Published online Feb 15, 2024. doi: 10.4251/wjgo.v16.i2.398
Peer-review started: October 27, 2023
First decision: November 6, 2023
Revised: November 23, 2023
Accepted: January 9, 2024
Article in press: January 9, 2024
Published online: February 15, 2024
Processing time: 98 Days and 7.4 Hours
Prohibitin 1 (PHB1) has been identified as an antiproliferative protein that is highly conserved and ubiquitously expressed, and it participates in a variety of essential cellular functions, including apoptosis, cell cycle regulation, prolifera
To investigate the effects of PHB1 on the proliferation and apoptosis of human HCC cells and the relevant mechanisms in vitro.
HCC patients and healthy individuals were enrolled in this study according to the inclusion and exclusion criteria; then, PHB1 levels in the sera and liver tissues of these participates were determined using ELISA, RT-PCR, and immunohistoche
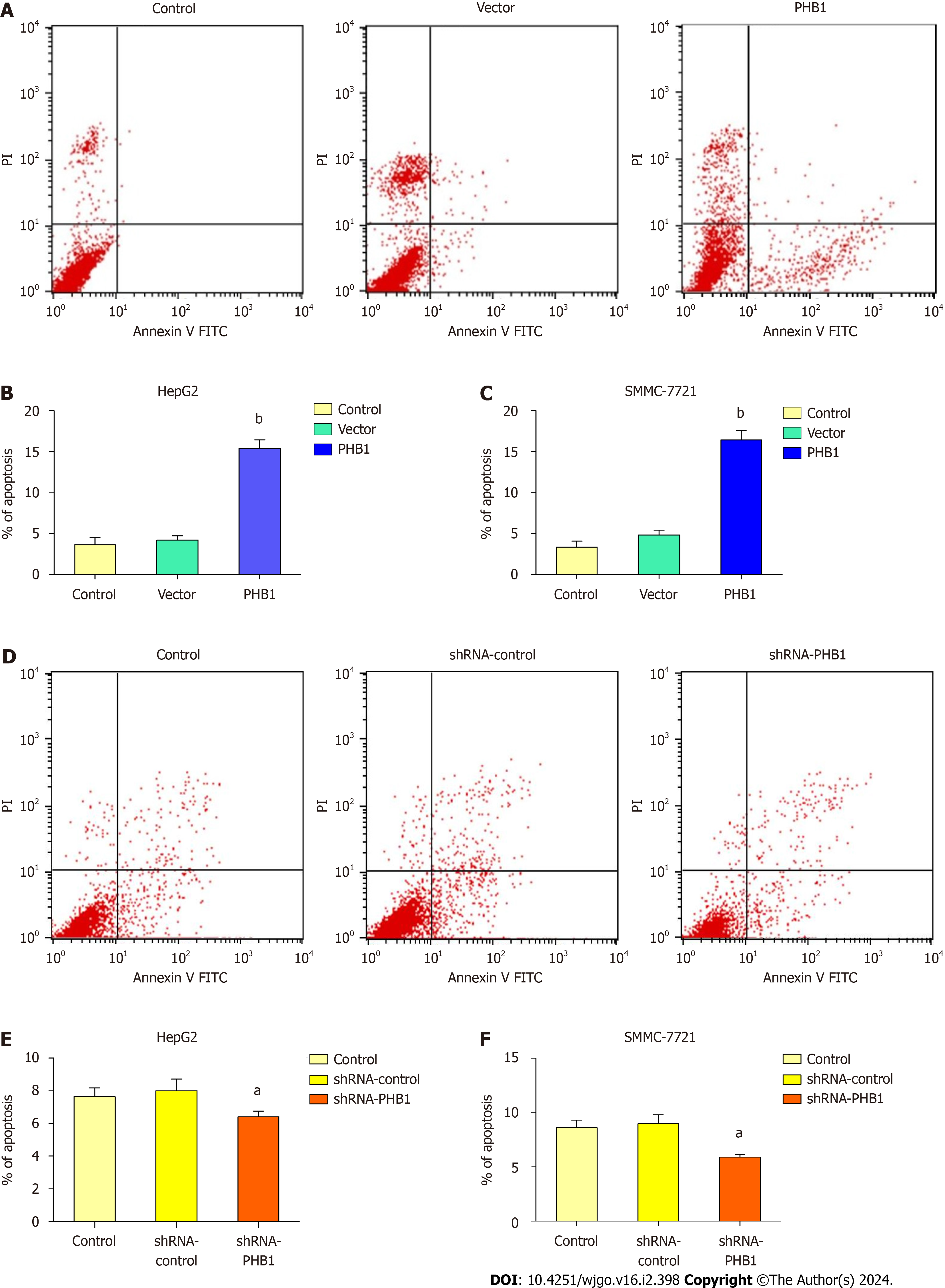
Decreased levels of PHB1 were found in the sera and liver tissues of HCC patients compared to those of healthy individuals, and decreased PHB1 was positively correlated with low differentiation, TNM stage III-IV, and alpha-fetoprotein ≥ 400 μg/L. Overexpression of PHB1 significantly inhibited human HCC cell proliferation in a time-dependent manner. FACS revealed that the overexpression of PHB1 arrested HCC cells in the G0/G1 phase of the cell cycle and induced apoptosis. The proportion of cells in the G0/G1 phase was significantly increased and the proportion of cells in the S phase was decreased in HepG2 cells that were transfected with pEGFP-PHB1 compared with untreated control and empty vector-transfected cells. The percentage of apoptotic HepG2 cells that were transfected with pEGFP-PHB1 was 15.41% ± 1.06%, which was significantly greater than that of apoptotic control cells (3.65% ± 0.85%, P < 0.01) and empty vector-transfected cells (4.21% ± 0.52%, P < 0.01). Similar results were obtained with SMMC-7721 cells. Furthermore, the mRNA and protein expression levels of p53, p21, Bax, caspase 3, and caspase 9 were increased while the mRNA and protein expression levels of Cyclin A2, Cy
PHB1 inhibits human HCC cell viability by arresting the cell cycle and inducing cell apoptosis via activation of the p53-mediated mitochondrial pathway.
Core Tip: Prohibitin 1 (PHB1) has been identified as an antiproliferative protein that may play an important role in the progression of hepatocellular carcinoma (HCC). However, the role of PHB1 in HCC is controversial. Here, we investigated the effect of PHB1 on HCC cells and the underlying molecular mechanisms. Our findings demonstrated that PHB1 is a unique tumour suppressor and that its overexpression inhibits human HCC cell viability by arresting the cell cycle and inducing cell apoptosis via activation of the p53-mediated mitochondrial pathway.
