Published online Jul 21, 2016. doi: 10.3748/wjg.v22.i27.6246
Peer-review started: March 23, 2016
First decision: April 14, 2016
Revised: May 19, 2016
Accepted: June 15, 2016
Article in press: June 15, 2016
Published online: July 21, 2016
AIM: To assess the distribution of proteins coded by genes reported as relevant for the molecular classification of hepatocellular carcinoma (HCC).
METHODS: In this retrospective cross-sectional study, the following clinicopathological data were analyzed in 80 autopsied HCC patients: sex, age, ethnicity, alcohol intake, infection with hepatitis B and/or C virus, infection with human immunodeficiency virus, prior treatment, basic and immediate causes of death, liver weight, presence of cirrhosis, number and size of nodules, gross pattern, histological grade and variants, architectural pattern, invasion of large veins, and presence and location of extrahepatic metastases. The protein products of genes known to be involved in molecular pathogenesis of HCC, including epidermal growth factor receptor (EGFR), MET, keratin 19 (K19), vimentin, beta-catenin, mechanistic target of rapamycin (mTOR), extracellular signaling-related kinase (ERK)1, ERK2, Ki67, cyclin D1, caspase 3 and p53, were detected by immunohistochemistry on tissue microarrays. The expression levels were scored and statistically assessed for correlation with HCC parameters.
RESULTS: Infection with hepatitis C virus was identified in 49% of the 80 autopsy patients, cirrhosis in 90%, advanced tumors in 95%, and extrahepatic metastases in 38%. Expression of K19, p53 and ERK1 correlated to high-grade lesions. Expression of ERK1, nuclear beta-catenin, cyclin D1 and ERK2 correlated to higher rates of cell proliferation as determined by Ki67. Expression of MET, EGFR (> 0) and caspase 3 correlated with lower histological grades. Expression of EGFR correlated to that of caspase 3, and overexpression of EGFR (≥ 200/300) was observed in low-grade tumors more frequently (grades 1 and 2: 67% vs grade 3: 27% and grade 4: 30%). Expression of ERK1 was associated with that of K19 and vimentin, whereas expression of ERK2 was associated with that of cyclin D1, MET and membrane beta-catenin. Expression of vimentin was strongly correlated with that of K19.
CONCLUSION: Expression of K19, p53, ERK1, ERK2, vimentin and nuclear beta-catenin was related to higher-grade markers, as opposed to expression/overexpression of EGFR, MET and caspase 3.
Core tip: This study assessed the immunohistochemistry-detected expression of several protein products of genes known to be involved in the molecular pathogenesis of hepatocellular carcinoma (HCC) in a retrospective autopsy cohort of patients with HCC. The data showed that expression profiles of these markers may be related to different pathways underlying HCC progression and metastasis, and that the Edmondson-Steiner’s tumor grade may reflect currently recognized molecular subclasses of HCC. This cross-sectional analysis supports the strategy of translating genomic data into panels of immunohistochemical markers for risk evaluation in HCC and also reinforces the paramount importance of histological grade in this context.
- Citation: Felipe-Silva A, Wakamatsu A, dos Santos Cirqueira C, Alves VAF. Immunohistochemistry panel segregates molecular types of hepatocellular carcinoma in Brazilian autopsy cases. World J Gastroenterol 2016; 22(27): 6246-6256
- URL: https://www.wjgnet.com/1007-9327/full/v22/i27/6246.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i27.6246
Hepatocellular carcinoma (HCC) has a high incidence in East Asia and Africa, where the rate of infection with hepatitis B virus (HBV) and intake of aflatoxin are more prevalent. Recent years have seen increasing rates of HCC incidence and mortality in western countries, including Brazil, where cirrhosis-related infection with the hepatitis C virus (HCV) predominates[1,2].
The recent revival of interest in autopsy studies for advanced neoplasms derives from the unique opportunity a corpse presents to assess morphological and molecular aspects of the progression and dissemination patterns of both primary and metastatic tumors[3-5].
Gene expression studies have proven useful for grouping HCC according to molecular profiles[6-8]. A meta-analysis of genetic studies by Hoshida et al[9] defined at least two groups of HCC. Subsequent analysis characterized more aggressive types of HCC (Hoshida’s S1/S2 subclasses) by the expression of keratin 19 (K19), p53 mutation, and/or regulation by the MET receptor[10]; moreover, this high-proliferation HCC group also appeared to be related to a stem-cell phenotype. A less aggressive type of HCC (Hoshida’s S3 subclass) retains the hepatocyte-like phenotype and includes the molecular categories featuring chromosome 7 polysomy [wherein the epidermal growth factor receptor (EGFR) gene and MET oncogene are located] and CTNNB1-mediated activation of the Wnt pathway[11,12]. Two other molecular categories feature activation of the interferon pathway and amplification of the VEGFA gene[13].
In the present study, we assessed the expression of protein products of genes that have been reported as relevant for the molecular classification of HCC using an autopsy cohort of patients with HCC.
Among the total 5836 medical autopsies performed in the Pathology Department of the University of Sao Paulo School of Medicine Hospital between the years of 2003 and 2009, 188 presented primary liver tumors. Excluding tumors that were determined to be cholangiocarcinomas (n = 65), combined hepatocellular-cholangiocarcinoma (n = 1), epithelioid hemangioendothelioma (n = 1) and other malignant non-HCC neoplasms and poorly preserved specimens (n = 13), 108 cases were confirmed as HCC. These cases, and all related data, were recorded consecutively in the paper files and electronic database of the Hospital das Clínicas of the University of Sao Paulo School Of Medicine, Brazil’s largest academic hospital, and were accessed for study in accordance with the investigative protocols of the hospital’s Ethics Committee for Research Project Analysis (CAPPesq). Archived pathological slides were reviewed for each case, and sufficient preserved tumor tissue and clinicopathological data was available for 80 of the 108 HCC cases reviewed.
