Published online Jul 28, 2015. doi: 10.3748/wjg.v21.i28.8527
Peer-review started: January 3, 2015
First decision: March 10, 2015
Revised: March 29, 2015
Accepted: June 15, 2015
Article in press: June 16, 2015
Published online: July 28, 2015
Processing time: 207 Days and 21.6 Hours
Pancreatic cancer remains difficult to treat and has a high mortality rate. It is difficult to diagnose early, mainly due to the lack of screening imaging modalities and specific biomarkers. Consequently, it is important to develop biomarkers that enable the detection of early stage tumors. Emerging evidence is accumulating that tumor cells release substantial amounts of RNA into the bloodstream that strongly resist RNases in the blood and are present at sufficient levels for quantitative analyses. These circulating RNAs are upregulated in the serum and plasma of cancer patients, including those with pancreatic cancer, compared with healthy controls. The majority of RNA biomarker studies have assessed circulating microRNAs (miRs), which are often tissue-specific. There are few reports of the tumor-specific upregulation of other types of small non-coding RNAs (ncRNAs), such as small nucleolar RNAs and Piwi-interacting RNAs. Long ncRNAs (lncRNAs), such as HOTAIR and MALAT1, in the serum/plasma of pancreatic cancer patients have also been reported as diagnostic and prognostic markers. Among tissue-derived RNAs, some miRs show increased expression even in pre-cancerous tissues, and their expression profiles may allow for the discrimination between a chronic inflammatory state and carcinoma. Additionally, some miRs and lncRNAs have been reported with significant alterations in expression according to disease progression, and they may thus represent potential candidate diagnostic or prognostic biomarkers that may be used to evaluate patients once detection methods in peripheral blood are well established. Furthermore, recent innovations in high-throughput sequencing techniques have enabled the discovery of unannotated tumor-associated ncRNAs and tumor-specific alternative splicing as novel and specific biomarkers of cancers. Although much work is required to clarify the release mechanism, origin of tumor-specific circulating RNAs, and selectivity of carrier complexes, and technical advances must also be achieved, such as creating a consensus normalization protocol for quantitative data analysis, circulating RNAs are largely unexplored and might represent novel clinical biomarkers.
Core tip: In this review, we summarize the latest findings on circulating RNAs in serum with a focus on their clinical use as novel diagnostic and prognostic biomarkers for pancreatic cancer. In addition, we summarize the current issues that need to be addressed to enable the clinical use of these circulating RNAs. This review will allow readers to concisely understand the current status and issues about the use of serum circulating RNAs as novel biomarkers for pancreatic cancer.
- Citation: Kishikawa T, Otsuka M, Ohno M, Yoshikawa T, Takata A, Koike K. Circulating RNAs as new biomarkers for detecting pancreatic cancer. World J Gastroenterol 2015; 21(28): 8527-8540
- URL: https://www.wjgnet.com/1007-9327/full/v21/i28/8527.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i28.8527
Pancreatic cancer remains an intractable disease and is the fourth leading cause of cancer death, with an increasing prevalence in the United States. The incidence and death rate of pancreatic cancer are high, and the 5-year survival rate remains only 6%[1]. More than half of pancreatic cancer patients are diagnosed at an advanced stage due to the lack of methods for its early detection. At present, we cannot expect a dramatic innovation in imaging modalities that will enable the detection of early stage cancer or pre-cancerous lesions using convenient, inexpensive methods[2], such as chest X-rays in lung cancer, mammography in breast cancer, or gastroendoscopy in gastric cancer.
Circulating biomarkers are useful screening tools because of the ease and relatively non-invasive nature of their collection. Among biochemical tests, serum CA19-9 is used widely as a circulating biomarker of pancreatic adenocarcinoma, but its utility is limited to monitoring the response to chemotherapy or surgical dissection because its sensitivity and specificity are insufficient for its use as a diagnostic biomarker of early stage pancreatic cancer[3]. Other serum protein markers have been suggested to enhance the predictability of pancreatic cancer, such as CEACAM1[4], MUC-1[5], REG4[6], TIMP-1[7], DJ-1[8], but none of them are in routine clinical use. Therefore, it might be useful to examine other molecules, such as nucleic acids, as novel biomarkers for the early detection of pancreatic cancer. Nucleic acids have several advantages as follows: their amplification is technically easy and they are less affected by degeneration or modification than protein- or carbohydrate-based tumor makers. Consequently, circulating nucleic acids have attracted increasing attention as novel tools in cancer diagnosis.
In the first study of circulating nucleic acids in cancer patients, Leon et al[9] detected free circulating DNAs in serum by radioimmunoassay. The polymerase chain reaction (PCR) technique has been used to isolate tumor-derived mutated sequences of the Kras and Nras genes from the serum and plasma of cancer patients[10,11]. Because Kras mutations are frequently observed in lung and gastrointestinal cancers in addition to nearly 100% of pancreatic cancers[12], many researchers are interested in the detection of mutated Kras in plasma[13]. More recently, genome-wide high-throughput sequencing of circulating DNA has been demonstrated as a potential detection tool, as well as a predictor of chemosensitivity[14,15].
Since the late 1990s, quantitative reverse transcriptase (qRT)-PCR has been used to detect circulating RNAs as cancer diagnostic markers. Tumor-derived circulating RNAs have been detected in whole blood, plasma, or serum from patients with pancreatic cancer, gastric cancer[16], nasopharyngeal carcinoma[17], and melanoma[18]. Although RNA species in the bloodstream have been considered to be more fragile than DNA because of the high RNase level in blood[19], plasma RNAs have been found to be unexpectedly stable against RNase degradation[20,21].
Since the early 2000s, various types of messenger RNAs (mRNAs) that are upregulated in cancer tissues have been detected in the peripheral blood of patients with lung cancer[22], breast cancer[23,24], melanoma[21], hepatocellular carcinoma[25], colorectal cancer[26-28], prostate cancer[29], gastric cancer[30], glioblastoma[31], chronic myelogenous leukemia[32], and pancreatic cancer[33-35]. In particular, many studies have reported circulating hTERT mRNA in the serum or plasma as a marker of various cancers[36-41], because telomerase activity, which maintains telomere length and prevents eukaryotic cells from senescence, is upregulated in a wide variety of cancers, whereas it is mostly suppressed in non-cancer tissues[42]. Some studies have achieved higher sensitivity and specificity by evaluating a combination of several tumor-specific mRNAs rather than circulating DNA or carbohydrate tumor markers[40,43,44].
In pancreatic cancer, Funaki et al[16] have detected carcinoembryonic antigen (CEA) mRNA by RT-PCR in the whole blood of pancreatic cancer patients. Clarke et al[33] have detected epidermal growth factor receptor (EGFR) mRNA in the serum and Ishizone et al[34] have detected alpha 1,4-N-Acetylglucosaminyltransferase (α4GnT) mRNA in the mononuclear cell fraction of peripheral blood from pancreatic cancer patients. Further, Kang et al[35] have recently demonstrated that serum type VI collagen (COL6A3) mRNA is a good marker of pancreatic cancer because it undergoes tumor-specific alternative splicing, which is expected to result in high specificity.
MicroRNAs (miRs) are small, single-stranded non-coding RNAs (ncRNA) consisting of 18-22 nucleotides that regulate the post-transcriptional expression of multiple genes[45]. Because miRs play important roles in controlling cell proliferation, differentiation, and apoptotic induction by targeting the mRNAs of various genes and a single miR is able to control the expression of hundreds of genes[46,47], miR dysregulation may affect cancer development, as well as the expression of oncogenes or onco-suppressor genes. There have been many reports on the aberrant over-expression or downregulation of miRs in various cancer tissues and subsequent alternations in the expression of their target genes, which are involved in proliferation and malignant transformation. In addition, miR expression is tissue specific[48], and alteration in specific miRs have been associated with cancer development[49,50]. Consequently, an altered miR expression profile could be a biomarker of malignant tumors, as well as an attractive therapeutic target in cancer[51,52].
In 2008, two studies demonstrated that miRs are released into the circulation in a remarkably stable form, even after freeze/thaw cycling or room temperature incubation, and suggested that circulating miRs carry disease-specific signatures that can be exploited as non-invasive biomarkers[53,54].