- Citation: Shi JJ, Wang YK, Wang MQ, Deng J, Gao N, Li M, Li YP, Zhang X, Jia XL, Liu XT, Dang SS, Wang WJ. Prohibitin 1 inhibits cell proliferation and induces apoptosis via the p53-mediated mitochondrial pathway in vitro. World J Gastrointest Oncol 2024; 16(2): 398-413
- URL: https://www.wjgnet.com/1948-5204/full/v16/i2/398.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i2.398
Prohibitin (PHB) proteins are antiproliferative proteins that are highly conserved and ubiquitously expressed in various eukaryotic cell types[1]. The PHB proteins include PHB1 and PHB2; these proteins are encoded by genes that are located on different chromosomes and are associated with a large eukaryotic mitochondrial complex (PHB complex), which con
The tumour suppressor protein p53 causes a variety of responses, such as cell cycle inhibition and apoptosis induction, when it is activated by numerous stress signals, including DNA damage, hypoxia, and heat shock[10]. P53 mediates cell apoptosis by activating mitochondrial apoptotic pathways that promote the release of cytochrome C (Cyt C) into the cytoplasm; the interaction of Cyt C with apoptosis protease-activating factor (Apaf-1) in the cytoplasm activates caspase signalling pathways that involve, for example, caspase 9, caspase 3, and caspase 7, and ultimately induces cell apoptosis[11]. Cyt C release and Bcl-2 family member activation are particularly essential steps in the mitochondrial pathway. The Bcl-2 family is predominantly composed of antiapoptotic proteins, such as Bcl-2 and Bcl-XL, and proapoptotic proteins, including Bax, Bad, and Bek[11,12]. The mitochondrial pathway is predominantly regulated by the p53 effectors Bcl-2 and Bax; p53 upregulates the expression of Bax and downregulates the expression of Bcl-2, resulting in the induction of apop
Although several functions of PHB1 have been described, little is known about the definitive functions and mecha
The participants included well-documented HCC patients who were randomly enrolled from The Second Affiliated Hospital of Xi’an Jiaotong Uni
The human HCC cell lines HepG2, SMMC-7721, and Huh 7 and the human normal liver cell line L-02 were obtained from the American Type Culture Collection (Manassas, VA, United States). The cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; HyClone, Logan, UT, United States) supplemented with 10% (v/v) foetal bovine serum (HyClone), 100 U/mL penicillin (Sigma-Aldrich, St. Louis, United States), and 100 μg/mL streptomycin sulfate (Sigma-Aldrich) at 37 °C in a 5% (v/v) CO2 humidified atmosphere. The cell culture medium was replaced every other day. When the cells reached 80% confluence, the cells were washed with phosphate-buffered saline (PBS; 137 mmol/L NaCl, 2.7 mmol/L KCl, 4.3 mmol/L Na2HPO4, 1.4 mmol/L KH2PO4, pH 7.4) and then digested using 0.25% (w/v) tryptase (HyClone). The cells were harvested using DMEM followed by centrifugation at 800 rpm for 5 min. Cells in the loga
The coding region of PHB1 was amplified by PCR, digested with Hind III and Kpn I (TAKARA, Japan), and directly cloned and inserted into the pEGFP-N1 plasmid (TAKARA, Japan) to generate the pEGFP-PHB1 plasmid. Briefly, the coding region of PHB1 (819 bp) was amplified using high-fidelity PCR (Thermo Fisher Scientific, United States). The co
The PHB1-specific shRNA plasmid (shRNA-PHB1), its positive control plasmid (shRNA-GAPDH), and its negative control plasmid (shRNA-control) were purchased from Biomics Biotechnologies Co., Ltd. (Jiangsu, China). The sequence of shRNA-PHB1 is 5’-GTAGCAAAGATTTACAGAA-3’, the sequence of shRNA-GAPDH is 5’-GTATGCAACAG
HepG2 and SMMC-7721 cells in the logarithmic growth phase were seeded in 6-well plates at a density of 2 × 105 cells/well in 2 mL of growth medium without antibiotics and incubated in 5% CO2 at 37 °C for 24 h. The cells were then trans
The levels of PHB1 in patient serum were determined using an ELISA kit (CUSABIO, Wuhan, China) according to the manufacturer’s protocols. Serum samples were added to a 96-well plate coated with the capture antibody, and the plate was incubated with 100 μL of horseradish peroxidase (HRP)-conjugated secondary antibody for 60 min at 37 °C. The cells were then washed five times with 400 μL of wash solution. Fifty microlitres of chromogen solution A and 50 μL of chro
The normal tissues (NT) from healthy individuals (n = 30) as well as peritumoral tissues (PT) and tumour tissues (TT) from HCC patients (n = 30) were fixed with 4% (w/v) paraformaldehyde for 15 min, washed five times with PBS, per
Cell viability was determined using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetraazolium bromide (MTT; Sigma-Aldrich) assay as previously described[17]. HepG2 and SMMC-7721 cells were plated in 96-well plates at a density of 6 × 103 cells/well and incubated for 24 h; then, they were transfected with pEGFP-PHB1 or shRNA-PHB1. After 24, 48, or 72 h of transfection, 5 mg/mL MTT solution was added to each well and incubated for 4 h. The culture medium was then replaced with 150 μL of dimethyl sulfoxide. After shaking the culture plates for 5 min, OD was measured using an ND-1000 spectrophotometer at a wavelength of 570 nm. The cell viability percentages were calculated as follows: (OD treated/ODcontrol) × 100%.
HepG2 and SMMC-7721 cells were incubated for 48 h after transfection, washed with ice-cold PBS twice, and fixed in 70% (v/v) ice-cold ethanol overnight at -20 °C. The cells were resuspended in Tris-HCl buffer (10 mmol/L Tris-HCl, pH 7.5) containing 20 mg/L ribonuclease A (RNase A; Sigma-Aldrich) for 15 min, followed by incubation with 50 mg/L propidium iodide (PI; Sigma-Aldrich) for 1 h at 37 °C in the dark. The cell cycle distribution was then determined using a FACscan flow cytometer (CALIBUR, BD, United States), and the data were analysed with ModFit LT 2.0 software (Verity Software House, United States).
Cell apoptosis was analysed using Annexin V-FITC and PI double staining (Sigma-Aldrich) as previously described[18]. Briefly, HepG2 and SMMC-7721 cells were incubated for 48 h after transfection and then harvested and stained with 10 μL of Annexin V-FITC and 5 μL of PI in cold binding buffer (50 mmol/L HEPES, 700 mmol/L NaCl, 12.5 mmol/L CaCl2, pH 7.4) for 15 min at room temperature in the dark. After washing, cell apoptosis was analysed using a FACscan flow cytometer (CALIBUR, BD, United States).