All corpses had been preserved by routine refrigeration. Standard immunostaining practices were used for detection of vimentin expression. Normal (non-cancerous) tissues were also obtained from the HCC patients for use as internal controls and examined to provide evidence of adequate tissue preservation prior to continuing with further immunohistochemistry procedures. Detailed pathological data were recorded. Clinical and demographic data were retrieved from medical records and autopsy reports. The primary objective of the histological review was to define the major architectural patterns and histological grading (1-4) according to the system set forth by Edmondson and Steiner[14]. Tumors showing heterogeneous histological grades were classified as the highest grade shown[15]. HCC nodules < 2 cm were considered incidental. Presence of multiple intrahepatic nodules (≥ 4)-regardless of either massive or diffuse type-prompted analysis of primary HCC and intrahepatic metastases as intrahepatic tumors. Paraffin-embedded tissue blocks of primary HCC, extrahepatic metastases and non-neoplastic liver were selected for use in construction of tissue microarray (TMA) and immunohistochemistry (IHC). Liver fibrosis was classified using a standard 0-4 system, wherein F0 indicated no fibrosis, F1 indicated portal fibrosis without septa, F2 indicated portal fibrosis with few septa, F3 indicated numerous septa without cirrhosis, and F4 indicated cirrhosis.
TMAs are produced by extracting the tissue cores from many paraffin-donor blocks and then re-embedding into a single recipient block at defined array coordinates. For this study, two or three 1.0 mm cores of each original sampled tissue-primary HCC, extrahepatic metastases, and non-neoplastic liver-were selected for TMA. Cores of the primary HCC and extrahepatic metastases samples were used to construct two (duplicate) TMAs each, and the cores of the non-neoplastic samples were used to construct one TMA. For heterogeneous tumors, different areas were cored separately. When more than one primary HCC was present in a single case, all tumors were sampled, but data were computed for the largest one. Available non-neoplastic liver samples were selected as far from the tumor border as possible, usually from a different paraffin block. All available extrahepatic metastases and large vein invasion samples were cored.
Table 1 summarizes the protocols of the IHC procedures and respective antibodies used in this study.
| Antibody | Manufacturer | Clone | Species | Dilution | Retrieval1 | Positive control | Staining pattern |
| EGFR PharmDx | Dako | 2-18C9 | Mouse | Pre-diluted | Proteinase K | Provided by manufacturer | Membrane |
| Ki67 Ag | Dako | MIB-1 | Mouse | 1:400 | Standard | Tonsil | Nucleus |
| Caspase 3 | DBS | 3C SP03 | Mouse | 1:400 | Standard | Tonsil/stomach | Cytoplasm |
| ERK1 | DBS | Polyclonal | Rabbit | 1:100 | Standard | Breast carcinoma | Cytoplasm |
| ERK2 | DBS | Polyclonal | Rabbit | 1:400 | Standard | Breast carcinoma | Cytoplasm |
| MET | Cell signaling | Polyclonal | Rabbit | 1:50 | Standard | Breast carcinoma | Cytoplasm |
| mTOR | Cell signaling | Polyclonal | Rabbit | 1:50 | Standard | Lymphoid tissue | Cytoplasm |
| K19 | Novocastra | b170 | Mouse | 1:300 | Standard | Cholangio-carcinoma | Cytoplasm |
| Vimentin | Dako | Vim3B4 | Mouse | 1:3000 | Standard | Kidney | Cytoplasm |
| p53 | Dako | DO7 | Mouse | 1:100 | Standard | Breast carcinoma | Nucleus |
| Beta-catenin | BD | 14Beta-catenin | Mouse | 1:800 | Standard | Tonsil | Membrane/nucleus |
| Cyclin D1 | Dako | SP4 | Rabbit | 1:100 | Steamer 10 mmol/L TRIS, 1 mmol/L EDTA, pH 9 | Tonsil/intestine | Nucleus |
In brief, the slide-mounted sections of the paraffin-embedded tissues were deparaffinized and rehydrated. Antigen retrieval consisted of submerging the slides in 10 mM citrate buffer (pH 6) and steam heating for 40 min. After washing with distilled water, blocking of endogenous peroxidase was carried out by incubating with 6% hydrogen peroxide solution in methanol at room temperature for 10 min and repeating twice. Universal protein blocking was carried out by incubating CASBlock™ solution (Invitrogen, United States) at 37 °C for 10 min. Antigen detection with the primary antibody was carried out by incubating first at 37 °C for 30 min and then at 4 °C for 18 h. With the exception of EGFR PharmDx, all signal amplifications were achieved by application of the Novolink™ polymer system (Novocastra, United States) with incubation at 37 °C for 30 min. The immunoreactive signal was visualized by first incubating with chromogen 3-3′-diaminobenzidine (60 mg/dL in a phosphate buffer pH 7.4) at 37 °C for 5 min, followed by washes with distilled water, counterstaining with Harris’ hematoxylin by incubation at room temperature for 1 min, dehydration in a progressive alcohol series, clearing in xylene, and mounting with Entellan™ (Merck, United States).
Assessment of EGFR was carried out by applying the EGFR PharmDx Kit (Dako, United States) according to the manufacturer’s instructions. EGFR membrane expression was classified according to staining intensity, with scoring from 0 to 3+, and according to distribution among the HCC cells (0% to 100%). A score from 0 (no staining) to 300 (100% strong staining) was assigned to each spot by two independent pathologists. For spots where the two pathologists gave differing scores, a consensus score was reached with the two working in tandem on a two-observing microscope. For each case, a final score was calculated as the arithmetical average of all spots for that sample. The score for metastatic disease was calculated as the arithmetical average of the available metastases. Cases or samples were classified as EGFR-expressing when the score was > 0 and subclassified as “EGFR-overexpressing” when the final score was ≥ 200.
