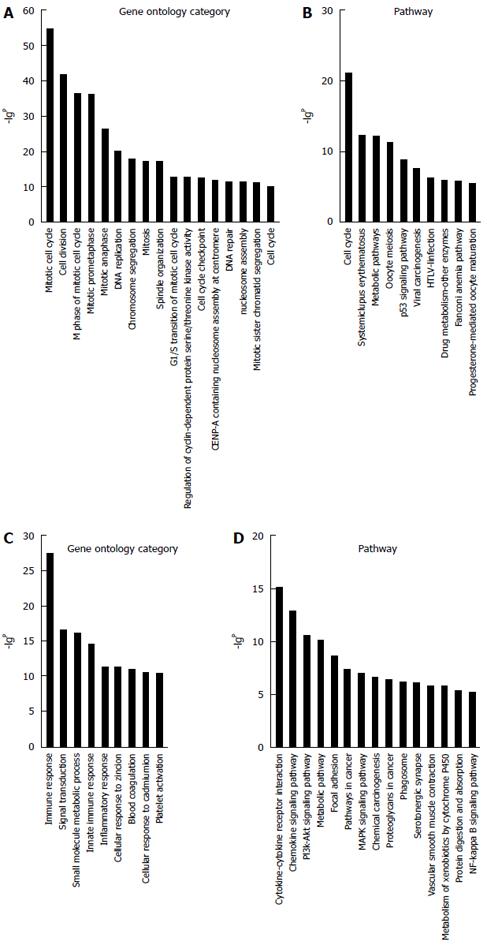
Figure 4 Gene ontology and pathway analyses.
A total of 1270 differentially expressed mRNAs were chosen with a fold change > 3; the column graphs represent the enrichment of these mRNAs. The (-lgP) value is a positive correlation with gene ontology (GO) (A); Signaling pathway enrichment for upregulated mRNAs (B); GO (C); Signaling pathway enrichment for downregulated mRNAs (D). The (-lgP) values above five are presented.
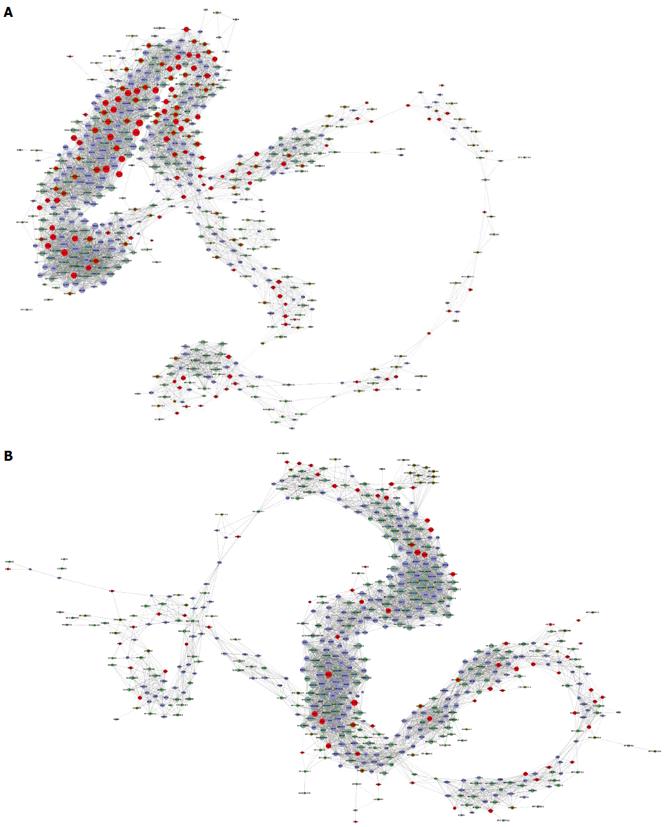
Figure 5 Long noncoding RNA-mRNA coexpression network.
A: The long noncoding (lnc)RNA-mRNA network containing the 349 filtered mRNAs and 392 filtered aberrant expressed lncRNAs in HCC tissues; B: The lncRNA-mRNA network containing the 349 filtered mRNAs and 392 filtered aberrant expressed lncRNAs in paired non-tumor tissues. Upregulated RNAs are shown in red, and downregulated RNAs are presented in blue. Nodes without a ring represent mRNAs, nodes with a ring represent lncRNAs, and node size represents the degree of centrality.
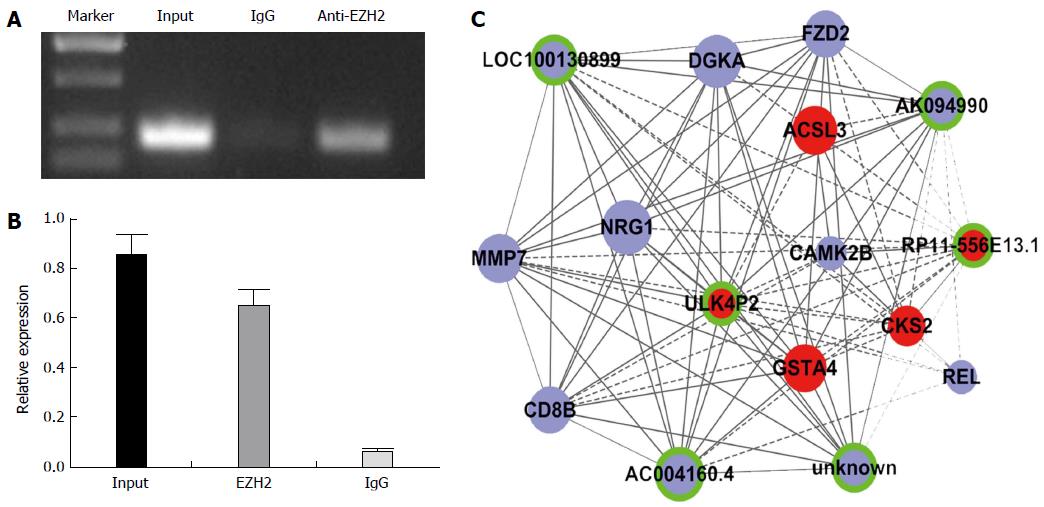
Figure 6 Large intergenic noncoding RNA ULK4P2 combines with enhancer of zeste homolog 2.
A, B: ULK4P2 combined with enhancer of zeste homolog 2 (EZH2) was measured in hepG2 cells via an RNA immunoprecipitation assay using an anti-EZH2 antibody (input = total RNA, IgG = control); C: ULK4P2 subnetwork in the hepatitis B virus-related hepatocellular carcinoma coexpression network. Upregulated RNAs are shown in red, downregulated RNAs are presented in blue, and nodes without a ring represent mRNAs, whereas nodes with a ring represent long noncoding RNAs. Node size represents the degree centrality.
- Citation: Yu TT, Xu XM, Hu Y, Deng JJ, Ge W, Han NN, Zhang MX. Long noncoding RNAs in hepatitis B virus-related hepatocellular carcinoma. World J Gastroenterol 2015; 21(23): 7208-7217
- URL: https://www.wjgnet.com/1007-9327/full/v21/i23/7208.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i23.7208