HepG2 and SMMC-7721 cells were incubated for 48 h after transfection and then harvested and resuspended in 400 μL of mitochondrial protein isolation buffer (AMERSCO, Ohio, United States). The cells were homogenized with a polytron ho
HepG2 and SMMC-7721 cells were incubated for 48 h after transfection and then harvested. Total RNA was isolated using an E.Z.N.A.® Total RNA Kit (Omega, Norcross, GA, United States) as previously described[19]. Briefly, 1 μg of each RNA sample was reverse transcribed to synthesize complementary DNA (cDNA) with an RT-PCR kit (TAKARA, Japan). Quantitative real-time PCR was performed using a SYBR® Premix Ex TaqTM II kit (TAKARA, Japan) to analyse the relative expression of PHB1, p53, p21, Cyclin A2, Cyclin E1, CDK2, Cyt C, Bax, Bcl-2, Caspase 3, and Caspase 9 according to the ma
HepG2 and SMMC-7721 cells were incubated for 48 h after transfection and then harvested. The cells were washed with ice-cold PBS, and cell lysates were prepared by lysing the cells with ice-cold RIPA lysis buffer [20 mmol/L Tris, 150 mmol/L NaCl, 1% (v/v) Triton X-100, 1% (w/v) digestive phosphatase inhibitors, 1% (w/v) protease inhibitors, and 1% (w/v) phenylmethyl sulfonylfluoride, pH 7.5 (Sigma-Aldrich)]. The cell lysates were centrifuged at 14000 × g at 4 °C for 15 min, after which the total protein concentrations in the supernatants were determined via the BCA assay (Kangweishiji, Beijing, China) according to the manufacturer’s protocol. Thirty micrograms of protein samples per lane was separated on 12% (w/v) SDS-PAGE gels (Sigma-Aldrich) and electrically transferred to polyvinylidene difluoride membranes (Sigma-Aldrich). The membranes were blocked with 5% (w/v) BSA (Sigma-Aldrich) in Tris-buffered saline (TBS; 0.1 M, pH 7.4) before incubation with primary antibodies overnight at 4 °C. The primary antibodies that were used included antibodies against PHB1 (1:5000 dilution, ab75766; Abcam), p53 (1:1000 dilution, ab31333; Abcam), p21 (1:1000 dilution, ab47452; Abcam), Cyclin A2 (1:1000 dilution, ab181591; Abcam), Cyclin E1 (1:1000 dilution, ab33911; Abcam), CDK2 (1:1000 dilution, ab32147; Abcam), Caspase 3 (1:500 dilution, ab13847; Abcam), Caspase 9 (1:1000 dilution, ab63488; Abcam), Cyt C (1:5000 dilution, ab133504; Abcam), Bax (1:1500 dilution, ab10813; Abcam), and Bcl-2 (1:1000 dilution, ab59348; Abcam). After washing, the membranes were incubated with HRP-conjugated goat anti-rabbit secondary antibodies (Santa Cruz) for 1 h at room temperature with agitation. The proteins were detected using the ECL Western Blotting Kit Reagent (Ther
Representative data are presented as the mean ± SD and were tested for normality and equal variance. The statistical analyses were performed using Student's t tests or one-way ANOVA with Bonferroni post hoc correction with SPSS 15.0 software (SPSS, Inc., United States). All the experiments were repeated at least three times with similar results. Diffe
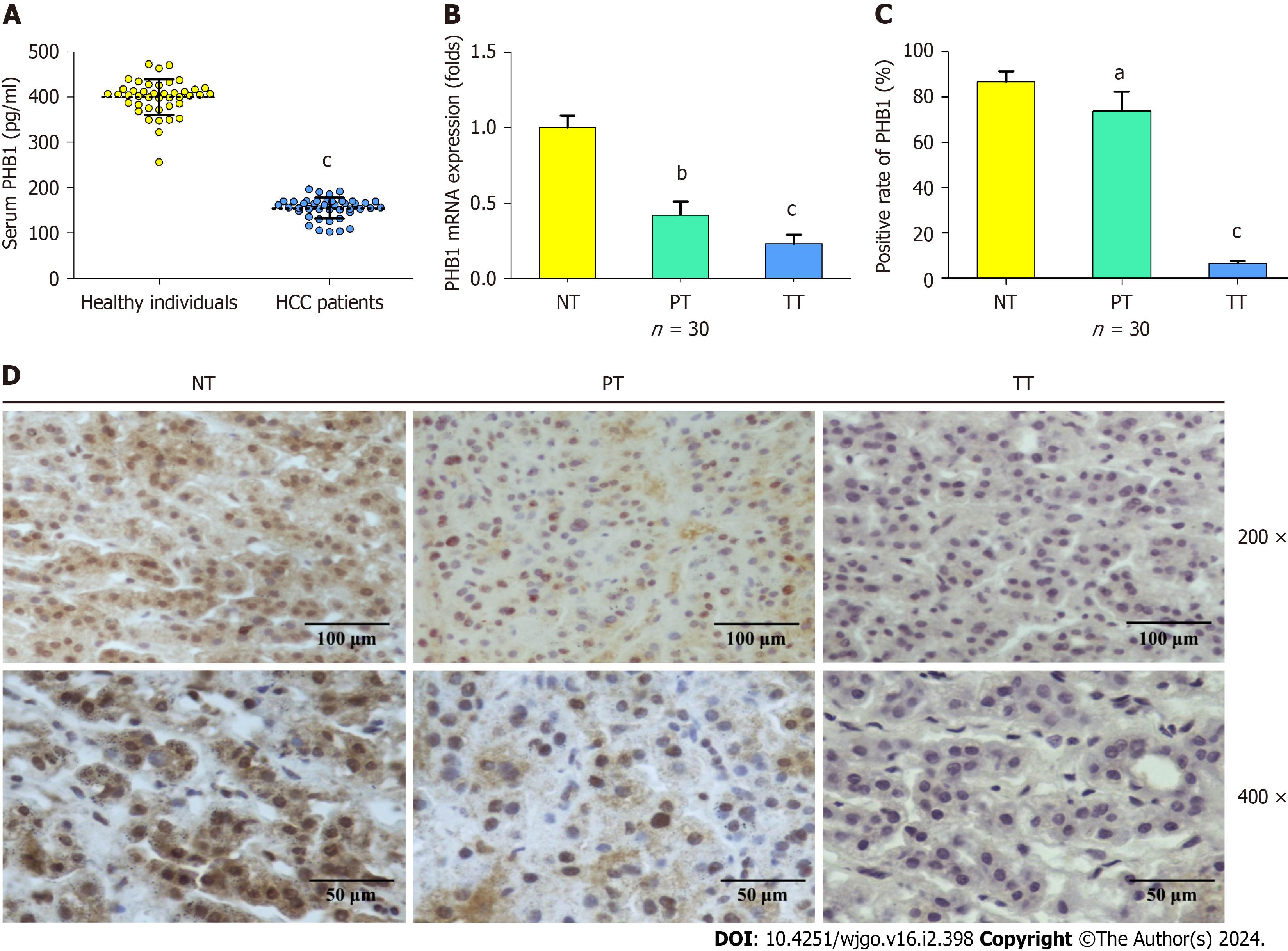
To investigate PHB1 expression in HCC patients, serum and liver tissue samples were harvested from HCC patients. The mean serum PHB1 levels were significantly lower in patients with HCC than in healthy individuals (154.86 ± 23.10 pg/mL vs 399.48 ± 39.1 pg/mL, P < 0.001; Figure 1A). Next, the mRNA expression levels of PHB1 in the liver samples of the HCC patients (n = 30) and healthy individuals (n = 30) were examined using RT-PCR. The PHB1 mRNA expression in the PT and TT of HCC patients was significantly lower than that in the NT of healthy individuals (P < 0.01 or P < 0.001; Figure 1B).