Membrane beta-catenin, mechanistic target of rapamycin (mTOR), MET, caspase 3, vimentin, extracellular signaling-related kinase (ERK)1 and ERK2 were analyzed using a scoring system similar to that described for EGFR, with both intensity and distribution of immunostaining being considered to obtain a score between 0 and 300. The cut-off values for expression and overexpression were set at 100 (of 300) and 200 (of 300), respectively, for membrane beta-catenin, mTOR, MET, ERK1 and ERK2. Any expression of vimentin in tumor cells was considered abnormal, with the cut-off being 0 (of 300). For caspase 3, an expression score < 10 (of 300) was considered to indicate a “loss of expression”.
Nuclear staining of Ki67, cyclin D1 and p53 was calculated for each spot by cell counting, with the result expressed as a percentage of the labeled cells. Nuclear K19 expression was estimated at 10% intervals of positive cells. Nuclear beta-catenin was semi-quantitatively scored at 0 (no staining), 1+ (weak or focal staining), 2+ (moderate staining) and 3+ (diffuse strong staining).
SPSS statistical software, version 15.0 (SPSS Inc., United States) was used for all statistical analyses. All numeric variables were tested by the Kolmogorov-Smirnov goodness-of-fit test to assess normal distribution. Frequency distributions of the clinicopathological data and the immunohistochemical categories were assessed using Fisher’s exact or χ2 tests, with the threshold of significance set at 0.05. If any difference was detected in a set of three variables, pairwise tests were used to detect outliers. Spearman’s correlation coefficient was used to assess correlations among scores and other categorical variables.
The clinicopathological data and organ distribution of metastases are summarized in Tables 2 and 3. Patients with cirrhosis (90.0%) accompanied by clinically-relevant non-incidental HCC predominated in this autopsy series (95.0%). HCV infection was a major factor in 39 patients (48.7%), followed by significant alcohol intake (30.0%) and HBV infection (18.7%). A history of chronic alcohol intake was present for male patients exclusively (42.9%, P < 0.0001).
| Feature | n (%) |
| Sex | |
| Male | 62 (77.5) |
| Female | 18 (22.5) |
| Age, yr | |
| Median | 59.5 |
| Range | 28-82 |
| With cirrhotic liver | 72 (90.0) |
| With incidental small HCC, < 2.0 cm | 4 (5.0) |
| Etiology | |
| HCV only | 27 (33.7) |
| HCV + alcohol | 9 (11.2) |
| HCV + HBV | 2 (2.5) |
| HCV + HBV + alcohol | 1 (1.3) |
| Alcohol only | 11 (13.8) |
| HBV only | 9 (11.2) |
| HBV + alcohol | 3 (3.8) |
| Hemochromatosis | 2 (2.5) |
| Cryptogenic | 10 (12.5) |
| Data not available | 6 (7.5) |
| Larger tumor size, cm | |
| Median | 4 |
| Range | 0.8-18.0 |
| Non-available/non-sizable cases | 22 (27.5) |
| Number of tumor nodules | |
| 1 | 24 (33.3) |
| 2 | 4 (5.6) |
| 3 | 1 (1.4) |
| ≥ 4 | 43 (53.8) |
| Non-available/uncountable cases | 8 (10.0) |
| Gross pattern | |
| Nodular | 31 (38.8) |
| Massive with satellite lesions | 25 (31.3) |
| Diffuse | 8 (10.0) |
| Non-available/non-sizable cases | 11 (13.8) |
| Histological grade, Edmondson-Steiner grade | |
| 1 + 2 | 22 (27.5) |
| 3 | 47 (58.8) |
| 4 | 11 (13.8) |
| Predominant histological pattern or variant | |
| Trabecular | 36 (45.0) |
| Acinar/pseudoacinar | 9 (11.2) |
| Solid/macrotrabecular | 24 (30.0) |
| Mixed | 9 (11.2) |
| Clear cell | 2 (2.5) |
| Metastasis feature | n (%) |
| Large vein invasion | 12 (15.0) |
| Extrahepatic metastases | 30 (37.5) |
| Extrahepatic metastases sites, cases by site | |
| Lungs | 21 (26.3) |
| Lymph node | 6 (7.5) |
| Adrenal, bone, spleen, diaphragm, peritoneum | 2 (2.5) |
| Small bowel, bladder, colon, pancreas, pituitaries, thyroid, pleura | 1 (1.3) |
The majority of females with HCC had HCV infection (72.2%), compared to less than one-half of the males (41.9%, P < 0.05). Females also had a smaller average tumor size (3.2 ± 2.0 cm) than the males (6.0 ± 4.4 cm, P < 0.01).
HCC was detected in 8 patients with non-cirrhotic livers (10%), consisting of 4 without fibrosis (F0), 2 with mild fibrosis (F1), 1 with bridging fibrosis (F2), and 1 classified as non-cirrhotic not otherwise specified.
Tumor sizes of ≥ 6.0 cm were found in 80% of the non-cirrhotic livers and in 30.2% of cirrhotic livers (P < 0.05). Non-cirrhotic cases also had a higher proportion of unknown risk factors for HCC (62.5%) than the cirrhotic cases (19.4%, P = 0.02).
Eleven of the HCC cases (14%) had undergone chemoembolization treatment prior to death, including 1 patient who received chemoembolization plus ethanol injection, 1 who received systemic chemotherapy in addition, and 1 who died of recurrent HCC after subsequent liver transplantation. Most of the HCC cases in our autopsy cohort (82%) had not received specific oncological treatment, mainly due to the acute presentation of advanced disease.
Four cases of incidental small HCC (5.0%) were detected in the cirrhotic livers, comprised of 1 patient with HCV, 1 with HBV, 1 with HBV + alcohol, and 1 of unknown cause; each cases had one or two nodules of size ranging between 0.8 cm and 1.5 cm.
Multiple intrahepatic tumors were detected in 31.6% (6/19) of cases with grade 2 HCC and in 69.8% (37/53) of the cases with grade 3 or 4 when combined (P < 0.01). Accordingly, the proportions of HCC cases classified as nodular, massive and diffuse among the grades 3 and 4 combined group were 67.7%, 88% and 75% respectively.