Regarding pancreatic cancer, several studies have described tumor-derived miRs in the circulation as diagnostic or prognostic biomarkers (Table 1). Wang et al[55] have analyzed the expression of miRs in the plasma of patients with pancreatic ductal adenocarcinoma and have identified miRs-21, miR-210, miR-155, and miR-196a, which have been reported to be upregulated in pancreatic cancer tissue and cell lines[56-67], as candidate biomarkers. Similarly, miR-200a/b, miR-18a, miR-221, and miR-196a/b have been found to be upregulated in the serum/plasma in parallel with cancer tissues[55,58-83]. Recently, comprehensive sequencing and microarray analyses have been performed to identify other circulating miRs, and combined analyses of the expression of several miRs have achieved high detectability with high sensitivity and specificity[73,75,77,78,81,82,84].
| Target candidate | Up/down | Sample | Number of patients | Extraction method | Quantification method | Target selection | Normalization | Potential value | Ref. |
| mRNA | |||||||||
| CEA mRNA | Up | Whole blood | 9 PK, 9 HC | AGPC | RT-PCR | Pre | ACTB | D | [16] |
| EGFR mRNA | Up | Serum | 11 PK, 23 HC | N/A | RT-PCR | Pre | B2M | D | [33] |
| 4GnT mRNA | Up | Whole blood | 55 PK, 10 CP, 70 HC | RNeasy Mini kit | RT-PCR | Pre | GAPDH | D | [34] |
| COL6A3 mRNA | Up | Serum | 44 PK, 46 BT, 30 HC | PureYield RNA Midiprep | qRT-PCR | Pre | GAPDH | D/P | [35] |
| miRNA | |||||||||
| mIR-155 | Up | Plasma | 49 PK, 36 HC | TRIzol LS | Taqman | Pre | miR-16 | D | [55] |
| miR-196a | Up | D | |||||||
| miR-21 | Up | D | |||||||
| miR-210 | Up | D | |||||||
| miR-155 | Up | Pancreatic juice | 16 PK, 5CP | mirVana PARIS kit | Taqman | Pre | miR-199 U6-snRNA | D | [68] |
| miR-21 | Up | D | |||||||
| miR-196a | Up | Serum | 35 PK, 15 CP, 15 HC | TRI Reagent BD | Taqman | Pre | cel-miR-39 | D/P | [69] |
| miR-200a | Up | Serum | 45 PK, 11 CP, 32 HC | mirVana miR isolation kit | Taqman | Pre | miR-16 | D | [70] |
| miR-200b | Up | D | |||||||
| miR-210 | Up | Plasma | 11 PK, 14 HC | Boiling | Taqman | Pre | cel-miR-54 | D | [71] |
| miR-18a | Up | Plasma | 36 PK, 30 HC | mirVana PARIS kit | Taqman | Pre | synthetic reference panel | D/T | [72] |
| miR-16 | Up | Plasma | 140 PK, 111 CP, 68 HC | TRI Reagent BD | Taqman | Pre | cel-miR-39 | D | [73] |
| miR-196a | Up | D | |||||||
| miR-185 | Up | Serum | 80 PK, 129 HC | TRIzol LS | Taqman | Sequence | serum volume | D | [74] |
| miR-191 | Up | D | |||||||
| miR-20a | Up | D | |||||||
| miR-21 | Up | D/P | |||||||
| miR-24 | Up | D | |||||||
| miR-25 | Up | D | |||||||
| miR-99a | Up | D | |||||||
| miR-1290 | Up | Serum | 41 PK, 38 BT, 35 CP, 19 HC | mirVana PARIS kit | Taqman | Taqman mA | miR-16 | D | [75] |
| miR-221 | Up | Plasma | 47 PK, 30 HC | mirVana PARIS kit | Taqman | Pre | synthetic reference panel | D/T/P | [76] |
| miR-375 | Down | D/T/P | |||||||
| miR-375 | Up | Plasma | 48 PK, 47 HC | Total RNA purification kit (Norgen) | Taqman | Affymetrix mA | cel-miR-39,cel-miR-54, cel-miR-238 | D | [77] |
| miR-27a3p | Up | Whole blood | 129 PK, 103 BT, 60HC | Trizol | Taqman | Sequence | U6 snRNA | D/S | [78] |
| miR-196a | Up | Serum | 19 PK, 10 CP, 20 BT, 10 HC | miReasy RNA extraction kit | Taqman | Pre | miR-24 | D | [79] |
| miR-196b | Up | D | |||||||
| miR-205 | Up | Pancreatic juice | 50 PK, 19 CP, 19 HC | TRIzol LS | Taqman | Agilent mA | U6 snRNA | D/P | [80] |
| miR-210 | Up | D/P | |||||||
| miR-492 | Up | D/P | |||||||
| miR-1427 | Up | D/P | |||||||
| miR-22 | Up | Plasma | 11 PK, 11 HR, 11 HC | TRI Reagent BD | Taqman | custom mA | miR-3196 | D | [81] |
| miR-642b | Up | D | |||||||
| miR-885-5p | Up | D | |||||||
| Multi gene index | Whole blood | 409 PK, 25 CP, 312 HC | PAXgene blood RNA | Taqman | Taqman mA | ath-miR159a | D | [82] | |
| miR-483-3p | Up | Plasma | 32 PK, 12 BT, 30 HC | mirVana PARIS kit | Taqman | Pre | miR-16 | D | [83] |
| miR-21 | Up | D | |||||||
| snRNA | |||||||||
| U2 snRNA | Up | Serum | 80 PK, 129 HC | mirVana miR isolation kit | qRT-PCR | Agilent mA | cel-miR-54 | D | [84] |
NcRNAs are divided into two families according to their lengths, small ncRNAs (up to 200 bases) and large ncRNAs (over 200 bases). Almost all of the studies examining circulating RNAs have focused on miRs because of the existence of established methods for quantifying their expression, such as microarray kits and qRT-PCR. There are currently few reports of other small ncRNA family members (Table 2), such as small nucleolar RNA (snoRNA)[85-88], small nuclear RNA (snRNA), and Piwi-RNA (piRNA)[89,90], in cancer tissues. SnoRNAs, which function as guide RNAs for the post-transcriptional modification of ribosomal RNAs and some spliceosomal RNAs[91], are deregulated in various cancer tissues and induce a tumor-promoting phenotype[85-88,92]. Using comprehensive next-generation sequence analysis, Liao et al[93] have found that the plasma snoRNA SNORD33/66/76 might serve as a diagnostic biomarker for non-small-cell lung cancer.
| Target candidate | Up/down | Cancer type | Sample | Number of patients | Extraction method | Quantification method | Normalization | Target selection | Ref. |
| Small ncRNA | |||||||||
| SNORD33/66/76 | Up | Lung | Plasma | 37 Ca, 26 HR, 22 HC | mirVana miR isolation kit | SYBR qRT-PCR | U6 snRNA | mA | [93] |
| piR-651 | Up | Gastric | Whole blood | 93 Ca, 32 HC | TRizol | SYBR qRT-PCR | U6 snRNA | Pre | [94] |
| piR-823 | Up | ||||||||
| U2snRNA | Up | Pancreatic | Serum | 80 Ca, 129 HC | mirVana miR isolation kit | qRT-PCR | cel-miR-54 | mA | [84] |
| U2snRNA | Up | Colorectal | 132 Ca, 129 HC | ||||||
| U2snRNA | Up | Lung | Serum | 62 Ca, 51 BT, 45 HC | miRNeasy mini kit | LNA qRT-PCR | cel-miR-39 | mA | [96] |
| U2snRNA | Up | Ovarian | Serum | 119 Ca, 35 HC | mirVana PARIS kit | qRT-PCR | cel-miR-54 | mA | [97] |
| Long ncRNA | |||||||||
| H19 | Up | Gastric | Plasma | 43 Ca, 34 HC | N/A | pre-amplification qRT-PCR | N/A | Pre | [107] |
| MALAT1 | Up | Prostate | Plasma | 87 Ca, 82 HR, 23HC | mirVana PARIS kit | Taqman | Input amount | Pre | [108] |
| HULC | Up | Hepato-cellular | Plasma | 30 Ca, 20 HC | TRIzol | qRT-PCR | GAPDH | Pre | [110] |
| TUG1 lincRNA | Up | Multiple myeloma | Plasma | 62 MM, 40 HC | TriPure | qRT-PCR | GAPDH | Pre | [111] |
| LincRNA-p21 | Down | CLL | 68 CLL, 40 HC |
PiRNAs interact with a subset of Argonaute proteins related to Piwi (Pelement induced wimpy testis in Drosophila) and maintain genomic integrity by epigenetically silencing transposons via DNA methylation, especially in germline stem cells. They have recently even been identified outside of the germline and in human cancer cells[89,90] and may be valuable biomarkers for detecting circulating gastric cancer cells[94].
SnRNAs are found within the splicing speckles of the cell nucleus and function as guides for pre-mRNA splicing in association with small nuclear ribonucleoproteins[95]. Several types of snRNA, including U2 snRNA and U6 snRNA, have been frequently used as housekeeping genes for normalization in qRT-PCR. However, it has also been reported that circulating U2 snRNA (RNU2-1) might be a useful diagnostic biomarker in pancreatic, colorectal, lung, and ovarian cancers[84,96,97].
High-throughput RNA sequencing techniques have revealed that long non-coding RNAs (lncRNAs), which are not translated into proteins, are transcribed as frequently as protein-coding mRNAs. Most lncRNAs are thought to have biological functions, and increasing numbers of cancer-associated lncRNAs, such as HOTAIR[98], MALAT-1[99], ANRIL[100], H19[101], PTCSC3[102], and PCA3[103,104], have been reported to be upregulated in various cancer cells and to play potential roles in both oncogenic and tumor-suppression pathways, functioning as epigenetic regulators, guides for alternative splicing, decoys of miRNAs, and a scaffolds for protein complexes[105,106].