To confirm the RT-PCR findings, we performed immunohistochemistry on TT and NT from the individuals mentioned above (n = 30). PHB1 was located mainly in the cytoplasm and nucleus of HCC cells. The PT and TT of HCC patients exhibited lower levels of PHB1 staining than did the NT of healthy individuals (P < 0.05 or P < 0.001; Figure 1C and D), and lower levels of PHB1 were positively correlated with low differentiation, TNM stage III-IV, and alpha-fetoprotein ≥ 400 μg/L (P < 0.05; Table 1).
| Variable | Total | PHB1 | χ2 value | P value | |
| Negative (%) | Positive (%) | ||||
| Gender | |||||
| Male | 21 | 18 (85.7) | 3 (14.3) | ||
| Female | 9 | 8 (88.9) | 1 (11.1) | 0.055 | 0.815 |
| Age (yr) | |||||
| < 50 | 11 | 9 (81.8) | 2 (18.2) | ||
| ≥ 50 | 19 | 17 (89.5) | 2 (10.5) | 0.353 | 0.552 |
| Tumor size | |||||
| ≤ 3 cm | 18 | 15 (83.3) | 3 (16.7) | ||
| > 3 cm | 12 | 11 (91.7) | 1 (8.3) | 0.433 | 0.511 |
| Tumor differentiation | |||||
| Poor | 23 | 22 (95.7) | 1 (4.3) | ||
| Well & moderate | 7 | 4 (57.1) | 3 (42.9) | 6.887 | 0.009 |
| TMN stage | |||||
| Ⅰ/Ⅱ | 9 | 6 (66.7) | 3 (33.3) | ||
| Ⅲ/Ⅳ | 21 | 20 (95.2) | 1 (4.8) | 4.451 | 0.035 |
| Extrahepatic metastases | |||||
| Yes | 17 | 14 (82.4) | 3 (17.6) | ||
| No | 13 | 12 (92.3) | 1 (7.7) | 0.632 | 0.427 |
| Portal vein tumor thrombus | |||||
| Yes | 16 | 14 (87.5) | 2 (12.5) | ||
| No | 14 | 12 (85.7) | 2 (14.3) | 0.021 | 0.886 |
| AFP (μg/L) | |||||
| < 400 | 8 | 5 (62.5) | 3 (37.5) | ||
| ≥ 400 | 22 | 21 (95.5) | 1 (4.5) | 5.514 | 0.019 |
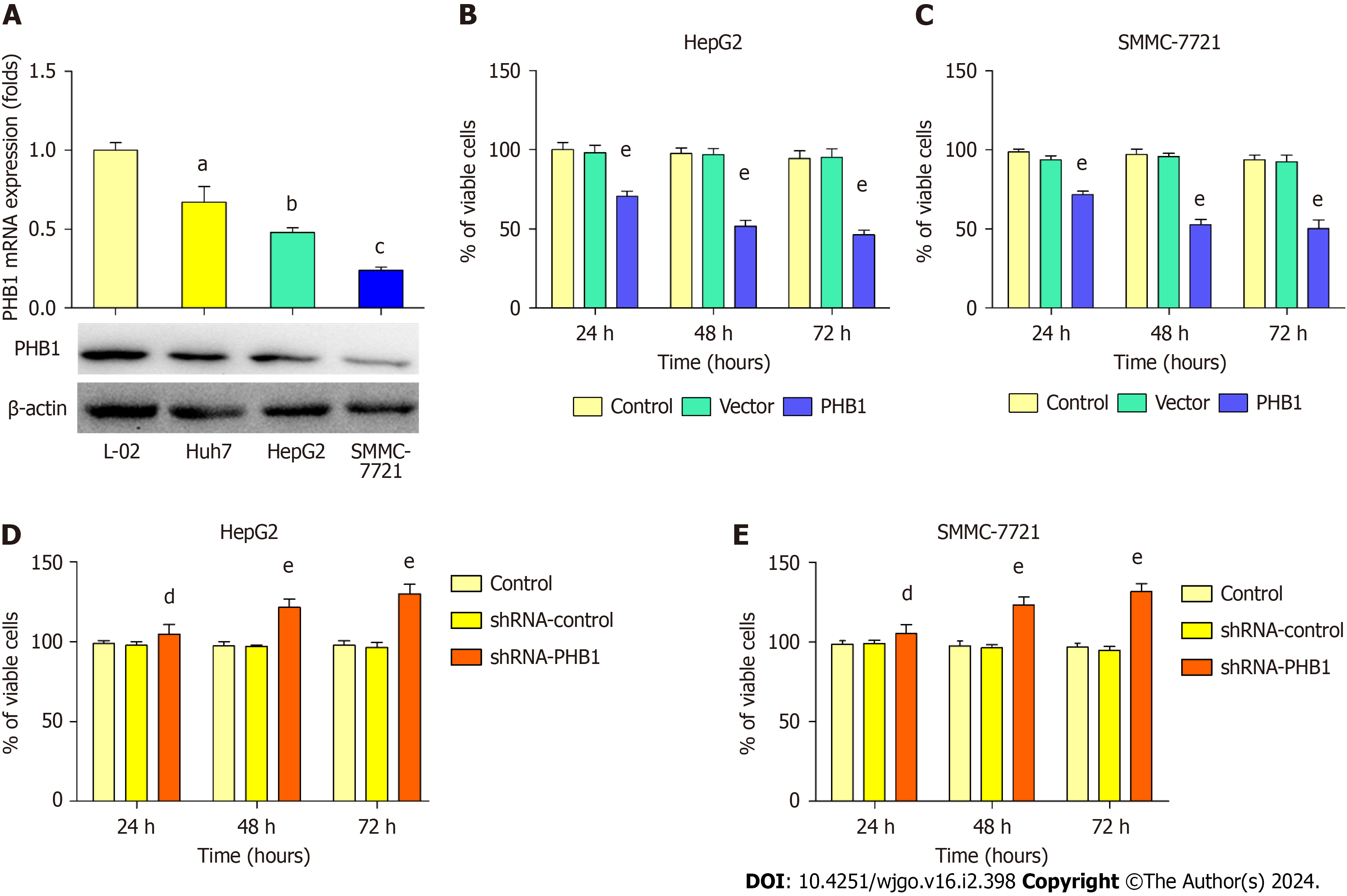
Because low expression levels of PHB1 were observed in the HCC patients, we next examined changes in PHB1 expre
HepG2 and SMMC-7721 cells were successfully transfected with the pEGFP-PHB1 plasmid and PHB1-specific short hairpin RNA (shRNA-PHB1). Compared with that of the control cells and the empty vector (pEGFP-N1)-transfected cells, the viability of the HCC cells was significantly decreased in a time-dependent manner after transfection with pEGFP-PHB1 for 24, 48, or 72 h (P < 0.01; Figure 2B and C). However, the HCC cell viability was significantly increased in a time-dependent manner when PHB1 was knocked down with shRNA-PHB1 (Figure 2D and E).