A trabecular pattern was predominant among the cases in the combined-group of patients with tumors of grades 1 and 2 (61.9%) and in patients with grade 3 tumors (47.8%). A solid pattern was seen in 90.9% of the group of patients with grade 4 HCC.
Extrahepatic metastases were detected in 53.5% of cases of multiple intrahepatic tumors, and in 10.3% of cases with one to three intrahepatic tumors (P < 0.001). Consistently, cases with extrahepatic metastases had increased liver weight (2388.3 ± 842.1 g) as compared to cases without extrahepatic metastases (1501.3 ± 625.5 g, P < 0.01).
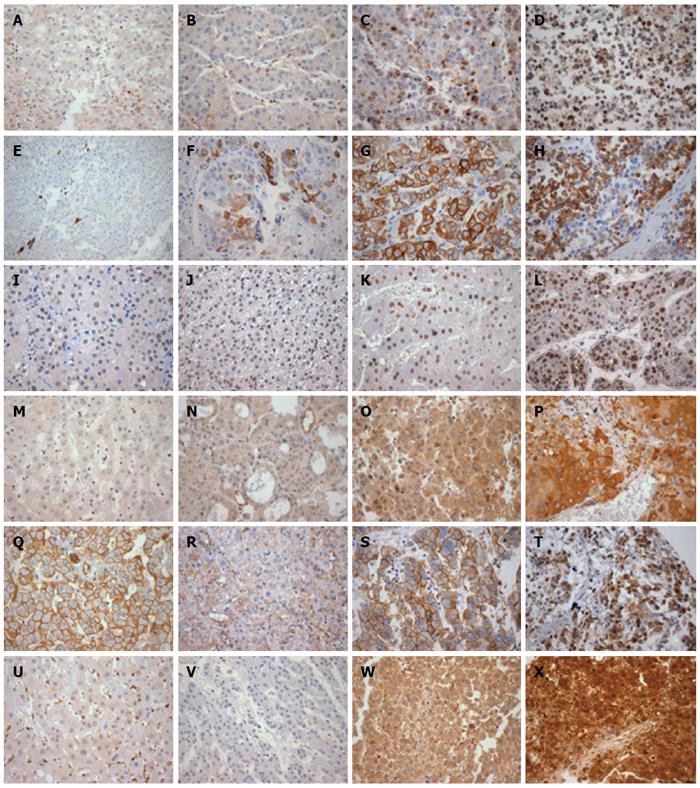
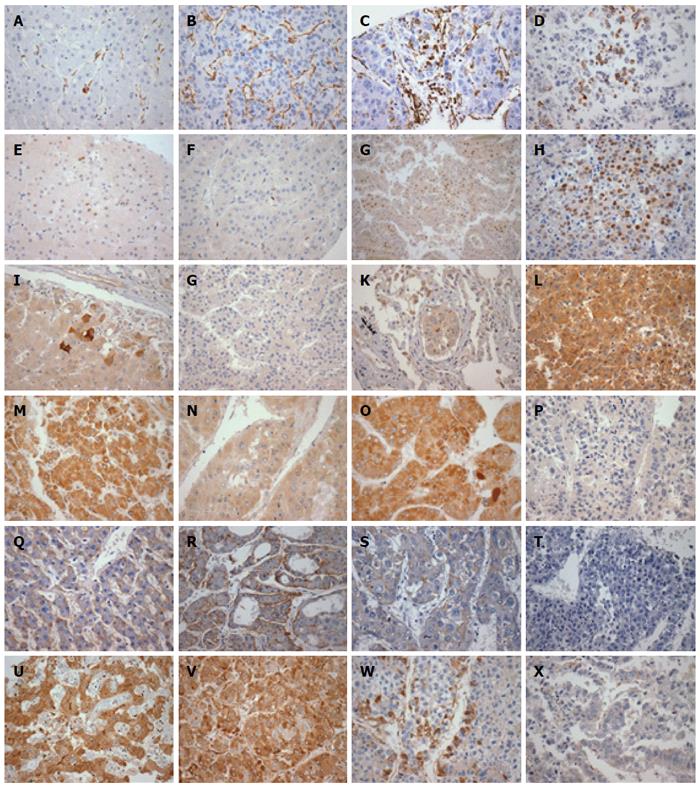
The expression patterns and cut-off points used for the different markers in primary HCC, metastases and non-neoplastic liver are summarized in Table 4 and shown in Figures 1 and 2. The discrepancy in numbers of cases presented in each table (Tables 4, 5 and 6) reflects the loss of some spots during TMA processing.
