Copyright
©2014 Baishideng Publishing Group Co.
World J Gastroenterol. Apr 7, 2014; 20(13): 3401-3409
Published online Apr 7, 2014. doi: 10.3748/wjg.v20.i13.3401
Published online Apr 7, 2014. doi: 10.3748/wjg.v20.i13.3401
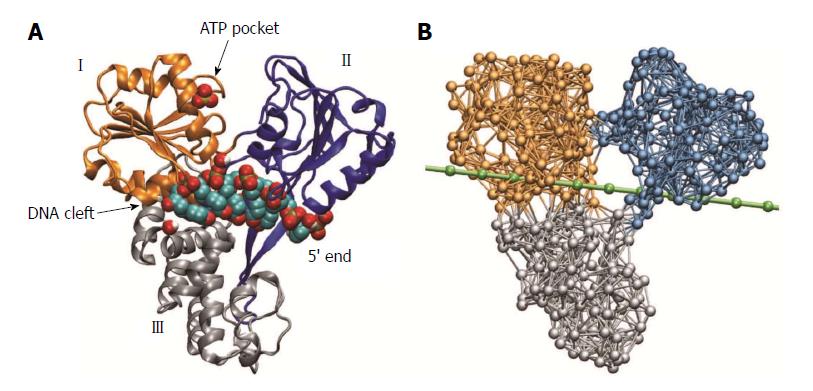
Figure 1 Molecular architecture of hepatitis C virus helicase.
A: Structure of hepatitis C virus (HCV) helicase in ribbon representation (Protein Data Bank code 1A1V). Motor domains I and II are coloured golden and blue, respectively. Domain III is shown in grey. A sulfate ion, shown as beads, is located at the surface of domain I in the proximity of the adenosine-triphosphate (ATP) binding pocket. The short DNA fragment (shown in bead representation) occupies a large cleft separating the motor domains from the third domain; B: Network representation of HCV helicase. The chain of beads representing a single DNA strand is shown in green.
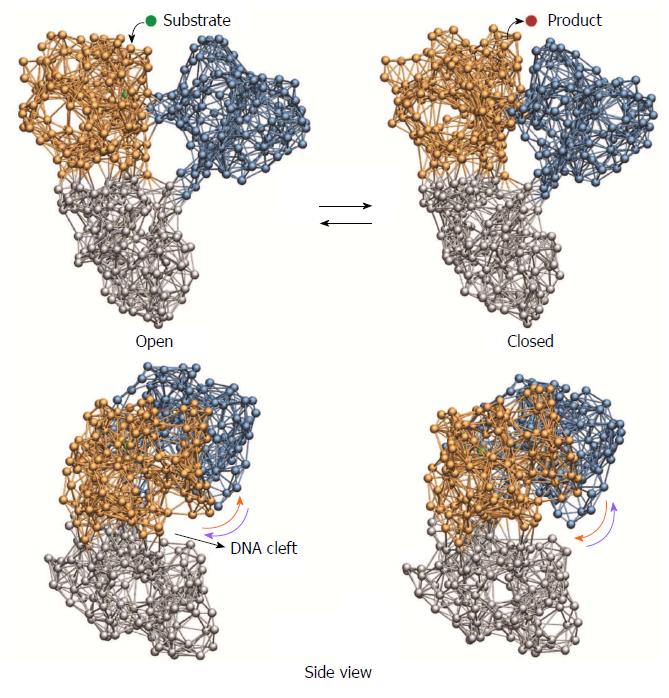
Figure 2 Cyclic ligand-induced conformational motions in hepatitis C virus helicase.
Top row: binding of the substrate ligand led to switching from open to closed conformation. Release of the product ligand induced motions back to the open form; Bottom row: motions of the motor domains relative to the nucleic acid binding cleft that are critical for grip control are indicated by arrows.
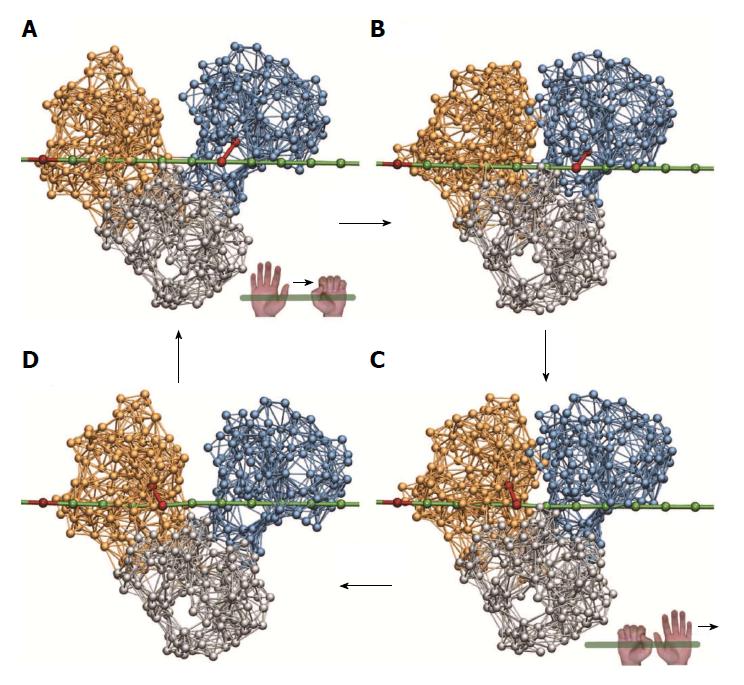
Figure 3 Ratcheting inchworm translocation.
Four consequent snapshots (A-D) within a single ligand-induced operation cycle are shown. Red bonds indicate connections between a motor domain and the DNA chain. After one cycle, the helicase is found in a similar open-shape conformation, but is propagated by one base along the DNA strand (compare A and D).
Figure 4 Unzipping of duplex DNA by hepatitis C virus helicase.
Shown are three consequent snapshots (A-C) from a simulation that demonstrates ratcheting inchworm translocation and mechanical strand separation.
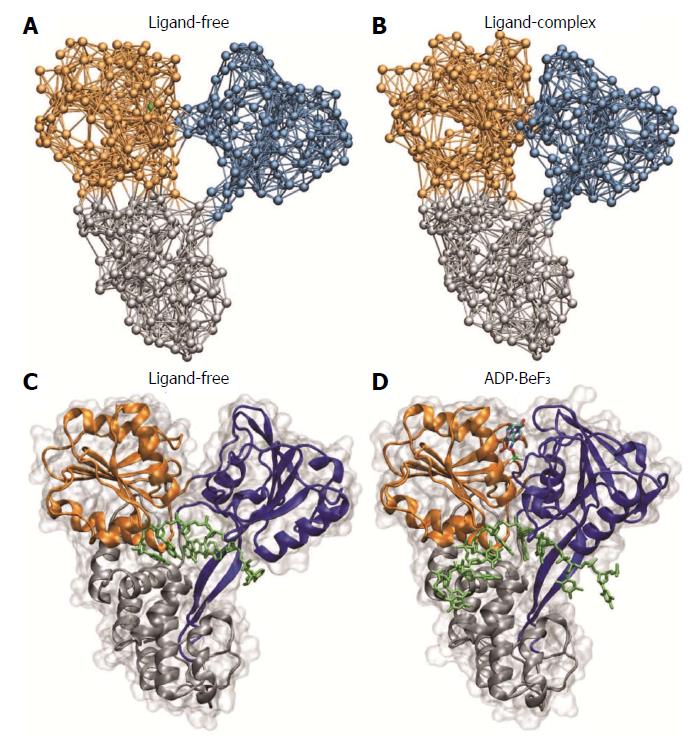
Figure 5 Comparison with novel hepatitis C virus crystal structures.
Structural changes between ligand-free conformation and ligand-bound complex as predicted by our simulations (A and B) are compared with structures determined in recent experiments (C and D, Protein Data Bank codes 3KQH and 3KQU; ADP.BeF3 was used as ATP analog). ATP: Adenosine-triphosphate; ADP: Adenosine-diphosphate.
- Citation: Flechsig H. Computational biology approach to uncover hepatitis C virus helicase operation. World J Gastroenterol 2014; 20(13): 3401-3409
- URL: https://www.wjgnet.com/1007-9327/full/v20/i13/3401.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i13.3401