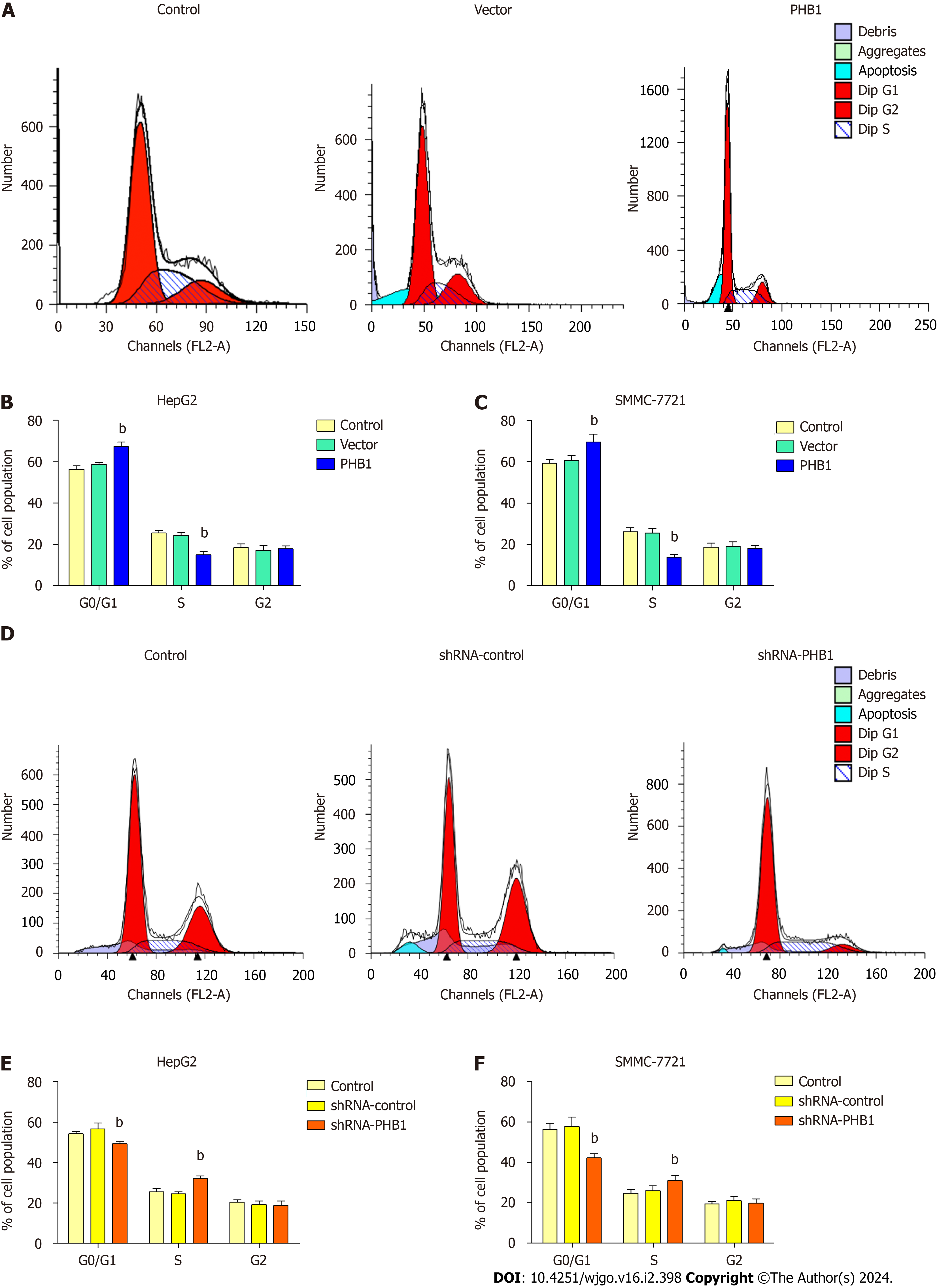
Because the proliferation of HCC cells was inhibited by PHB1 in a time-dependent manner, we investigated whether PHB1 plays a role in the cell cycle (Figure 3). HepG2 cells were transfected with pEGFP-PHB1 for 48 h, and an increase in the proportion of cells in the G0/G1 (67.27% ± 2.13% vs 56.25% ± 1.60% or 58.60% ± 0.91%) and a decrease in the pro
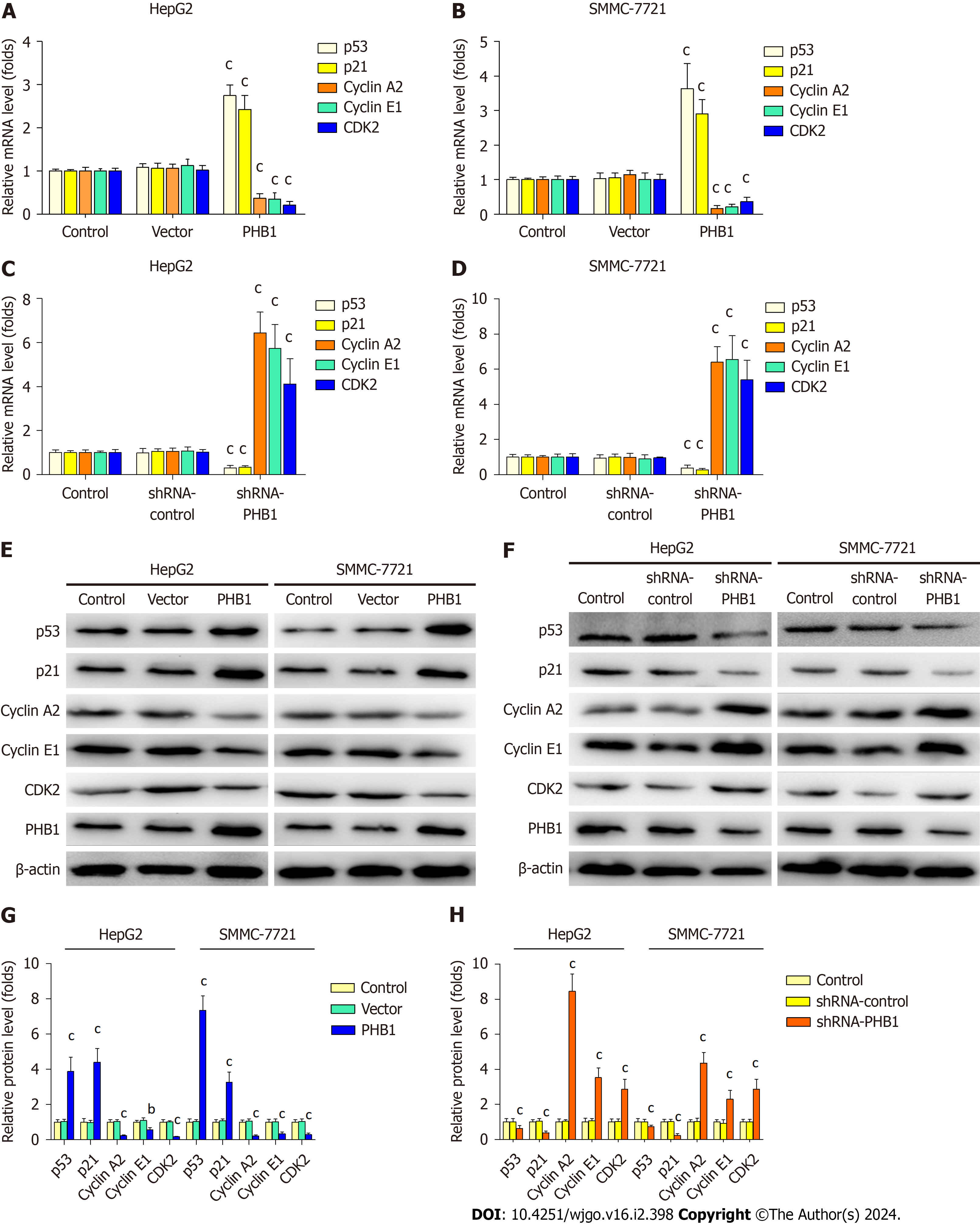
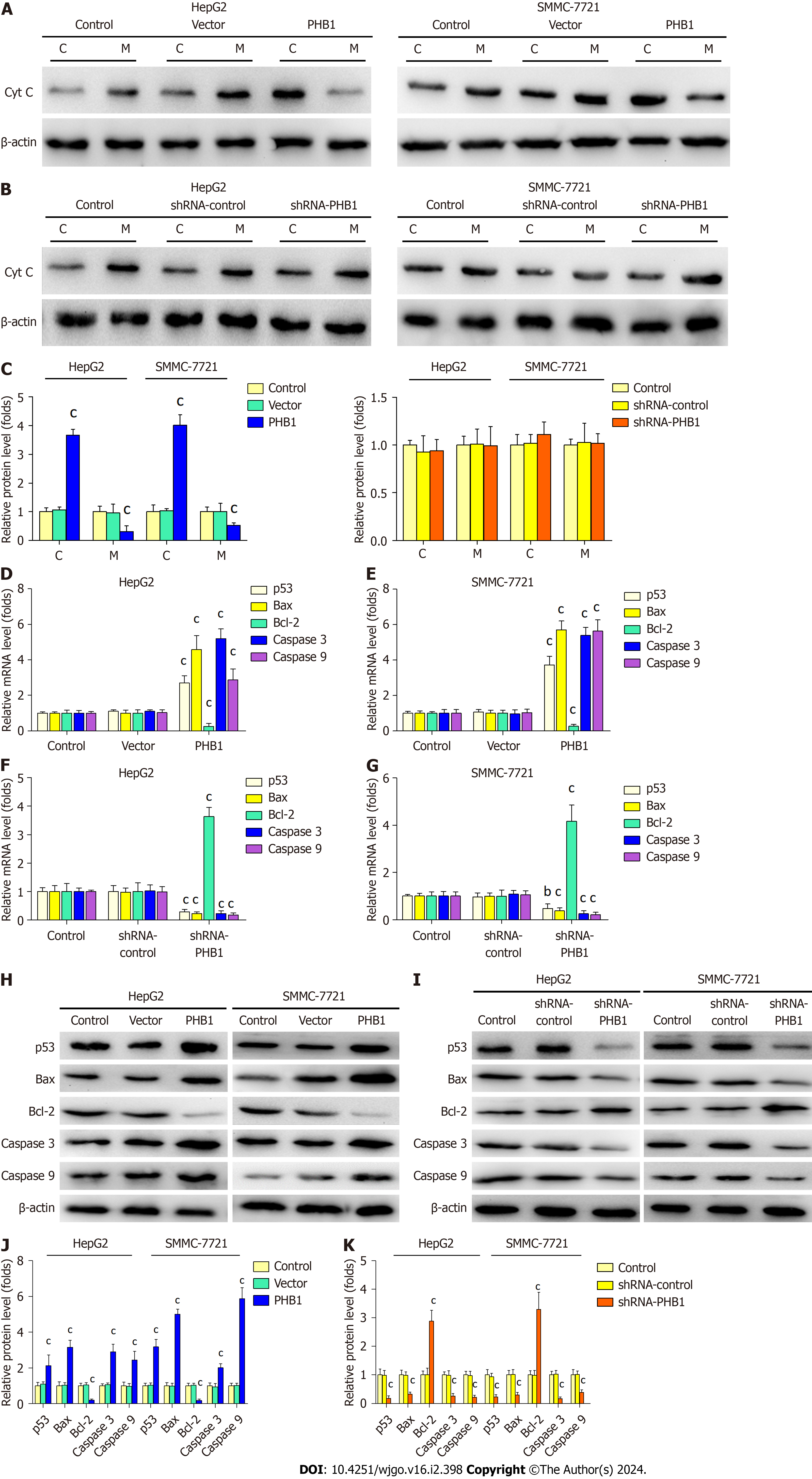
Because the overexpression of PHB1 arrested the cell cycle in the G0/G1 phase, we investigated the expression levels of cell cycle-related molecules in HCC cells (Figure 4). RT-PCR and Western blot analyses revealed that the p21 and p53 mRNA and protein expression levels were significantly increased but the Cyclin A2, Cyclin E1, and CDK2 mRNA and protein expression levels were significantly decreased in PHB1-overexpressing HepG2 (Figure 4A, E, and G) and SMMC-7721 cells (Figure 4B, E, and G). Conversely, the p21 and p53 mRNA and protein expression levels were lower and the Cyclin A2, Cyclin E1, and CDK2 mRNA and protein expression levels were higher in PHB1-knockdown HepG2 (Figure 4C, F, and H) and SMMC-7721 (Figure 4D, F, and H) cells.
Because the proliferation of HCC cells was inhibited by PHB1, we investigated whether PHB1 plays a role in the cell apoptosis (Figure 5). HepG2 cells were transfected with pEGFP-PHB1 for 48 h, and the percentage of apoptotic cells was 15.41% ± 1.06%, which was significantly higher than those of apoptotic untreated control cells (3.65% ± 0.85%, P < 0.01) and empty vector-transfected cells (4.21% ± 0.52%, P < 0.01; Figure 5A and B). However, the percentage of apoptotic cells decreased (6.41% ± 0.36% vs 7.65% ± 0.55% and 8.01% ± 0.72%, P < 0.05) in the PHB1-knockdown HepG2 cells (Figure 5D and E). Similar results were also obtained with SMMC-7721 cells (Figure 5C and F).
To determine the mechanism underlying PHB1-mediated apoptosis, we performed RT-PCR and Western blot to investi