| IHC marker | Cut-off | Non-neoplastic | Primary HCC | Metastases | P value |
| 26 cases | 75 cases | 17 cases | |||
| EGFR overexpression | ≥ 200/3001 | 17 (65) | 29 (39) | 6 (38)2 | 0.054 |
| Ki67 | ≥ 0.1% | 2 (8)a | 45 (63)4 | 13 (76) | < 0.001a |
| K19 | > 1% | 0 (0)a | 12 (16)3 | 5 (29) | 0.011a |
| Vimentin | > 0/300 | 0 (0) | 4 (6)3 | 3 (18) | 0.064 |
| Caspase 3 loss | ≤ 10/300 | 2 (8)a | 26 (36)3 | 7 (41) | 0.009a |
| Cyclin D1 | > 0.1% | 4 (15) | 16 (23)5 | 9 (53)a | 0.018a |
| mTOR expression | ≥ 100/3001 | 8 (31) | 14 (21)5 | 5 (31)2 | 0.533 |
| MET expression | ≥ 100/3001 | 10 (38) | 12 (17)4 | 5 (29) | 0.068 |
| ERK1 overexpression | ≥ 200/3001 | 0 (0)a | 10 (14)4 | 5 (33)3 | 0.006a |
| ERK2 overexpression | ≥ 200/3001 | 0 (0)a | 10 (14)4 | 4 (24) | 0.029a |
| p53 | ≥ 10% | 2 (8)a | 28 (42)5 | 8 (50)2 | 0.002a |
| Beta-catenin, membrane | ≥ 100/3001 | 10 (38) | 42 (61)5 | 11 (65) | 0.113 |
| Beta-catenin, nucleus | 2 or 3+ | 0 (0) | 9 (9)5 | 3 (18) | 0.070 |
| IHC marker | Cut-off | Cases/valid samples | P value | ||
| Grades 1 + 2 | Grade 3 | Grade 4 | |||
| EGFR overexpression | ≥ 200/3001 | 14 (67)a | 12 (27) | 3 (30) | 0.008a |
| Ki67 | ≥ 0.1% | 6 (30)a | 31 (74) | 8 (80) | 0.002a |
| K19 | > 1% | 0 (0) | 7 (17) | 5 (45)a | 0.005a |
| Vimentin | > 0/300 | 0 (0) | 3 (7) | 1 (9) | 0.465 |
| Caspase 3 loss | ≤ 10/300 | 3 (15) | 16 (38) | 8 (73)a | 0.006a |
| Cyclin D1 | > 0.1% | 4 (20) | 9 (21) | 3 (30) | 0.847 |
| mTOR expression | ≥ 100/3001 | 7 (37) | 17 (45) | 1 (10) | 0.134 |
| MET expression | ≥ 100/3001 | 8 (42)a | 6 (14) | 0 (0) | 0.012a |
| ERK1 overexpression | ≥ 200/3001 | 0 (0) | 8 (20) | 2 (18) | 0.081 |
| ERK2 overexpression | ≥ 200/3001 | 1 (5) | 6 (14) | 3 (30) | 0.148 |
| p53 | ≥ 10% | 5 (28) | 16 (42) | 7 (64) | 0.164 |
| Beta-catenin, membrane | ≥ 100/3001 | 13 (65) | 25 (64) | 4 (40) | 0.343 |
| Beta-catenin, nucleus | 2 or 3+ | 0 (0) | 1 (3) | 1 (10) | 0.352 |
| Grade | Ki67 | K19 | p53 | ERK1 | BcatN | ERK2 | Vim | CKD1 | BcatM | mTOR | MET | EGFR | Group No. | |
| Ki67 | 0.43a | Group 1 | ||||||||||||
| K19 | 0.37b | 0.03 | ||||||||||||
| p53 | 0.36c | 0.29d | 0.08 | |||||||||||
| ERK1 | 0.34c | 0.39b | 0.37b | 0.17 | ||||||||||
| BcatN | 0.23 | 0.44a | 0.13 | 0.24 | 0.18 | |||||||||
| ERK2 | 0.19 | 0.63a | 0.22 | -0.10 | 0.36c | 0.24 | ||||||||
| Vim | 0.14 | 0.11 | 0.51a | 0.02 | 0.31c | 0.08 | 0.14 | |||||||
| CKD1 | 0.03 | 0.46a | -0.32c | -0.10 | 0.03 | 0.26d | 0.29d | -0.19 | Group 2 | |||||
| BcatM | -0.08 | 0.26d | -0.01 | -0.23 | 0.17 | -0.07 | 0.42a | 0.01 | 0.25d | |||||
| mTOR | -0.24d | -0.02 | -0.20 | -0.21 | 0.25d | -0.03 | 0.13 | -0.13 | 0.17 | 0.44a | ||||
| MET | -0.32c | 0.16 | -0.20 | -0.35c | -0.01 | 0.22 | 0.39b | -0.18 | 0.38b | 0.18 | 0.40b | |||
| EGFR | -0.34c | 0.00 | -0.14 | -0.13 | -0.04 | -0.24 | 0.22 | -0.07 | 0.13 | 0.20 | 0.11 | 0.23 | Group 3 | |
| Casp3 | -0.42a | -0.18 | -0.25d | -0.46a | -0.19 | -0.06 | 0.06 | -0.27d | 0.19 | 0.16 | 0.32b | 0.41a | 0.35c |
The non-neoplastic liver tissues, mostly representative of cirrhosis, showed higher rates of EGFR overexpression, higher caspase 3 expression (P = 0.009), lower p53 expression (P = 0.002) and lower rates of Ki67-evidenced cell proliferation (P < 0.001) than the primary HCC samples. K19 expression was below the threshold of detection for all non-neoplastic hepatocytes examined. High-level expression of ERK1 (P = 0.006) and ERK2 (P = 0.029) was only detected in tumor samples, and predominantly in metastases samples. Cyclin D1 was more frequently expressed in metastases samples than in other samples (P = 0.018). There was a trend towards increased expression of vimentin and nuclear beta-catenin in tumor samples and metastases when compared to non-neoplastic liver samples. The expression of mTOR, MET, ERK1 and ERK2 was higher in metastases samples than in intrahepatic liver tumor samples, although the difference between the two did not reach statistical significance.
Table 5 summarizes the relation of histological grade and expression of the immunohistochemical markers of HCC. While EGFR overexpression and MET expression were more common in the combined-group of patients with grades 1 and 2 HCC, K19 expression and loss of caspase 3 were more frequent in the group with grade 4 HCC. The combined-group of patients with grades 1 and 2 HCC showed a trend towards very low Ki67 index (< 0.1%).
Table 6 presents the association coefficients for the scores or values of the immunohistochemical markers of HCC and the grade of primary HCC. Although the association coefficients were mostly moderate or weak, some clear segregation existed between markers that were positively associated with the histological grade and cell proliferation (such as p53, K19, and nuclear beta-catenin-in group 1) vs markers with the opposite profile (e.g., MET, EGFR and caspase 3-in group 3). Vimentin was significantly associated with K19. In group 2, the immunohistochemical markers of mTOR, membrane beta-catenin and cyclin D1 showed intermediate characteristics, and were less associated with histological grade, but still showed association with Ki67-evidenced cell proliferation. ERK1 and ERK2 were both associated with Ki67-evidenced cell proliferation and with each other’s expression. However, ERK1 was associated with K19 and vimentin, while ERK2 was associated with cyclin D1, MET and membrane beta-catenin.