Although there is little data on the use of circulating lncRNAs as cancer-detecting markers, they could be used as novel potential biomarkers for diagnostic, prognostic, and therapeutic purposes (Table 2). For example, the secretion of H19 lncRNA in the plasma is increased in gastric cancer patients[107]. In addition, serum MALAT-1 RNA was upregulated in prostate cancer[108] and plasma TUC339 and HULC lncRNA were identified as novel markers for the detection of hepatocellular carcinoma[109,110]. Most recently, Isin et al[111] have reported tumor-type specific changes in TUG1 and LincRNA-p21 expression in the plasma in association with B-cell neoplasm.
There are no reports of upregulated lncRNAs in the circulation in pancreatic cancer patients at present. However, some reports have shown significant changes in the expression of lncRNA in pancreatic cancer tissues and cell lines, which have been previously reported in other types of cancer[112-117] (Table 3). Recently, novel candidate genes have been found to be diagnostic and prognostic markers of pancreatic cancer using comprehensive microarray or RNA sequence analyses (Table 3)[118-121]. In particular, the expression of HSATII RNA, which is a highly repetitive transcript from centromeric heterochromatic regions, differs significantly in cancer and pre-cancer tissues compared with normal pancreatic tissue. Although it is necessary to examine whether circulating lncRNAs are as stable as miRs, which are resistant to RNases, these tumor-specific lncRNAs might be useful detection markers in the serum/plasma with high sensitivity and specificity.
| Target candidate | Up/down | Sample | Number of patients | Extraction method | Quantification method | Normalization | Target selection | Potential value | Ref. |
| HOTAIR | Up | Tissue | 36 PK, 36 HC | mirVana RNA isolation kit | qRT-PCR | GAPDH | Pre | P | [112] |
| MALAT-1 | Up | Tissue | 126 PK, 15 HC | TRIzol | qRT-PCR | N/A | Pre | D/P/S | [113] |
| MEG3 | Up | Tissue | 31 PNET, 7 HC | RNeasy | qRT-PCR | GAPDH | Pre | D | [114] |
| Gas5 | Up | Tissue | 23 PK, 23 AN | TRIzol | qRT-PCR | ACTB | Pre | D | [115] |
| HULC | Up | 304 PK, 304 AN | TRIzol | qRT-PCR | GAPDH | Pre | D/P | [116] | |
| LOC285194 | Down | Tissue | 85 PK, 85 AN | TRIzol | qRT-PCR | GAPDH | Pre | D/P | [117] |
| PPP3CB intronic lncRNA | Up | Tissue | 11 PK, 7 metastasis | TRIzol | qRT-PCR | HMBS | mA | D/S | [118] |
| MAP3K1 | Up | ||||||||
| DAPK1 | Up | ||||||||
| BC008363 | Up | Tissue | 30PK, 30 AN | mA | D/P | [119] | |||
| ENST00000480739 | Down | 35 PK, 35 AN | TRIzol | qRT-PCR | ACTB | mA | D/P | [120] | |
| HSATII | Up | Tissue | 11 PK, 2 HC | TRIzol | Sequence | Input amount | Sequence | D/P | [121] |
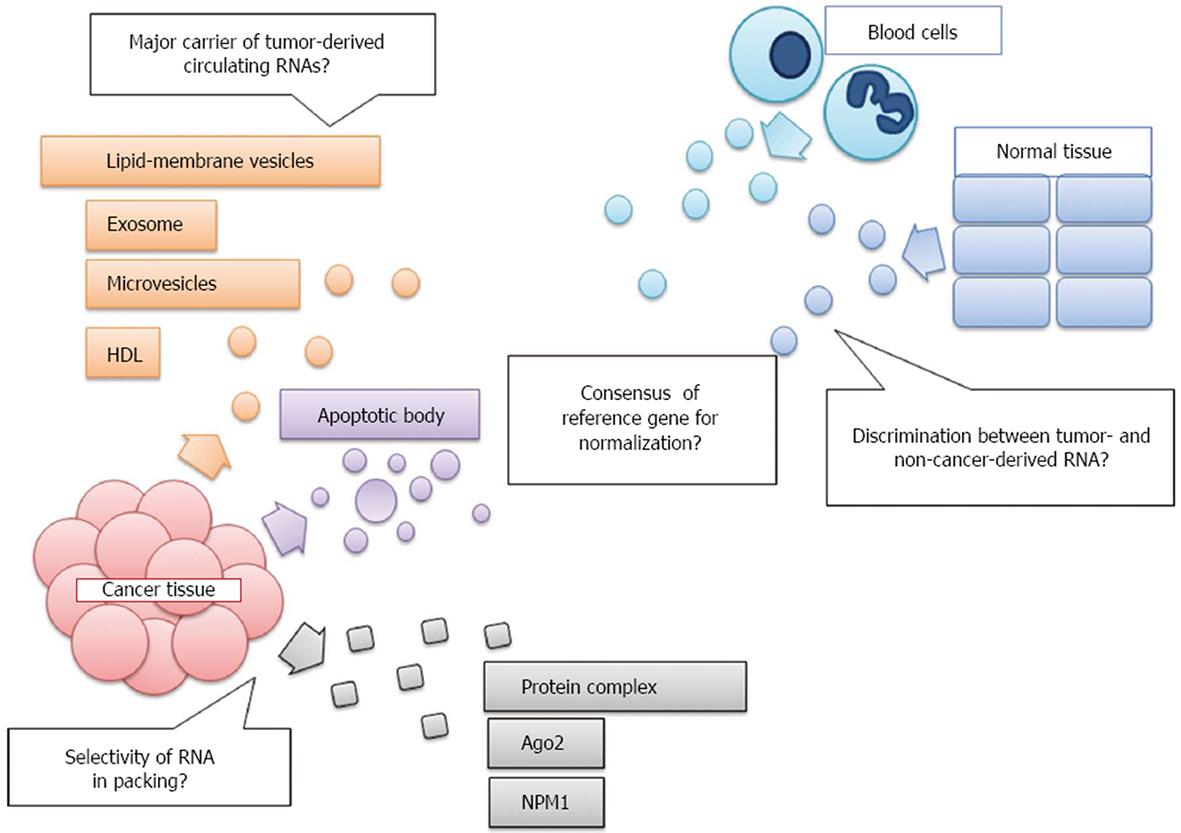
The origin and manner of release of extracellular cancer-associated RNAs are unclear but could potentially affect the significance of results. It is not clear whether the cancer-associated RNAs detected in the circulation result from tumor cell death and lysis or whether they are actively secreted by tumor cells (Figure 1). While living cells actively release RNA encapsulated in large lipoprotein complexes, such as exosomes or microvesicles (MVs), RNA from dead or dying cells found in blood is associated with apoptotic bodies or protein complexes[122]. An exosome is a 40 to 140-nm-diameter lipoprotein membranous vesicle of endocytic origin that is formed from the fusion of multivesicular bodies (MVB) with the plasma membrane and released into extracellular spaces. MVs are larger than exosomes, with diameters ranging from 100-1000 nm and heterogeneous morphologies, and they originate from the plasma membrane via direct outward budding into the extracellular space. Apoptotic bodies are membrane vesicles that are heterogeneous in shape, range from 50-500 nm in diameter, contain organelles, and are released via outward protrusion of the plasma membrane during the late phase of apoptosis[123-125].
Many studies have shown that tumors specifically secrete exosomes or MVs, which selectively contain specific miRs[126,127]. However, Arroyo et al[128] and Turchinovich et al[129] have asserted that miRs in circulation are principally found in association with the Ago2 ribonucleoprotein complex and not vesicles, indicating that the detected miRs are derived predominantly via apoptotic and necrotic processes, which occur frequently in tumor cells. Some reports have suggested that high- and low- density lipoproteins (HDLs and LDLs, respectively) contain circulating RNA[130] and that certain miRs are specifically encapsulated within them. At present, it is unclear whether cancer cells choose specific carriers for particular miRs depending on particular biological functions.
Moreover, circulating miRs in cancer patient are derived from multiple sources, including not only circulating and primary tumor cells, but also immune cells and other blood cells and cancer-adjacent non-cancerous cells. The mechanisms regulating RNA secretion and the determination of selectivity as well as the functional roles of secreted RNAs are poorly understood. Limited studies have been performed to distinguish the efficiencies of antibodies between tumor-derived exosomes and other tissue derived exosomes[131-133]. These studies have distinguished tumor-derived exosomes from other tissue derived exosomes by magnetic-activated cell sorting and flow cytometry using an antibody for tumor-specific exosome surface antigens, such as EpCAM, and CD39/73[131-133]. Precise analyses of these “purified” circulating RNA may reveal a more specific expression signature of primary tumors. On the other hand, cancer-adjacent tissues, such as stromal cells, also release aberrantly expressing RNAs and transmit intercellular signals to cancer cells, such as chemoresistance signals[134], indicating that alternations in RNA expression signatures of cancer-adjacent tissues may also have some potential as cancer diagnostic markers.
In addition to the stability of miRs in various body fluids, their levels can be easily measured by qRT-PCR, which allows for high-precision signal amplification using the TaqMan PCR method with stem-looped RT primers or oligo-dT primers after the polyadenylation of templates. Nevertheless, there is still significant variability among microarray analysis reports. One of the reasons for this validation is that the total amount of RNA in serum and plasma is small compared to the amount of RNA extracted from tissues or cell lines. Several novel detection methods have been developed to allow for rapid quantification of RNA, improve the sensitivity and specificity of detection and reducing the number of amplification steps required before detection[135]. Detection strategies range from the use of simple molecular beacons[136] and enzymatic luminescence[137] to the use of different nanoparticle-based probes[138-140] and different forms of electrophoresis, such as capillary isotachophoresis[141] and circular exponential amplification[142].