Abnormal expression of proteins, especially the PHB1 protein, may be closely related to the progression of HCC[20]. Several studies have indicated that PHB1 mRNA and protein levels are higher in liver-specific Phb1 KO (Mat1a knockout) mice and in normal human hepatocytes, and liver-specific PHB1 deficiency results in marked liver injury, fibrosis, and HCC[21]. Further research has shown that PHB1 acts as a negative regulator in murine liver and human HCC cells via the WNT-beta-catenin signalling pathway[22]. In MHCC97-H cells, which are highly metastatic liver cancer cells, the overexpression of PHB1 inhibits cell proliferation and increases migration[23]. However, whether PHB1 is overexpressed in HCC patients is still largely unknown. We examined PHB protein levels in the sera and liver tissues of HCC patients and healthy participants and first found that PHB1 protein expression was significantly higher in HCC patients than in healthy individuals, which is consistent with previous results in patients with gastric cancer[24,25]. Because low levels of PHB1 expression were observed in HCC patients, we next observed overexpression of PHB1 in HCC cells, which is consistent with previous results from other cancer cells[14,15,20].
PHB1 plays critical roles in cell survival and apoptosis, but these effects are complex in terms of cell proliferation and apoptosis. Several previous studies have reported that silencing PHB1 induces or accelerates apoptosis in transformed cell lines, which contradicts the conclusions of other studies[26-29]. Interestingly, PHB1 knockdown blocks cell proliferation and induces apoptosis in human hepatoma PLC/PRF/5 cells via the NF-κB signalling pathway[30]. However, PHB1 deficiency results in spontaneous HCC formation in mice, and the knockdown of PHB1 in the mouse normal hepatocyte cell line AML12 increases cyclin D1 and E2F transcription factor expression to accelerate proliferation[21]. Therefore, we investigated the effects of PHB1 knockdown and overexpression in HepG2 and SMMC-7721 cells. In the present study, the overexpression of PHB1 inhibited cell proliferation in a time-dependent manner and altered cell cycle progression, but these effects were reversed when PHB1 was knocked down with shRNA-PHB1 in HepG2 and SMMC-7721 cells.