A few recent studies have sought to translate genomic data into panels of immunohistochemical markers, aiming to generate risk models for survival and prognostic evaluation of HCC[16,17]. Although immunohistochemical studies do not always directly reflect the underlying molecular mechanisms of a given tumor, this cross-sectional analysis in a large post-mortem series supports the feasibility of such a strategy and reinforces the paramount importance of the histological grade.
Overexpression of EGFR has been reported in HCC and the surrounding chronically inflamed tissue. In our autopsy cohort, EGFR overexpression was found to be related to more differentiated HCC. Previous studies have presented conflicting results on this topic, either reporting detection of higher EGFR expression in high-grade tumors[18,19] or demonstrating a complete absence of any association[20,21]. Our findings are similar to those described by Morimitsu et al[22], who showed a progressive loss of EGFR expression in less differentiated HCC in surgical specimens.
Our study also found that expression of Ki67, cyclin D1 and caspase 3 indicate higher proliferative activity and lower rates of apoptosis in more aggressive tumor populations, especially in metastases. However, these findings may not accurately reflect the histological grade since the metastases samples examined showed morphological similarities to the intrahepatic tumor samples examined. Similarly, lower expression of caspase 3 has been previously reported as present in less differentiated prostate adenocarcinomas[23]. Conversely, Persad et al[24] showed increased expression of caspase 3 in 52% of resected HCC samples, a distinguishing feature from the surrounding non-HCC tissue. The results from our study, presented herein, probably reflect a larger sample of poorly differentiated HCC with a higher proportion of caspase 3 loss.
The higher rates of K19 and vimentin expression we observed in metastatic HCC suggest a more aggressive behavior of K19-positive HCC; this finding could serve to reinforce previous evidence that K19 expression is a “progenitor cell feature”, while vimentin expression could denote epithelial-mesenchymal transition[25-27].
A subpopulation of HCC has been reported as presenting constitutively increased expression of cyclin D1. Activation of the cyclin D1 promoter has also been characterized as one of the prime targets of the beta-catenin pathway. Joo et al[26] linked the overexpression of cyclin D1 to well-differentiated HCC that shows a low proliferation index as evidenced by detection of Ki67. The authors, however, did not identify an association between the expression of cyclin D1 and p53, as we observed in the current study. Instead, our results are similar to those described by Schmitt-Graeff et al[28], in which an association was found to exist between the expression of cyclin D1 and cell proliferation, but not with histological grade. Unlike that finding and those reported by Prange et al[29], we identified a weak correlation between the expression of cyclin D1 and the nuclear or membrane expression of beta-catenin. Our finding is in accordance with another study that identified an association between aberrant nuclear expression of beta-catenin and the Ki67 index[30]. Dysregulation of the Wnt pathway may be more relevant in a type of HCC with a higher rate of cell proliferation, but which is distinct from tumors with stemness features.
Our study also showed that expression of ERK1 associated with histological grade, as well as with the expression of K19 and vimentin. On the other hand, expression of ERK2 correlated to that of cyclin D1, membrane beta-catenin and MET, but not to histological grade. These findings suggest that ERK1 is preferentially expressed in HCC that has a stem-cell phenotype, while ERK2 is preferentially expressed in HCC that has a dysregulated Wnt pathway and/or dysregulated MET pathway. It has been reported that activation of kinases in HCC indicates aggressive behavior and may represent an independent prognostic factor for progression to HCC in HCV-infected individuals[31]. This activation seems to be related to the mammalian sterile-20-like kinase 4, which functions as an enhancer of cell proliferation and invasion through its ability to promote the process of epithelial-mesenchymal transition[32]. ERK2 may play a more prominent role in the proliferation of hepatocytes, while ERK1 may play a pro-apoptotic role[33]. An HCC type having a progenitor cell phenotype may present mechanisms of escaping the pro-apoptotic action of ERK1, which are presumably related to the mutation of p53. The overexpression of ERK1/ERK2 kinases in this study’s series of advanced cases was not related to EGFR overexpression.
Autopsy studies may have limitations that should be considered when interpreting results, particularly those related to autolytic changes that may affect immunoreactivity. In our institution, corpses are refrigerated and medical autopsies are performed as soon as all technical and legal procedures are addressed and resolved. Nevertheless, we sought to minimize the effects of autolytic changes by excluding samples with morphological features of poor preservation, specifically by examining vimentin staining (comparing to positive and negative controls) to assure that protein preservation was good enough to yield reliable immunohistochemical studies[34].
In conclusion, in the present autopsy cohort we found relevant associations between immunohistochemical markers with morphological features of tumors, especially that of histological grading of HCC[35]. The expression of K19, p53, ERK1, ERK2, vimentin and nuclear beta-catenin showed an association with pathological markers of higher-grade tumors, as opposed to cases expressing/over-expressing EGFR, MET and caspase 3. Further studies in the clinical setting are warranted to assess the prognostic value of this approach not only in advanced cases but also in early stages of HCC, especially in surgical resections or in explants.
Hepatocellular carcinoma (HCC) is an increasingly prevalent cancer and many patients are still diagnosed in the advanced stages, and usually as a complication of other chronic liver disease. In Brazil, chronic viral hepatitis and alcohol intake are the major related causes of HCC. Autopsy studies of advanced tumors provide a unique opportunity to assess important pathological aspects of progression and dissemination patterns of HCC.
Recent studies have sought to translate genomic data into panels of immunohistochemical markers, aiming to generate risk models for survival and prognostic evaluation of HCC. Morphological and immunohistochemical data may be incorporated in clinicopathological indices to predict HCC molecular classification and prognosis. The primary objective of the current study was to assess the distribution of proteins coded by genes reported as relevant to molecular classification of HCC based upon a detailed clinicopathological analysis of an autopsy cohort.
This retrospective cross-sectional analysis of a large post-mortem cohort series shows that a panel of immunohistochemical markers may be related to different pathways underlying progression and metastasis of HCC, and that the Edmondson-Steiner’s tumor grade may reflect currently recognized molecular subclasses of the disease. It also provides a detailed description of clinicopathological data in a unique autopsy series of HCC patients in Brazil.
The findings from this study form a foundation for further investigations of morpho-molecular correlations, not only in advanced cases of HCC but also in early stage cases and especially in surgical resections or explants.
Edmondson-Steiner’s tumor grade classifies HCC on a scale from 1 to 4, based on nuclear and cellular atypia; described in 1954, this system is the most widely accepted for tumor grading of HCC. Immunohistochemistry technique is used to detect cell or tissue antigens in two phases: (1) slide preparation, which involves specimen fixation, tissue processing and the immunohistochemical reaction (antigen retrieval, blocking of non-specific interaction sites, blocking of endogenous peroxidase, incubation with primary antibody, detection of immunoreactivity and counterstaining, as well as slide mounting); and (2) interpretation and quantification of the detected expression. Tissue microarrays contain many small representative cylindrical cores of tissue from different cases assembled on a single paraffin block and correspondent histological slide, thereby facilitating high-throughput analysis of multiple specimens simultaneously; it is produced by extracting the tissue cores from different (hundreds of) paraffin-donor blocks and re-embedding these into a single recipient block at defined array coordinates, thereby permitting simultaneous analysis of protein expression under standardized conditions on a single glass slide and also providing maximal preservation and use of limited and irreplaceable archival tissue samples.
The authors have conducted a detailed and elegantly illustrated histological study showing that groups of markers may be related to different pathways underlying HCC progression and metastasis, and that Edmondson-Steiner´s tumor grade may reflect currently recognized molecular subclasses of HCC.
Manuscript source: Unsolicited manuscript
P- Reviewer: Niu ZS, Peng SY S- Editor: Ma YJ L- Editor: A E- Editor: Wang CH
| 1. | Mittal S, El-Serag HB. Epidemiology of hepatocellular carcinoma: consider the population. J Clin Gastroenterol. 2013;47 Suppl:S2-S6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 770] [Cited by in F6Publishing: 832] [Article Influence: 75.6] [Reference Citation Analysis (0)] |
| 2. | Carrilho FJ, Kikuchi L, Branco F, Goncalves CS, Mattos AA. Clinical and epidemiological aspects of hepatocellular carcinoma in Brazil. Clinics (Sao Paulo). 2010;65:1285-1290. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 59] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 3. | Matsukuma S, Sato K. Peritoneal seeding of hepatocellular carcinoma: clinicopathological characteristics of 17 autopsy cases. Pathol Int. 2011;61:356-362. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 4. | Subhawong AP, Nassar H, Halushka MK, Illei PB, Vang R, Argani P. Heterogeneity of Bcl-2 expression in metastatic breast carcinoma. Mod Pathol. 2010;23:1089-1096. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 5. | Cimino-Mathews A, Hicks JL, Illei PB, Halushka MK, Fetting JH, De Marzo AM, Park BH, Argani P. Androgen receptor expression is usually maintained in initial surgically resected breast cancer metastases but is often lost in end-stage metastases found at autopsy. Hum Pathol. 2012;43:1003-1011. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 6. | Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, Durnez A, Demetris AJ, Thorgeirsson SS. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667-676. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 677] [Cited by in F6Publishing: 652] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 7. | Kim SM, Leem SH, Chu IS, Park YY, Kim SC, Kim SB, Park ES, Lim JY, Heo J, Kim YJ. Sixty-five gene-based risk score classifier predicts overall survival in hepatocellular carcinoma. Hepatology. 2012;55:1443-1452. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 100] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 8. | Nault JC, De Reyniès A, Villanueva A, Calderaro J, Rebouissou S, Couchy G, Decaens T, Franco D, Imbeaud S, Rousseau F. A hepatocellular carcinoma 5-gene score associated with survival of patients after liver resection. Gastroenterology. 2013;145:176-187. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 251] [Cited by in F6Publishing: 247] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 9. | Hoshida Y, Toffanin S, Lachenmayer A, Villanueva A, Minguez B, Llovet JM. Molecular classification and novel targets in hepatocellular carcinoma: recent advancements. Semin Liver Dis. 2010;30:35-51. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 232] [Cited by in F6Publishing: 243] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 10. | Villanueva A, Hoshida Y, Battiston C, Tovar V, Sia D, Alsinet C, Cornella H, Liberzon A, Kobayashi M, Kumada H. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology. 2011;140:1501-1512.e2. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 294] [Cited by in F6Publishing: 306] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 11. | Kondo S, Ojima H, Tsuda H, Hashimoto J, Morizane C, Ikeda M, Ueno H, Tamura K, Shimada K, Kanai Y. Clinical impact of c-Met expression and its gene amplification in hepatocellular carcinoma. Int J Clin Oncol. 2013;18:207-213. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 56] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 12. | Keng VW, Sia D, Sarver AL, Tschida BR, Fan D, Alsinet C, Solé M, Lee WL, Kuka TP, Moriarity BS. Sex bias occurrence of hepatocellular carcinoma in Poly7 molecular subclass is associated with EGFR. Hepatology. 2013;57:120-130. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 13. | Chiang DY, Villanueva A, Hoshida Y, Peix J, Newell P, Minguez B, LeBlanc AC, Donovan DJ, Thung SN, Solé M. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68:6779-6788. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 655] [Reference Citation Analysis (0)] |