At present, there is no consensus on using endogenous reference RNAs to normalize circulating miRs levels for analyzing qRT-PCR results (Figure 1). It has been suggested that some miRs, such as miR-16, miR-223, and let-7, are highly and constantly expressed in the serum/plasma and are correlated with the number of blood cells[55,70,75]. However, other reports have claimed that miRs that have been recommended as controls are significantly altered in certain pathological states[87,143]. Because gene expression can vary within a population, to identify miR signatures related to diseases, it is important to determine the range of normal variability across demographic populations[144]. As another alternative to small RNAs, 5S rRNA, and U6 snRNA are commonly used as reference genes when examining cell-extracted miR levels[68,80,93]. However, some reports have suggested that they are very unstable because they are degraded by RNases in the plasma/serum, unlike miRs[54]. At present, external control normalization with a spiked mycetogenic gene or normalization to the total amount of input RNA is widely performed (Tables 1 and 2).
Recently, microarray chip analysis of lncRNAs has been exponentially developed in gene number and has become a convenient measuring tool. However, comprehensive RNA sequence analysis by next generation sequencing can yield more information about unannotated genes and alternative spliced transcript signatures that may be possible candidates for novel detecting markers[145]. Several robust RNA sequencing protocols have been developed that are applicable even with a minute amount of templates and at the single cell level[146,147]. In fact, tumor-specific alternative splicing has already been reported in pancreatic cancer[35,148-151]. These alternatively spliced transcripts are considered more specific because they are unaffected by transcripts produced by the same gene in normal cells. A candidate gene with a splicing pattern that is distinct between cancer and normal cells may represent a highly specific marker, in addition to tumor-specific mutations of circulating DNA.
Despite the rapidly increasing number of reports, the biological significance of circulating RNAs in cancer remains unclear. It believed that cancer cells communicate with surrounding non-cancerous cells, such as stromal cells and immune cells, via extracellular RNAs that are protected by their carriers. This intercellular connection contributes to their uncontrolled proliferation, the malignant transformation of surrounding cells, the recruitment of new blood vessels, and the stimulation or escape from the response to cancer cells[31,152,153]. Understanding the mechanisms by which cellular RNAs are selectively loaded into a specific carriers is important, not only for the more precise quantification of tumor-specific circulating RNAs as powerful detection markers but also to improve novel therapeutic strategies, such as RNA delivery to target tissues[154].
It is difficult to detect emerging cancerous lesions of pre-cancerous cystic tumors, such as intraductal papillary mucinous neoplasms (IPMNs), and to distinguish a focal cancer lesion from mass-forming pancreatitis, even when patients are extensively examined, for example, by endoscopic retrograde cholangiopancreatography (ERCP), and endoscopic ultrasonography (EUS). Prophylactic surgical resection of a pre-cancerous tumor is not always acceptable due to its capacity for invading the pancreas and surrounding organs, in contrast with colon and gastric adenomas, which are easily resectable by endoscopic procedures. Consequently, a specific biomarker is desperately needed to detect pre-cancerous lesions or to discriminate between malignant tumors and inflammatory lesions. In this regard, some studies have indicated that the tissue specificity of miRs and the dramatic changes that occur in their expression profiles over the course of oncogenesis should enable the detection pre-cancerous lesions, such as pancreatic intraepithelial neoplasia (PanIN)[58,62-65,155] and IPMN[156], in pancreatic tissues. In addition, some miRs expression changes in aspirated cystic fluid from IPMN have been shown to predict the presence of cancer[157]. It is difficult to obtain surgically resected pancreatic pre-cancerous lesion specimens. In particular, PanIN lesions are very small and are commonly only identifiable at the microscopic level; therefore, they are normally obtained by laser capture microdissection from resected chronic pancreatitis and mainly pancreatic cancer specimens. Furthermore, it is difficult to collect blood samples from patients with a pre-cancerous lesion because current screening examinations rarely detect high-risk patients in advance.
It is accepted that the progression of pancreatic ductal adenocarcinoma is associated with the accumulation of genetic alternation, as well as the adenoma-carcinoma sequence, in colorectal cancer[12]. As genetic alterations accumulate in normal pancreatic epithelial cells, pre-cancerous lesions called PanINs emerge and advance in stage, eventually progressing to invasive cancer. According to this multistep carcinogenesis hypothesis, several oncogenic mouse models have been established[158]. Mice that express a constitutively active mutant KrasG12D protein in a pancreas epithelium-specific manner, demonstrate gradual PanIN progression. The combination of endogenous KrasG12D expression and dysfunction of tumor suppressor genes, such as p16INK4a/Arf knockout, mutant p53 expression, p53 knockout, or transforming growth factor-β receptor 2 (Tgfbr2) knockout, result in the progression of invasive pancreatic ductal adenocarcinoma, which is histologically closed to human cancer tissue. These pre-cancer and progressive cancer mouse models may have advantages over human samples in terms of genetic simplicity and the limited influence of environmental factors, such as inflammation, for the evaluation of mutation-induced transcription alternations. In fact, we have performed microarray analysis of miRs from mouse normal pancreata, a pre-cancerous PanIN-like tumor and massive ductal adenocarcinoma and have found that the majority of miRs upregulated in cancer, such as miR-21 and miR-192, are already increased at the pre-cancerous stage in Kras-mutated PanIN tissues (unpublished data). Moreover, collecting blood samples from these genetically engineered mice is important to identify cancer- or pre-cancer-associated circulating RNAs that may be present in humans[159]. The evaluation of these circulating RNAs is also important in distinguishing cancer from inflammation because human pancreatic cancer tissues consistently accompany chronic inflammation.
On the other hand, pancreatic cancer has a high rate of metastasis and dissemination, which cause postoperative recurrence and the dysfunction of other organs, such as gastrointestinal obstruction, and eventually result in poor prognosis. One possible avenue for improving prognosis is to predict the presence of undetectable metastasis by clinical imaging and to select the best-suited treatment under preoperative or postoperative conditions. The levels of several individual miRs, including miR-10b, 21, 31, 126, 335, and 373, have been correlated with metastatic outcome in carcinoma patients[160]. Additionally, other classes of ncRNAs, such as HOTAIR[98], have been proposed as putative biomarkers for metastatic potential in human breast tumors. These results suggest their potential utilities as biomarkers for metastatic propensity. Similar strategies may be applicable for the evaluation of pancreatic carcinoma.
Although their release mechanisms and biological significance require further study, circulating RNAs are significantly beneficial to the pancreatic cancer field, representing possible novel diagnostic and/or prognostic markers. Improvements in extraction and amplification procedures and growing availability of next-generation sequencing technologies may allow for the discovery of more specific and sensitive RNA markers in the near future.