Cell cycle arrest driven by p53 requires the transcription of GADD45, 14-3-3σ, and p21, which are downstream target genes of p53[31,32]. Specifically, the p21 gene encodes a cyclin-dependent kinase inhibitor, and its transcription is in
Furthermore, we found that PHB1 induced apoptosis in HepG2 and SMMC-7721 cells. To further investigate the mechanism underlying PHB1-induced apoptosis, we measured the expression levels of apoptosis-related molecules in HCC cells. Because PHB1 plays an important role in the stability of mitochondrial proteins, we speculated that PHB1 induced cell apoptosis via the mitochondrial signalling pathway. Our studies indicated that the overexpression of PHB1 increased p53, Bax, caspase 3, and caspase 9 mRNA and protein expression but decreased Bcl-2 mRNA and protein ex
In conclusion, we have showed that PHB1 inhibits the viability of human HCC cells by arresting the cell cycle and inducing cell apoptosis via activation of the p53-mediated mitochondrial pathway. We propose a working model of the role of PHB1 in inducing apoptosis and activating the p53-mediated mitochondrial pathway. In this model, PHB1 induces the expression of p53 in the nucleus and upregulates Bax expression to activate the mitochondrial signalling pathway, which further enhances Cyt C release into the cytosol and activates caspase 9 and caspase 3. Although additional studies are needed to determine the precise mechanisms by which PHB1 regulates cell apoptosis, our findings reveal a formerly undescribed function of PHB1 in the induction of apoptosis in human HCC cells.
Prohibitin 1 (PHB1) has been identified as an antiproliferative protein that is highly conserved and ubiquitously expre
This study aimed to provide a theoretical basis for the use of PHB1 as a possible therapeutic target for liver cancer.
To investigate the effects of PHB1 on cell proliferation, cell cycle progression, and apoptosis in human HCC cells as well as the relevant mechanisms.
ELISA, immunohistochemistry, flow cytometry, quantitative real-time PCR, Western blot, and transfection assays were used in this study.
Low expression of PHB1 was associated with tumour progression in HCC patients. PHB1 inhibited the viability of human HCC cells by arresting the cell cycle. PHB1 induced p53 expression and upregulated Bax expression to activate the mitochondrial signalling pathway, which further enhanced cytochrome C release into the cytosol and activated caspase 9 and caspase 3 to induce apoptosis.
PHB1 inhibits the viability of human HCC cells by arresting the cell cycle and inducing cell apoptosis via activation of the p53-mediated mitochondrial pathway. Our results revealed a novel mechanism by which PHB1 inhibits HCC and showed that the intrinsic mitochondrial pathway is involved in PHB1 overexpression-induced apoptosis in human HCC cells.
Although our findings reveal a formerly undescribed function of PHB1 in the induction of apoptosis in human HCC cells, additional studies are needed to understand the precise mechanisms by which PHB1 regulates cell apoptosis.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single-blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Grassi G, Italy; Wang L, China S-Editor: Lin C L-Editor: Wang TQ P-Editor: Xu ZH
| 1. | Mishra S, Murphy LC, Murphy LJ. The Prohibitins: emerging roles in diverse functions. J Cell Mol Med. 2006;10:353-363. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 207] [Cited by in F6Publishing: 231] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 2. | Nijtmans LG, Artal SM, Grivell LA, Coates PJ. The mitochondrial PHB complex: roles in mitochondrial respiratory complex assembly, ageing and degenerative disease. Cell Mol Life Sci. 2002;59:143-155. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 237] [Cited by in F6Publishing: 228] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 3. | Schulte U, den Brave F, Haupt A, Gupta A, Song J, Müller CS, Engelke J, Mishra S, Mårtensson C, Ellenrieder L, Priesnitz C, Straub SP, Doan KN, Kulawiak B, Bildl W, Rampelt H, Wiedemann N, Pfanner N, Fakler B, Becker T. Mitochondrial complexome reveals quality-control pathways of protein import. Nature. 2023;614:153-159. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 47] [Article Influence: 47.0] [Reference Citation Analysis (0)] |
| 4. | Peng YT, Chen P, Ouyang RY, Song L. Multifaceted role of prohibitin in cell survival and apoptosis. Apoptosis. 2015;20:1135-1149. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 136] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 5. | Yanran W, Jung S, Ko KS. Saturated Fatty Acid-Induced Impairment of Hepatic Lipid Metabolism Is Worsened by Prohibitin 1 Deficiency in Hepatocytes. J Med Food. 2022;25:845-852. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 6. | Signorile A, Sgaramella G, Bellomo F, De Rasmo D. Prohibitins: A Critical Role in Mitochondrial Functions and Implication in Diseases. Cells. 2019;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 124] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 7. | Oyang L, Ouyang L, Yang L, Lin J, Xia L, Tan S, Wu N, Han Y, Yang Y, Li J, Chen X, Tang Y, Su M, Luo X, Xiong W, Zeng Z, Liao Q, Zhou Y. LPLUNC1 reduces glycolysis in nasopharyngeal carcinoma cells through the PHB1-p53/c-Myc axis. Cancer Sci. 2023;114:870-884. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 8. | Wang S, Nath N, Adlam M, Chellappan S. Prohibitin, a potential tumor suppressor, interacts with RB and regulates E2F function. Oncogene. 1999;18:3501-3510. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 175] [Cited by in F6Publishing: 188] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 9. | Najem A, Krayem M, Sabbah S, Pesetti M, Journe F, Awada A, Désaubry L, Ghanem GE. Targeting Prohibitins to Inhibit Melanoma Growth and Overcome Resistance to Targeted Therapies. Cells. 2023;12. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 10. | Bacher S, Schmitz ML. Understanding and Therapeutic Targeting of the p53 Network. Cancers (Basel). 2023;15. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 11. | Tong X, Tang R, Xiao M, Xu J, Wang W, Zhang B, Liu J, Yu X, Shi S. Targeting cell death pathways for cancer therapy: recent developments in necroptosis, pyroptosis, ferroptosis, and cuproptosis research. J Hematol Oncol. 2022;15:174. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 265] [Article Influence: 132.5] [Reference Citation Analysis (0)] |
| 12. | Czabotar PE, Garcia-Saez AJ. Mechanisms of BCL-2 family proteins in mitochondrial apoptosis. Nat Rev Mol Cell Biol. 2023;24:732-748. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 87] [Article Influence: 87.0] [Reference Citation Analysis (0)] |
| 13. | Zeng J, Hills SA, Ozono E, Diffley JFX. Cyclin E-induced replicative stress drives p53-dependent whole-genome duplication. Cell. 2023;186:528-542.e14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 34] [Reference Citation Analysis (0)] |