| 14. | Edmondson HA, Steiner PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer. 1954;7:462-503. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 15. | Han DH, Choi GH, Kim KS, Choi JS, Park YN, Kim SU, Park JY, Ahn SH, Han KH. Prognostic significance of the worst grade in hepatocellular carcinoma with heterogeneous histologic grades of differentiation. J Gastroenterol Hepatol. 2013;28:1384-1390. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 45] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 16. | Srivastava S, Wong KF, Ong CW, Huak CY, Yeoh KG, Teh M, Luk JM, Salto-Tellez M. A morpho-molecular prognostic model for hepatocellular carcinoma. Br J Cancer. 2012;107:334-339. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 17. | Niu ZS, Niu XJ, Wang M. Management of hepatocellular carcinoma: Predictive value of immunohistochemical markers for postoperative survival. World J Hepatol. 2015;7:7-27. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 18. | Ito Y, Takeda T, Sakon M, Tsujimoto M, Higashiyama S, Noda K, Miyoshi E, Monden M, Matsuura N. Expression and clinical significance of erb-B receptor family in hepatocellular carcinoma. Br J Cancer. 2001;84:1377-1383. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 198] [Cited by in F6Publishing: 216] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 19. | Tsiambas E, Manaios L, Papanikolopoulos C, Rigopoulos DN, Tsounis D, Karameris A, Soultati A, Koliopoulou A, Kravvaritis C, Sergentanis T. Chromogenic in situ hybridization analysis of Epidermal Growth Factor Receptor gene/chromosome 7 numerical aberrations in hepatocellular carcinoma based on tissue microarrays. Pathol Oncol Res. 2009;15:511-520. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 20. | Kira S, Nakanishi T, Suemori S, Kitamoto M, Watanabe Y, Kajiyama G. Expression of transforming growth factor alpha and epidermal growth factor receptor in human hepatocellular carcinoma. Liver. 1997;17:177-182. [PubMed] [Cited in This Article: ] |
| 21. | Kannangai R, Sahin F, Torbenson MS. EGFR is phosphorylated at Ty845 in hepatocellular carcinoma. Mod Pathol. 2006;19:1456-1461. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 35] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 22. | Morimitsu Y, Hsia CC, Kojiro M, Tabor E. Nodules of less-differentiated tumor within or adjacent to hepatocellular carcinoma: relative expression of transforming growth factor-alpha and its receptor in the different areas of tumor. Hum Pathol. 1995;26:1126-1132. [PubMed] [Cited in This Article: ] |
| 23. | Winter RN, Kramer A, Borkowski A, Kyprianou N. Loss of caspase-1 and caspase-3 protein expression in human prostate cancer. Cancer Res. 2001;61:1227-1232. [PubMed] [Cited in This Article: ] |
| 24. | Persad R, Liu C, Wu TT, Houlihan PS, Hamilton SR, Diehl AM, Rashid A. Overexpression of caspase-3 in hepatocellular carcinomas. Mod Pathol. 2004;17:861-867. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 73] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 25. | Kim H, Choi GH, Na DC, Ahn EY, Kim GI, Lee JE, Cho JY, Yoo JE, Choi JS, Park YN. Human hepatocellular carcinomas with “Stemness”-related marker expression: keratin 19 expression and a poor prognosis. Hepatology. 2011;54:1707-1717. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 249] [Cited by in F6Publishing: 251] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 26. | Joo M, Kang YK, Kim MR, Lee HK, Jang JJ. Cyclin D1 overexpression in hepatocellular carcinoma. Liver. 2001;21:89-95. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 76] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 27. | Govaere O, Roskams T. Pathogenesis and prognosis of hepatocellular carcinoma at the cellular and molecular levels. Clin Liver Dis. 2015;19:261-276. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 28. | Schmitt-Graeff A, Ertelt-Heitzmann V, Allgaier HP, Olschewski M, Nitschke R, Haxelmans S, Koelble K, Behrens J, Blum HE. Coordinated expression of cyclin D1 and LEF-1/TCF transcription factor is restricted to a subset of hepatocellular carcinoma. Liver Int. 2005;25:839-847. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 22] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 29. | Prange W, Breuhahn K, Fischer F, Zilkens C, Pietsch T, Petmecky K, Eilers R, Dienes HP, Schirmacher P. Beta-catenin accumulation in the progression of human hepatocarcinogenesis correlates with loss of E-cadherin and accumulation of p53, but not with expression of conventional WNT-1 target genes. J Pathol. 2003;201:250-259. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 94] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 30. | Joo M, Lee HK, Kang YK. Expression of beta-catenin in hepatocellular carcinoma in relation to tumor cell proliferation and cyclin D1 expression. J Korean Med Sci. 2003;18:211-217. [PubMed] [Cited in This Article: ] |
| 31. | Schmitz KJ, Wohlschlaeger J, Lang H, Sotiropoulos GC, Malago M, Steveling K, Reis H, Cicinnati VR, Schmid KW, Baba HA. Activation of the ERK and AKT signalling pathway predicts poor prognosis in hepatocellular carcinoma and ERK activation in cancer tissue is associated with hepatitis C virus infection. J Hepatol. 2008;48:83-90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 236] [Cited by in F6Publishing: 248] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 32. | Lin ZH, Wang L, Zhang JB, Liu Y, Li XQ, Guo L, Zhang B, Zhu WW, Ye QH. MST4 promotes hepatocellular carcinoma epithelial-mesenchymal transition and metastasis via activation of the p-ERK pathway. Int J Oncol. 2014;45:629-640. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 33. | Guégan JP, Frémin C, Baffet G. The MAPK MEK1/2-ERK1/2 Pathway and Its Implication in Hepatocyte Cell Cycle Control. Int J Hepatol. 2012;2012:328372. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 34. | Alves VA, Pinheiro C, Morais-Santos F, Felipe-Silva A, Longatto-Filho A, Baltazar F. Characterization of monocarboxylate transporter activity in hepatocellular carcinoma. World J Gastroenterol. 2014;20:11780-11787. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 27] [Cited by in F6Publishing: 25] [Article Influence: 2.5] [Reference Citation Analysis (2)] |
| 35. | Tan PS, Nakagawa S, Goossens N, Venkatesh A, Huang T, Ward SC, Sun X, Song WM, Koh A, Canasto-Chibuque C. Clinicopathological indices to predict hepatocellular carcinoma molecular classification. Liver Int. 2016;36:108-118. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 66] [Article Influence: 8.3] [Reference Citation Analysis (0)] |