P- Reviewer: Crea F, Kleeff J, Martinez-Zorzano VS, Ogura T, Shi C S- Editor: Ma YJ L- Editor: A E- Editor: Zhang DN
| 1. | Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9215] [Cited by in RCA: 9847] [Article Influence: 820.6] [Reference Citation Analysis (4)] |
| 2. | Costello E, Greenhalf W, Neoptolemos JP. New biomarkers and targets in pancreatic cancer and their application to treatment. Nat Rev Gastroenterol Hepatol. 2012;9:435-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 158] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 3. | Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS, Somerfield MR, Hayes DF, Bast RC. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313-5327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1057] [Cited by in RCA: 1109] [Article Influence: 58.4] [Reference Citation Analysis (0)] |
| 4. | Simeone DM, Ji B, Banerjee M, Arumugam T, Li D, Anderson MA, Bamberger AM, Greenson J, Brand RE, Ramachandran V. CEACAM1, a novel serum biomarker for pancreatic cancer. Pancreas. 2007;34:436-443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 112] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 5. | Gold DV, Modrak DE, Ying Z, Cardillo TM, Sharkey RM, Goldenberg DM. New MUC1 serum immunoassay differentiates pancreatic cancer from pancreatitis. J Clin Oncol. 2006;24:252-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 100] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 6. | Takayama R, Nakagawa H, Sawaki A, Mizuno N, Kawai H, Tajika M, Yatabe Y, Matsuo K, Uehara R, Ono K. Serum tumor antigen REG4 as a diagnostic biomarker in pancreatic ductal adenocarcinoma. J Gastroenterol. 2010;45:52-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 7. | Brand RE, Nolen BM, Zeh HJ, Allen PJ, Eloubeidi MA, Goldberg M, Elton E, Arnoletti JP, Christein JD, Vickers SM. Serum biomarker panels for the detection of pancreatic cancer. Clin Cancer Res. 2011;17:805-816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 192] [Cited by in RCA: 184] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 8. | He XY, Liu BY, Yao WY, Zhao XJ, Zheng Z, Li JF, Yu BQ, Yuan YZ. Serum DJ-1 as a diagnostic marker and prognostic factor for pancreatic cancer. J Dig Dis. 2011;12:131-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 9. | Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646-650. [PubMed] |
| 10. | Sorenson GD, Pribish DM, Valone FH, Memoli VA, Bzik DJ, Yao SL. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol Biomarkers Prev. 1994;3:67-71. [PubMed] |
| 11. | Vasioukhin V, Anker P, Maurice P, Lyautey J, Lederrey C, Stroun M. Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia. Br J Haematol. 1994;86:774-779. [PubMed] |
| 12. | Bardeesy N, DePinho RA. Pancreatic cancer biology and genetics. Nat Rev Cancer. 2002;2:897-909. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 848] [Cited by in RCA: 844] [Article Influence: 36.7] [Reference Citation Analysis (1)] |
| 13. | Sorenson GD. Detection of mutated KRAS2 sequences as tumor markers in plasma/serum of patients with gastrointestinal cancer. Clin Cancer Res. 2000;6:2129-2137. [PubMed] |
| 14. | Chan KC, Jiang P, Zheng YW, Liao GJ, Sun H, Wong J, Siu SS, Chan WC, Chan SL, Chan AT. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem. 2013;59:211-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 369] [Cited by in RCA: 388] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 15. | Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, Parkinson C, Chin SF, Kingsbury Z, Wong AS. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1288] [Cited by in RCA: 1306] [Article Influence: 108.8] [Reference Citation Analysis (0)] |
| 16. | Funaki NO, Tanaka J, Kasamatsu T, Ohshio G, Hosotani R, Okino T, Imamura M. Identification of carcinoembryonic antigen mRNA in circulating peripheral blood of pancreatic carcinoma and gastric carcinoma patients. Life Sci. 1996;59:2187-2199. [PubMed] |
| 17. | Lo KW, Lo YM, Leung SF, Tsang YS, Chan LY, Johnson PJ, Hjelm NM, Lee JC, Huang DP. Analysis of cell-free Epstein-Barr virus associated RNA in the plasma of patients with nasopharyngeal carcinoma. Clin Chem. 1999;45:1292-1294. [PubMed] |
| 18. | Kopreski MS, Benko FA, Kwak LW, Gocke CD. Detection of tumor messenger RNA in the serum of patients with malignant melanoma. Clin Cancer Res. 1999;5:1961-1965. [PubMed] |
| 19. | Reddi KK, Holland JF. Elevated serum ribonuclease in patients with pancreatic cancer. Proc Natl Acad Sci USA. 1976;73:2308-2310. [PubMed] |
| 20. | Tsui NB, Ng EK, Lo YM. Stability of endogenous and added RNA in blood specimens, serum, and plasma. Clin Chem. 2002;48:1647-1653. [PubMed] |
| 21. | Hasselmann DO, Rappl G, Tilgen W, Reinhold U. Extracellular tyrosinase mRNA within apoptotic bodies is protected from degradation in human serum. Clin Chem. 2001;47:1488-1489. [PubMed] |
| 22. | Fleischhacker M, Beinert T, Ermitsch M, Seferi D, Possinger K, Engelmann C, Jandrig B. Detection of amplifiable messenger RNA in the serum of patients with lung cancer. Ann N Y Acad Sci. 2001;945:179-188. [PubMed] |
| 23. | Gal S, Fidler C, Lo YM, Chin K, Moore J, Harris AL, Wainscoat JS. Detection of mammaglobin mRNA in the plasma of breast cancer patients. Ann N Y Acad Sci. 2001;945:192-194. [PubMed] |
| 24. | Silva JM, Dominguez G, Silva J, Garcia JM, Sanchez A, Rodriguez O, Provencio M, España P, Bonilla F. Detection of epithelial messenger RNA in the plasma of breast cancer patients is associated with poor prognosis tumor characteristics. Clin Cancer Res. 2001;7:2821-2825. [PubMed] |
| 25. | Ng EK, Tsui NB, Lam NY, Chiu RW, Yu SC, Wong SC, Lo ES, Rainer TH, Johnson PJ, Lo YM. Presence of filterable and nonfilterable mRNA in the plasma of cancer patients and healthy individuals. Clin Chem. 2002;48:1212-1217. [PubMed] |
| 26. | Silva JM, Rodriguez R, Garcia JM, Muñoz C, Silva J, Dominguez G, Provencio M, España P, Bonilla F. Detection of epithelial tumour RNA in the plasma of colon cancer patients is associated with advanced stages and circulating tumour cells. Gut. 2002;50:530-534. [PubMed] |
| 27. | Wong SC, Lo SF, Cheung MT, Ng KO, Tse CW, Lai BS, Lee KC, Lo YM. Quantification of plasma beta-catenin mRNA in colorectal cancer and adenoma patients. Clin Cancer Res. 2004;10:1613-1617. [PubMed] |
| 28. | Garcia V, García JM, Peña C, Silva J, Domínguez G, Hurtado A, Alonso I, Rodriguez R, Provencio M, Bonilla F. Thymidylate synthase messenger RNA expression in plasma from patients with colon cancer: prognostic potential. Clin Cancer Res. 2006;12:2095-2100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 29. | Chu DC, Chuang CK, Liou YF, Tzou RD, Lee HC, Sun CF. The use of real-time quantitative PCR to detect circulating prostate-specific membrane antigen mRNA in patients with prostate carcinoma. Ann N Y Acad Sci. 2004;1022:157-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 30. | Xu W, Zhou H, Qian H, Bu X, Chen D, Gu H, Zhu W, Yan Y, Mao F. Combination of circulating CXCR4 and Bmi-1 mRNA in plasma: A potential novel tumor marker for gastric cancer. Mol Med Rep. 2009;2:765-771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 31. | Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470-1476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4100] [Cited by in RCA: 3922] [Article Influence: 230.7] [Reference Citation Analysis (0)] |
| 32. | Narita M, Saito A, Kojima A, Iwabuchi M, Satoh N, Uchiyama T, Yamahira A, Furukawa T, Sone H, Takahashi M. Quantification of BCR-ABL mRNA in plasma/serum of patients with chronic myelogenous leukemia. Int J Med Sci. 2012;9:901-908. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 33. | Clarke LE, Leitzel K, Smith J, Ali SM, Lipton A. Epidermal growth factor receptor mRNA in peripheral blood of patients with pancreatic, lung, and colon carcinomas detected by RT-PCR. Int J Oncol. 2003;22:425-430. [PubMed] |
| 34. | Ishizone S, Yamauchi K, Kawa S, Suzuki T, Shimizu F, Harada O, Sugiyama A, Miyagawa S, Fukuda M, Nakayama J. Clinical utility of quantitative RT-PCR targeted to alpha1,4-N-acetylglucosaminyltransferase mRNA for detection of pancreatic cancer. Cancer Sci. 2006;97:119-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 22] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 35. | Kang CY, Wang J, Axell-House D, Soni P, Chu ML, Chipitsyna G, Sarosiek K, Sendecki J, Hyslop T, Al-Zoubi M. Clinical significance of serum COL6A3 in pancreatic ductal adenocarcinoma. J Gastrointest Surg. 2014;18:7-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 36. | Chen XQ, Bonnefoi H, Pelte MF, Lyautey J, Lederrey C, Movarekhi S, Schaeffer P, Mulcahy HE, Meyer P, Stroun M. Telomerase RNA as a detection marker in the serum of breast cancer patients. Clin Cancer Res. 2000;6:3823-3826. [PubMed] |
| 37. | Dasí F, Lledó S, García-Granero E, Ripoll R, Marugán M, Tormo M, García-Conde J, Aliño SF. Real-time quantification in plasma of human telomerase reverse transcriptase (hTERT) mRNA: a simple blood test to monitor disease in cancer patients. Lab Invest. 2001;81:767-769. [PubMed] |
| 38. | Miura N, Maeda Y, Kanbe T, Yazama H, Takeda Y, Sato R, Tsukamoto T, Sato E, Marumoto A, Harada T. Serum human telomerase reverse transcriptase messenger RNA as a novel tumor marker for hepatocellular carcinoma. Clin Cancer Res. 2005;11:3205-3209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 56] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 39. | Dasí F, Martínez-Rodes P, March JA, Santamaría J, Martínez-Javaloyas JM, Gil M, Aliño SF. Real-time quantification of human telomerase reverse transcriptase mRNA in the plasma of patients with prostate cancer. Ann N Y Acad Sci. 2006;1075:204-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 40. | Tani N, Ichikawa D, Ikoma D, Tomita H, Sai S, Ikoma H, Okamoto K, Ochiai T, Ueda Y, Otsuji E. Circulating cell-free mRNA in plasma as a tumor marker for patients with primary and recurrent gastric cancer. Anticancer Res. 2007;27:1207-1212. [PubMed] |
| 41. | March-Villalba JA, Martínez-Jabaloyas JM, Herrero MJ, Santamaria J, Aliño SF, Dasí F. Cell-free circulating plasma hTERT mRNA is a useful marker for prostate cancer diagnosis and is associated with poor prognosis tumor characteristics. PLoS One. 2012;7:e43470. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 69] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 42. | Paradis V, Dargère D, Laurendeau I, Benoît G, Vidaud M, Jardin A, Bedossa P. Expression of the RNA component of human telomerase (hTR) in prostate cancer, prostatic intraepithelial neoplasia, and normal prostate tissue. J Pathol. 1999;189:213-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 43. | Miura N, Nakamura H, Sato R, Tsukamoto T, Harada T, Takahashi S, Adachi Y, Shomori K, Sano A, Kishimoto Y. Clinical usefulness of serum telomerase reverse transcriptase (hTERT) mRNA and epidermal growth factor receptor (EGFR) mRNA as a novel tumor marker for lung cancer. Cancer Sci. 2006;97:1366-1373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 67] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 44. | Silva J, Silva JM, García V, García JM, Domínguez G, Bonilla F. RNA is more sensitive than DNA in identification of breast cancer patients bearing tumor nucleic acids in plasma. Genes Chromosomes Cancer. 2002;35:375-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 45. | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14460] [Cited by in RCA: 16008] [Article Influence: 1000.5] [Reference Citation Analysis (2)] |
| 46. | Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1694] [Cited by in RCA: 1731] [Article Influence: 91.1] [Reference Citation Analysis (0)] |
| 47. | Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8847] [Cited by in RCA: 9260] [Article Influence: 463.0] [Reference Citation Analysis (0)] |
| 48. | Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 761] [Cited by in RCA: 837] [Article Influence: 46.5] [Reference Citation Analysis (0)] |
| 49. | Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7723] [Cited by in RCA: 7362] [Article Influence: 368.1] [Reference Citation Analysis (0)] |
| 50. | Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S, Levy A. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462-469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 728] [Cited by in RCA: 719] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 51. | Papaconstantinou IG, Lykoudis PM, Gazouli M, Manta A, Polymeneas G, Voros D. A review on the role of microRNA in biology, diagnosis, and treatment of pancreatic adenocarcinoma. Pancreas. 2012;41:671-677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 52. | Ajit SK. Circulating microRNAs as biomarkers, therapeutic targets, and signaling molecules. Sensors (Basel). 2012;12:3359-3369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 128] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 53. | Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513-10518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5636] [Cited by in RCA: 6284] [Article Influence: 369.6] [Reference Citation Analysis (0)] |
| 54. | Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997-1006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3218] [Cited by in RCA: 3536] [Article Influence: 208.0] [Reference Citation Analysis (0)] |
| 55. | Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL, Frazier ML, Killary AM, Sen S. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila). 2009;2:807-813. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 434] [Cited by in RCA: 440] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 56. | Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, Labourier E, Hahn SA. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442-4452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 483] [Cited by in RCA: 536] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 57. | Gironella M, Seux M, Xie MJ, Cano C, Tomasini R, Gommeaux J, Garcia S, Nowak J, Yeung ML, Jeang KT. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci USA. 2007;104:16170-16175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 419] [Cited by in RCA: 430] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 58. | Dillhoff M, Liu J, Frankel W, Croce C, Bloomston M. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J Gastrointest Surg. 2008;12:2171-2176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 313] [Cited by in RCA: 330] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 59. | Moriyama T, Ohuchida K, Mizumoto K, Yu J, Sato N, Nabae T, Takahata S, Toma H, Nagai E, Tanaka M. MicroRNA-21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Mol Cancer Ther. 2009;8:1067-1074. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 265] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 60. | Ikenaga N, Ohuchida K, Mizumoto K, Yu J, Kayashima T, Sakai H, Fujita H, Nakata K, Tanaka M. MicroRNA-203 expression as a new prognostic marker of pancreatic adenocarcinoma. Ann Surg Oncol. 2010;17:3120-3128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 98] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 61. | Yu J, Ohuchida K, Mizumoto K, Fujita H, Nakata K, Tanaka M. MicroRNA miR-17-5p is overexpressed in pancreatic cancer, associated with a poor prognosis, and involved in cancer cell proliferation and invasion. Cancer Biol Ther. 2010;10:748-757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 96] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 62. | Ryu JK, Hong SM, Karikari CA, Hruban RH, Goggins MG, Maitra A. Aberrant MicroRNA-155 expression is an early event in the multistep progression of pancreatic adenocarcinoma. Pancreatology. 2010;10:66-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 112] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 63. | Yu J, Li A, Hong SM, Hruban RH, Goggins M. MicroRNA alterations of pancreatic intraepithelial neoplasias. Clin Cancer Res. 2012;18:981-992. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 169] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 64. | Su A, He S, Tian B, Hu W, Zhang Z. MicroRNA-221 mediates the effects of PDGF-BB on migration, proliferation, and the epithelial-mesenchymal transition in pancreatic cancer cells. PLoS One. 2013;8:e71309. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 65. | Xue Y, Abou Tayoun AN, Abo KM, Pipas JM, Gordon SR, Gardner TB, Barth RJ, Suriawinata AA, Tsongalis GJ. MicroRNAs as diagnostic markers for pancreatic ductal adenocarcinoma and its precursor, pancreatic intraepithelial neoplasm. Cancer Genet. 2013;206:217-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 58] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 66. | Song S, Zhou J, He S, Zhu D, Zhang Z, Zhao H, Wang Y, Li D. Expression levels of microRNA-375 in pancreatic cancer. Biomed Rep. 2013;1:393-398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 67. | Ma MZ, Kong X, Weng MZ, Cheng K, Gong W, Quan ZW, Peng CH. Candidate microRNA biomarkers of pancreatic ductal adenocarcinoma: meta-analysis, experimental validation and clinical significance. J Exp Clin Cancer Res. 2013;32:71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 76] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 68. | Sadakari Y, Ohtsuka T, Ohuchida K, Tsutsumi K, Takahata S, Nakamura M, Mizumoto K, Tanaka M. MicroRNA expression analyses in preoperative pancreatic juice samples of pancreatic ductal adenocarcinoma. JOP. 2010;11:587-592. [PubMed] |
| 69. | Kong X, Du Y, Wang G, Gao J, Gong Y, Li L, Zhang Z, Zhu J, Jing Q, Qin Y. Detection of differentially expressed microRNAs in serum of pancreatic ductal adenocarcinoma patients: miR-196a could be a potential marker for poor prognosis. Dig Dis Sci. 2011;56:602-609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 122] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 70. | Li A, Omura N, Hong SM, Vincent A, Walter K, Griffith M, Borges M, Goggins M. Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Res. 2010;70:5226-5237. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 233] [Cited by in RCA: 240] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 71. | Ho AS, Huang X, Cao H, Christman-Skieller C, Bennewith K, Le QT, Koong AC. Circulating miR-210 as a Novel Hypoxia Marker in Pancreatic Cancer. Transl Oncol. 2010;3:109-113. [PubMed] |
| 72. | Morimura R, Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, Nagata H, Konishi H, Shiozaki A, Ikoma H, Okamoto K. Novel diagnostic value of circulating miR-18a in plasma of patients with pancreatic cancer. Br J Cancer. 2011;105:1733-1740. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 189] [Cited by in RCA: 212] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 73. | Liu J, Gao J, Du Y, Li Z, Ren Y, Gu J, Wang X, Gong Y, Wang W, Kong X. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int J Cancer. 2012;131:683-691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 231] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 74. | Liu R, Chen X, Du Y, Yao W, Shen L, Wang C, Hu Z, Zhuang R, Ning G, Zhang C. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin Chem. 2012;58:610-618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 312] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 75. | Li A, Yu J, Kim H, Wolfgang CL, Canto MI, Hruban RH, Goggins M. MicroRNA array analysis finds elevated serum miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls. Clin Cancer Res. 2013;19:3600-3610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 239] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 76. | Kawaguchi T, Komatsu S, Ichikawa D, Morimura R, Tsujiura M, Konishi H, Takeshita H, Nagata H, Arita T, Hirajima S. Clinical impact of circulating miR-221 in plasma of patients with pancreatic cancer. Br J Cancer. 2013;108:361-369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 175] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 77. | Carlsen AL, Joergensen MT, Knudsen S, de Muckadell OB, Heegaard NH. Cell-free plasma microRNA in pancreatic ductal adenocarcinoma and disease controls. Pancreas. 2013;42:1107-1113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 78. | Wang WS, Liu LX, Li GP, Chen Y, Li CY, Jin DY, Wang XL. Combined serum CA19-9 and miR-27a-3p in peripheral blood mononuclear cells to diagnose pancreatic cancer. Cancer Prev Res (Phila). 2013;6:331-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 65] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 79. | Slater EP, Strauch K, Rospleszcz S, Ramaswamy A, Esposito I, Klöppel G, Matthäi E, Heeger K, Fendrich V, Langer P. MicroRNA-196a and -196b as Potential Biomarkers for the Early Detection of Familial Pancreatic Cancer. Transl Oncol. 2014;7:464-471. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 69] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 80. | Wang J, Raimondo M, Guha S, Chen J, Diao L, Dong X, Wallace MB, Killary AM, Frazier ML, Woodward TA. Circulating microRNAs in Pancreatic Juice as Candidate Biomarkers of Pancreatic Cancer. J Cancer. 2014;5:696-705. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 94] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 81. | Ganepola GA, Rutledge JR, Suman P, Yiengpruksawan A, Chang DH. Novel blood-based microRNA biomarker panel for early diagnosis of pancreatic cancer. World J Gastrointest Oncol. 2014;6:22-33. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 88] [Cited by in RCA: 89] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 82. | Schultz NA, Dehlendorff C, Jensen BV, Bjerregaard JK, Nielsen KR, Bojesen SE, Calatayud D, Nielsen SE, Yilmaz M, Holländer NH. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA. 2014;311:392-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 342] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 83. | Abue M, Yokoyama M, Shibuya R, Tamai K, Yamaguchi K, Sato I, Tanaka N, Hamada S, Shimosegawa T, Sugamura K. Circulating miR-483-3p and miR-21 is highly expressed in plasma of pancreatic cancer. Int J Oncol. 2015;46:539-547. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 157] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 84. | Baraniskin A, Nöpel-Dünnebacke S, Ahrens M, Jensen SG, Zöllner H, Maghnouj A, Wos A, Mayerle J, Munding J, Kost D. Circulating U2 small nuclear RNA fragments as a novel diagnostic biomarker for pancreatic and colorectal adenocarcinoma. Int J Cancer. 2013;132:E48-E57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 110] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 85. | Dong XY, Guo P, Boyd J, Sun X, Li Q, Zhou W, Dong JT. Implication of snoRNA U50 in human breast cancer. J Genet Genomics. 2009;36:447-454. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 159] [Cited by in RCA: 159] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 86. | Mei YP, Liao JP, Shen J, Yu L, Liu BL, Liu L, Li RY, Ji L, Dorsey SG, Jiang ZR. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene. 2012;31:2794-2804. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 186] [Cited by in RCA: 212] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 87. | Gee HE, Buffa FM, Camps C, Ramachandran A, Leek R, Taylor M, Patil M, Sheldon H, Betts G, Homer J. The small-nucleolar RNAs commonly used for microRNA normalisation correlate with tumour pathology and prognosis. Br J Cancer. 2011;104:1168-1177. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 205] [Cited by in RCA: 229] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 88. | Valleron W, Laprevotte E, Gautier EF, Quelen C, Demur C, Delabesse E, Agirre X, Prósper F, Kiss T, Brousset P. Specific small nucleolar RNA expression profiles in acute leukemia. Leukemia. 2012;26:2052-2060. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 94] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 89. | Qiao D, Zeeman AM, Deng W, Looijenga LH, Lin H. Molecular characterization of hiwi, a human member of the piwi gene family whose overexpression is correlated to seminomas. Oncogene. 2002;21:3988-3999. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 221] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 90. | Cheng J, Guo JM, Xiao BX, Miao Y, Jiang Z, Zhou H, Li QN. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin Chim Acta. 2011;412:1621-1625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 222] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 91. | Williams GT, Farzaneh F. Are snoRNAs and snoRNA host genes new players in cancer? Nat Rev Cancer. 2012;12:84-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 272] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 92. | Xu G, Yang F, Ding CL, Zhao LJ, Ren H, Zhao P, Wang W, Qi ZT. Small nucleolar RNA 113-1 suppresses tumorigenesis in hepatocellular carcinoma. Mol Cancer. 2014;13:216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 87] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 93. | Liao J, Yu L, Mei Y, Guarnera M, Shen J, Li R, Liu Z, Jiang F. Small nucleolar RNA signatures as biomarkers for non-small-cell lung cancer. Mol Cancer. 2010;9:198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 190] [Cited by in RCA: 215] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 94. | Cui L, Lou Y, Zhang X, Zhou H, Deng H, Song H, Yu X, Xiao B, Wang W, Guo J. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using piRNAs as markers. Clin Biochem. 2011;44:1050-1057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 120] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 95. | Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 653] [Cited by in RCA: 581] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 96. | Mazières J, Catherinne C, Delfour O, Gouin S, Rouquette I, Delisle MB, Prévot G, Escamilla R, Didier A, Persing DH. Alternative processing of the U2 small nuclear RNA produces a 19-22nt fragment with relevance for the detection of non-small cell lung cancer in human serum. PLoS One. 2013;8:e60134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 30] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 97. | Kuhlmann JD, Baraniskin A, Hahn SA, Mosel F, Bredemeier M, Wimberger P, Kimmig R, Kasimir-Bauer S. Circulating U2 small nuclear RNA fragments as a novel diagnostic tool for patients with epithelial ovarian cancer. Clin Chem. 2014;60:206-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 51] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 98. | Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071-1076. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4202] [Cited by in RCA: 4202] [Article Influence: 280.1] [Reference Citation Analysis (0)] |
| 99. | Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031-8041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1584] [Cited by in RCA: 1788] [Article Influence: 81.3] [Reference Citation Analysis (0)] |
| 100. | Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956-1962. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 830] [Cited by in RCA: 810] [Article Influence: 57.9] [Reference Citation Analysis (0)] |
| 101. | Gabory A, Ripoche MA, Yoshimizu T, Dandolo L. The H19 gene: regulation and function of a non-coding RNA. Cytogenet Genome Res. 2006;113:188-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 187] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 102. | Fan M, Li X, Jiang W, Huang Y, Li J, Wang Z. A long non-coding RNA, PTCSC3, as a tumor suppressor and a target of miRNAs in thyroid cancer cells. Exp Ther Med. 2013;5:1143-1146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 136] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 103. | Hessels D, Schalken JA. The use of PCA3 in the diagnosis of prostate cancer. Nat Rev Urol. 2009;6:255-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 170] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 104. | Day JR, Jost M, Reynolds MA, Groskopf J, Rittenhouse H. PCA3: from basic molecular science to the clinical lab. Cancer Lett. 2011;301:1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 62] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 105. | Crea F, Clermont PL, Parolia A, Wang Y, Helgason CD. The non-coding transcriptome as a dynamic regulator of cancer metastasis. Cancer Metastasis Rev. 2014;33:1-16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 73] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 106. | Ling H, Vincent K, Pichler M, Fodde R, Berindan-Neagoe I, Slack FJ, Calin GA. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene. 2015;Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 236] [Cited by in RCA: 271] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 107. | Arita T, Ichikawa D, Konishi H, Komatsu S, Shiozaki A, Shoda K, Kawaguchi T, Hirajima S, Nagata H, Kubota T. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 2013;33:3185-3193. [PubMed] |
| 108. | Ren S, Wang F, Shen J, Sun Y, Xu W, Lu J, Wei M, Xu C, Wu C, Zhang Z. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur J Cancer. 2013;49:2949-2959. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 260] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 109. | Kogure T, Yan IK, Lin WL, Patel T. Extracellular Vesicle-Mediated Transfer of a Novel Long Noncoding RNA TUC339: A Mechanism of Intercellular Signaling in Human Hepatocellular Cancer. Genes Cancer. 2013;4:261-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 262] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 110. | Xie H, Ma H, Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. Biomed Res Int. 2013;2013:136106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 144] [Cited by in RCA: 219] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 111. | Isin M, Ozgur E, Cetin G, Erten N, Aktan M, Gezer U, Dalay N. Investigation of circulating lncRNAs in B-cell neoplasms. Clin Chim Acta. 2014;431:255-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 132] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 112. | Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616-1625. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 631] [Cited by in RCA: 684] [Article Influence: 57.0] [Reference Citation Analysis (0)] |
| 113. | Pang EJ, Yang R, Fu XB, Liu YF. Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol. 2015;36:2403-2407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 159] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 114. | Modali SD, Parekh VI, Kebebew E, Agarwal SK. Epigenetic regulation of the lncRNA MEG3 and its target c-MET in pancreatic neuroendocrine tumors. Mol Endocrinol. 2015;29:224-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 95] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 115. | Lu X, Fang Y, Wang Z, Xie J, Zhan Q, Deng X, Chen H, Jin J, Peng C, Li H. Downregulation of gas5 increases pancreatic cancer cell proliferation by regulating CDK6. Cell Tissue Res. 2013;354:891-896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 125] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 116. | Peng W, Gao W, Feng J. Long noncoding RNA HULC is a novel biomarker of poor prognosis in patients with pancreatic cancer. Med Oncol. 2014;31:346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 127] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 117. | Huang C, Yu W, Wang Q, Cui H, Wang Y, Zhang L, Han F, Huang T. Increased expression of the lncRNA PVT1 is associated with poor prognosis in pancreatic cancer patients. Minerva Med. 2015;106:143-149. [PubMed] |
| 118. | Tahira AC, Kubrusly MS, Faria MF, Dazzani B, Fonseca RS, Maracaja-Coutinho V, Verjovski-Almeida S, Machado MC, Reis EM. Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Mol Cancer. 2011;10:141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 135] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 119. | Li J, Liu D, Hua R, Zhang J, Liu W, Huo Y, Cheng Y, Hong J, Sun Y. Long non-coding RNAs expressed in pancreatic ductal adenocarcinoma and lncRNA BC008363 an independent prognostic factor in PDAC. Pancreatology. 2014;14:385-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 120. | Sun YW, Chen YF, Li J, Huo YM, Liu DJ, Hua R, Zhang JF, Liu W, Yang JY, Fu XL. A novel long non-coding RNA ENST00000480739 suppresses tumour cell invasion by regulating OS-9 and HIF-1α in pancreatic ductal adenocarcinoma. Br J Cancer. 2014;111:2131-2141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 95] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 121. | Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, Contino G, Deshpande V, Iafrate AJ, Letovsky S. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331:593-596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 364] [Cited by in RCA: 416] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 122. | Ma R, Jiang T, Kang X. Circulating microRNAs in cancer: origin, function and application. J Exp Clin Cancer Res. 2012;31:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 140] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 123. | Samos J, García-Olmo DC, Picazo MG, Rubio-Vitaller A, García-Olmo D. Circulating nucleic acids in plasma/serum and tumor progression: are apoptotic bodies involved? An experimental study in a rat cancer model. Ann N Y Acad Sci. 2006;1075:165-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 124. | Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, Hristov M, Köppel T, Jahantigh MN, Lutgens E. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. 2009;2:ra81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 959] [Cited by in RCA: 1048] [Article Influence: 65.5] [Reference Citation Analysis (0)] |
| 125. | Sato-Kuwabara Y, Melo SA, Soares FA, Calin GA. The fusion of two worlds: non-coding RNAs and extracellular vesicles--diagnostic and therapeutic implications (Review). Int J Oncol. 2015;46:17-27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 186] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 126. | Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2012;7:e30679. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 712] [Cited by in RCA: 850] [Article Influence: 65.4] [Reference Citation Analysis (2)] |
| 127. | Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442-17452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1382] [Cited by in RCA: 1581] [Article Influence: 105.4] [Reference Citation Analysis (0)] |
| 128. | Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003-5008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2345] [Cited by in RCA: 2621] [Article Influence: 187.2] [Reference Citation Analysis (0)] |
| 129. | Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223-7233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1356] [Cited by in RCA: 1513] [Article Influence: 108.1] [Reference Citation Analysis (0)] |
| 130. | Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423-433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2292] [Cited by in RCA: 2172] [Article Influence: 155.1] [Reference Citation Analysis (0)] |
| 131. | Rupp AK, Rupp C, Keller S, Brase JC, Ehehalt R, Fogel M, Moldenhauer G, Marmé F, Sültmann H, Altevogt P. Loss of EpCAM expression in breast cancer derived serum exosomes: role of proteolytic cleavage. Gynecol Oncol. 2011;122:437-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 221] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 132. | Clayton A, Al-Taei S, Webber J, Mason MD, Tabi Z. Cancer exosomes express CD39 and CD73, which suppress T cells through adenosine production. J Immunol. 2011;187:676-683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 363] [Cited by in RCA: 434] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 133. | Oksvold MP, Kullmann A, Forfang L, Kierulf B, Li M, Brech A, Vlassov AV, Smeland EB, Neurauter A, Pedersen KW. Expression of B-cell surface antigens in subpopulations of exosomes released from B-cell lymphoma cells. Clin Ther. 2014;36:847-862.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 122] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 134. | Boelens MC, Wu TJ, Nabet BY, Xu B, Qiu Y, Yoon T, Azzam DJ, Twyman-Saint Victor C, Wiemann BZ, Ishwaran H. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell. 2014;159:499-513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 537] [Cited by in RCA: 653] [Article Influence: 65.3] [Reference Citation Analysis (0)] |
| 135. | Saikumar J, Ramachandran K, Vaidya VS. Noninvasive micromarkers. Clin Chem. 2014;60:1158-1173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 32] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 136. | Baker MB, Bao G, Searles CD. In vitro quantification of specific microRNA using molecular beacons. Nucleic Acids Res. 2012;40:e13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 96] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 137. | Sun Y, Gregory KJ, Chen NG, Golovlev V. Rapid and direct microRNA quantification by an enzymatic luminescence assay. Anal Biochem. 2012;429:11-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 138. | Dong H, Jin S, Ju H, Hao K, Xu LP, Lu H, Zhang X. Trace and label-free microRNA detection using oligonucleotide encapsulated silver nanoclusters as probes. Anal Chem. 2012;84:8670-8674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 140] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 139. | Tu Y, Wu P, Zhang H, Cai C. Fluorescence quenching of gold nanoparticles integrating with a conformation-switched hairpin oligonucleotide probe for microRNA detection. Chem Commun (Camb). 2012;48:10718-10720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 140. | Wang L, Cheng Y, Wang H, Li Z. A homogeneous fluorescence sensing platform with water-soluble carbon nanoparticles for detection of microRNA and nuclease activity. Analyst. 2012;137:3667-3672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 141. | Ban E, Song EJ. Capillary electrophoresis methods for microRNAs assays: a review. Anal Chim Acta. 2014;852:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 142. | Wang J, Yi X, Tang H, Han H, Wu M, Zhou F. Direct quantification of microRNA at low picomolar level in sera of glioma patients using a competitive hybridization followed by amplified voltammetric detection. Anal Chem. 2012;84:6400-6406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 93] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 143. | McDonald JS, Milosevic D, Reddi HV, Grebe SK, Algeciras-Schimnich A. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem. 2011;57:833-840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 492] [Cited by in RCA: 519] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 144. | Duttagupta R, Jiang R, Gollub J, Getts RC, Jones KW. Impact of cellular miRNAs on circulating miRNA biomarker signatures. PLoS One. 2011;6:e20769. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 248] [Cited by in RCA: 238] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 145. | Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1788] [Cited by in RCA: 2120] [Article Influence: 212.0] [Reference Citation Analysis (0)] |
| 146. | Saliba AE, Westermann AJ, Gorski SA, Vogel J. Single-cell RNA-seq: advances and future challenges. Nucleic Acids Res. 2014;42:8845-8860. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 510] [Cited by in RCA: 562] [Article Influence: 51.1] [Reference Citation Analysis (0)] |
| 147. | Ramsköld D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777-782. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1078] [Cited by in RCA: 1166] [Article Influence: 97.2] [Reference Citation Analysis (0)] |
| 148. | Tsai YS, Dominguez D, Gomez SM, Wang Z. Transcriptome-wide identification and study of cancer-specific splicing events across multiple tumors. Oncotarget. 2015;6:6825-6839. [PubMed] |
| 149. | Sebestyén E, Zawisza M, Eyras E. Detection of recurrent alternative splicing switches in tumor samples reveals novel signatures of cancer. Nucleic Acids Res. 2015;43:1345-1356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 137] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 150. | Bonomi S, Gallo S, Catillo M, Pignataro D, Biamonti G, Ghigna C. Oncogenic alternative splicing switches: role in cancer progression and prospects for therapy. Int J Cell Biol. 2013;2013:962038. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 85] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 151. | Zhu Y, Zhang JJ, Xie KL, Tang J, Liang WB, Zhu R, Zhu Y, Wang B, Tao JQ, Zhi XF. Specific-detection of clinical samples, systematic functional investigations, and transcriptome analysis reveals that splice variant MUC4/Y contributes to the malignant progression of pancreatic cancer by triggering malignancy-related positive feedback loops signaling. J Transl Med. 2014;12:309. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 152. | Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8246] [Cited by in RCA: 9715] [Article Influence: 539.7] [Reference Citation Analysis (0)] |
| 153. | Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, Sun F, Lu J, Yin Y, Cai X. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39:133-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 891] [Cited by in RCA: 961] [Article Influence: 64.1] [Reference Citation Analysis (0)] |
| 154. | Rachagani S, Macha MA, Heimann N, Seshacharyulu P, Haridas D, Chugh S, Batra SK. Clinical implications of miRNAs in the pathogenesis, diagnosis and therapy of pancreatic cancer. Adv Drug Deliv Rev. 2015;81:16-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 82] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 155. | Bauer AS, Keller A, Costello E, Greenhalf W, Bier M, Borries A, Beier M, Neoptolemos J, Büchler M, Werner J. Diagnosis of pancreatic ductal adenocarcinoma and chronic pancreatitis by measurement of microRNA abundance in blood and tissue. PLoS One. 2012;7:e34151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 95] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 156. | Habbe N, Koorstra JB, Mendell JT, Offerhaus GJ, Ryu JK, Feldmann G, Mullendore ME, Goggins MG, Hong SM, Maitra A. MicroRNA miR-155 is a biomarker of early pancreatic neoplasia. Cancer Biol Ther. 2009;8:340-346. [PubMed] |
| 157. | Maker AV, Carrara S, Jamieson NB, Pelaez-Luna M, Lennon AM, Dal Molin M, Scarpa A, Frulloni L, Brugge WR. Cyst fluid biomarkers for intraductal papillary mucinous neoplasms of the pancreas: a critical review from the international expert meeting on pancreatic branch-duct-intraductal papillary mucinous neoplasms. J Am Coll Surg. 2015;220:243-253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 158. | Ijichi H. Genetically-engineered mouse models for pancreatic cancer: Advances and current limitations. World J Clin Oncol. 2011;2:195-202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 17] [Cited by in RCA: 23] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 159. | Yabushita S, Fukamachi K, Tanaka H, Sumida K, Deguchi Y, Sukata T, Kawamura S, Uwagawa S, Suzui M, Tsuda H. Circulating microRNAs in serum of human K-ras oncogene transgenic rats with pancreatic ductal adenocarcinomas. Pancreas. 2012;41:1013-1018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 160. | Valastyan S, Weinberg RA. MicroRNAs: Crucial multi-tasking components in the complex circuitry of tumor metastasis. Cell Cycle. 2009;8:3506-3512. [PubMed] |