| 14. | Zhang L, Ji Q, Ni ZH, Sun J. Prohibitin induces apoptosis in BGC823 gastric cancer cells through the mitochondrial pathway. Asian Pac J Cancer Prev. 2012;13:3803-3807. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 15. | Kong B, Han CY, Kim SI, Patten DA, Han Y, Carmona E, Shieh DB, Cheung AC, Mes-Masson AM, Harper ME, Song YS, Tsang BK. Prohibitin 1 interacts with p53 in the regulation of mitochondrial dynamics and chemoresistance in gynecologic cancers. J Ovarian Res. 2022;15:70. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 16. | Liu YH, Peck K, Lin JY. Involvement of prohibitin upregulation in abrin-triggered apoptosis. Evid Based Complement Alternat Med. 2012;2012:605154. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 17. | Shi JJ, Jia XL, Li M, Yang N, Li YP, Zhang X, Gao N, Dang SS. Guggulsterone induces apoptosis of human hepatocellular carcinoma cells through intrinsic mitochondrial pathway. World J Gastroenterol. 2015;21:13277-13287. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 20] [Cited by in F6Publishing: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 18. | Sun MZ, Dang SS, Wang WJ, Jia XL, Zhai S, Zhang X, Li M, Li YP, Xun M. Cytokeratin 8 is increased in hepatitis C virus cells and its ectopic expression induces apoptosis of SMMC7721 cells. World J Gastroenterol. 2013;19:6178-6187. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 4] [Cited by in F6Publishing: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 19. | Dang SS, Sun MZ, Yang E, Xun M, Ma L, Jia ZS, Wang WJ, Jia XL. Prohibitin is overexpressed in Huh-7-HCV and Huh-7.5-HCV cells harboring in vitro transcribed full-length hepatitis C virus RNA. Virol J. 2011;8:424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 20. | Barbier-Torres L, Lu SC. Prohibitin 1 in liver injury and cancer. Exp Biol Med (Maywood). 2020;245:385-394. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 21. | Ko KS, Tomasi ML, Iglesias-Ara A, French BA, French SW, Ramani K, Lozano JJ, Oh P, He L, Stiles BL, Li TW, Yang H, Martínez-Chantar ML, Mato JM, Lu SC. Liver-specific deletion of prohibitin 1 results in spontaneous liver injury, fibrosis, and hepatocellular carcinoma in mice. Hepatology. 2010;52:2096-2108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 102] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 22. | Mavila N, Tang Y, Berlind J, Ramani K, Wang J, Mato JM, Lu SC. Prohibitin 1 Acts As a Negative Regulator of Wingless/Integrated-Beta-Catenin Signaling in Murine Liver and Human Liver Cancer Cells. Hepatol Commun. 2018;2:1583-1600. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 23. | Xu Z, Wu J, Zha X. Up-regulation of prohibitin 1 is involved in the proliferation and migration of liver cancer cells. Sci China Life Sci. 2011;54:121-127. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 24. | Ren H, Du N, Liu G, Hu HT, Tian W, Deng ZP, Shi JS. Analysis of variabilities of serum proteomic spectra in patients with gastric cancer before and after operation. World J Gastroenterol. 2006;12:2789-2792. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 28] [Cited by in F6Publishing: 26] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 25. | Kang X, Zhang L, Sun J, Ni Z, Ma Y, Chen X, Sheng X, Chen T. Prohibitin: a potential biomarker for tissue-based detection of gastric cancer. J Gastroenterol. 2008;43:618-625. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 44] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 26. | Gregory-Bass RC, Olatinwo M, Xu W, Matthews R, Stiles JK, Thomas K, Liu D, Tsang B, Thompson WE. Prohibitin silencing reverses stabilization of mitochondrial integrity and chemoresistance in ovarian cancer cells by increasing their sensitivity to apoptosis. Int J Cancer. 2008;122:1923-1930. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 62] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 27. | Wu Q, Wu S. Lipid rafts association and anti-apoptotic function of prohibitin in ultraviolet B light-irradiated HaCaT keratinocytes. Exp Dermatol. 2012;21:640-642. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 28. | Zhong N, Cui Y, Zhou X, Li T, Han J. Identification of prohibitin 1 as a potential prognostic biomarker in human pancreatic carcinoma using modified aqueous two-phase partition system combined with 2D-MALDI-TOF-TOF-MS/MS. Tumour Biol. 2015;36:1221-1231. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 29. | He P, Liu Y, Qi J, Zhu H, Wang Y, Zhao J, Cheng X, Wang C, Zhang M. Prohibitin promotes apoptosis of promyelocytic leukemia induced by arsenic sulfide. Int J Oncol. 2015;47:2286-2295. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 30. | Sánchez-Quiles V, Santamaría E, Segura V, Sesma L, Prieto J, Corrales FJ. Prohibitin deficiency blocks proliferation and induces apoptosis in human hepatoma cells: molecular mechanisms and functional implications. Proteomics. 2010;10:1609-1620. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 31. | Hassin O, Oren M. Drugging p53 in cancer: one protein, many targets. Nat Rev Drug Discov. 2023;22:127-144. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 222] [Article Influence: 222.0] [Reference Citation Analysis (0)] |
| 32. | Lataster L, Huber HM, Böttcher C, Föller S, Takors R, Radziwill G. Cell Cycle Control by Optogenetically Regulated Cell Cycle Inhibitor Protein p21. Biology (Basel). 2023;12. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 33. | Raut GK, Chakrabarti M, Pamarthy D, Bhadra MP. Glucose starvation-induced oxidative stress causes mitochondrial dysfunction and apoptosis via Prohibitin 1 upregulation in human breast cancer cells. Free Radic Biol Med. 2019;145:428-441. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 34. | Takagi H, Moyama C, Taniguchi K, Ando K, Matsuda R, Ando S, Ii H, Kageyama S, Kawauchi A, Chouha N, Désaubry L, Nakata S. Fluorizoline Blocks the Interaction between Prohibitin-2 and γ-Glutamylcyclotransferase and Induces p21(Waf1/Cip1) Expression in MCF7 Breast Cancer Cells. Mol Pharmacol. 2022;101:78-86. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 35. | Callahan KP, Minhajuddin M, Corbett C, Lagadinou ED, Rossi RM, Grose V, Balys MM, Pan L, Jacob S, Frontier A, Grever MR, Lucas DM, Kinghorn AD, Liesveld JL, Becker MW, Jordan CT. Flavaglines target primitive leukemia cells and enhance anti-leukemia drug activity. Leukemia. 2014;28:1960-1968. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 36. | Meinhardt AL, Munkhbaatar E, Höckendorf U, Dietzen M, Dechant M, Anton M, Jacob A, Steiger K, Weichert W, Brcic L, McGranahan N, Branca C, Kaufmann T, Dengler MA, Jost PJ. The BCL-2 family member BOK promotes KRAS-driven lung cancer progression in a p53-dependent manner. Oncogene. 2022;41:1376-1382. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 1] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 37. | Wei H, Wang H, Wang G, Qu L, Jiang L, Dai S, Chen X, Zhang Y, Chen Z, Li Y, Guo M, Chen Y. Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity. Nat Commun. 2023;14:4300. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 14] [Reference Citation Analysis (0)] |
| 38. | Wang S, Zhou M, Ouyang J, Geng Z, Wang Z. Tetraarsenictetrasulfide and Arsenic Trioxide Exert Synergistic Effects on Induction of Apoptosis and Differentiation in Acute Promyelocytic Leukemia Cells. PLoS One. 2015;10:e0130343. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |