Published online Feb 28, 2012. doi: 10.3748/wjg.v18.i8.814
Revised: April 26, 2011
Accepted: May 3, 2011
Published online: February 28, 2012
AIM: To investigate the protein expression profile of mismatch repair (MMR) genes in suspected cases of Lynch syndrome and to characterize the associated germline mutations.
METHODS: Immunohistochemical analysis of tumor samples was performed to determine the protein expression profile of MMR protein. Germline mutation screening was carried out on peripheral blood samples. The entire exon regions of MLH1 and MSH2 genes were amplified by polymerase chain reaction, screened by denaturing high performance liquid chromatography (dHPLC) and analyzed by DNA sequencing to characterize the germline mutations.
RESULTS: Three out of 34 tissue samples (8.8%) and four out of 34 tissue samples (11.8%) showed loss of nuclear staining by immunohistochemistry, indicating the absence of MLH1 and MSH2 protein expression in carcinoma cells, respectively. dHPLC analysis followed by DNA sequencing showed these samples to have germline mutations of MSH2 gene. However, no deleterious mutations were identified in any of the 19 exons or coding regions of MLH1 gene, but we were able to identify MLH1 promoter polymorphism, -93G > A (rs1800734), in 21 out of 34 patients (61.8%). We identified one novel mutation, transversion mutation c.2005G > C, which resulted in a missense mutation (Gly669Arg), a transversion mutation in exon 1, c.142G > T, which resulted in a nonsense mutation (Glu48Stop) and splice-site mutation, c.2006-6T > C, which was adjacent to exon 13 of MSH2 gene.
CONCLUSION: Germline mutations were identified in four Malaysian Lynch syndrome patients. Immunohistochemical analysis of tumor tissue proved to be a good pre-screening test before proceeding to germline mutation analysis of DNA MMR genes.
-
Citation: Zahary MN, Kaur G, Abu Hassan MR, Singh H, Naik VR, Ankathil R. Germline mutation analysis of
MLH1 andMSH2 in Malaysian Lynch syndrome patients. World J Gastroenterol 2012; 18(8): 814-820 - URL: https://www.wjgnet.com/1007-9327/full/v18/i8/814.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i8.814
Colorectal cancer (CRC) is the most common gastrointestinal cancer in Malaysia and Lynch syndrome accounts for approximately 1%-5% of all colorectal cancers[1-3]. Lynch syndrome is generally divided into type I which describes families presenting with colonic cancers at an early age in the absence of multiple colonic adenomas, whereas type II describes similar families who also develop extracolonic cancers particularly those of the female reproductive tract[4]. Lynch syndrome is characterized by autosomal dominant inheritance with multiple members affected in families, inheritance of susceptibility genes (mismatch repair genes) with incomplete penetrance (80%-90%), and early onset of CRC and/or extracolonic cancers such as cancers of the endometrium, ovary, stomach, small intestine, hepatobiliary tract, upper urinary tract, brain and skin[5,6].
Germline mutations in a group of DNA mismatch repair (MMR) genes (MLH1, MSH2, MSH6, PMS1 and PMS2) have been found to be associated with susceptibility to Lynch syndrome[5-7]. Germline mutations of MLH1 and MSH2 genes account for more than 90% of the mutations identified in Lynch syndrome families[5,8]. The loss of MMR function will lead to a phenomenon known as microsatellite instability (MSI) which is the hallmark of Lynch syndrome.
In addition to tumor MSI testing which is available to evaluate the MSI status and germline mutation analysis using peripheral blood, immunohistochemical testing of tumor tissue for the expression status of MMR protein has emerged as another option available in the genetic testing of Lynch syndrome. Data on germline mutations of MMR genes are available from different parts of the world in varying frequencies, however, no reports are available from Malaysia. Hence, this study was undertaken to unravel the spectrum of germline mutations in DNA MMR genes in Malaysian Lynch syndrome patients. We investigated the protein expression profile of MMR genes in suspected cases of Lynch syndrome and characterized the associated germline mutations. This report focuses on the protein expression profile and germline mutations identified in MLH1 and MSH2 gene.
This study was approved by the Research and Ethics Committee, School of Medical Sciences, Universiti Sains Malaysia and National Institutes of Health for conducting research under the Ministry of Health, Malaysia. Thirty-four patients who fulfilled any of the following revised Bethesda Guidelines were recruited from three collaborating centres in Malaysia namely Hospital Universiti Sains Malaysia, Hospital Alor Star, Kedah and Hospital Queen Elizabeth, Kota Kinabalu, Sabah. (1) CRC with age less than 50 years old; (2) Presence of synchronous or metachronous colorectal or other hereditary nonpolyposis colorectal cancer (HNPCC)-associated tumors regardless of age; (3) CRC with MSI-positive morphology with age less than 60 years old; (4) CRC with one or more first-degree relatives with CRC or other HNPCC-related tumor, with one of the cancers with age less than 50 years old; and (5) CRC with two or more first- or second-degree relatives with CRC or other HNPCC-related tumor (regardless of age), including cancers (endometrial, stomach, ovarian, cervical, esophageal, leukemia, thyroid, bladder, ureter and renal pelvis, biliary tract, small bowel, breast, pancreas, liver, larynx, bronchus, lung and brain (glioblastoma), sebaceous gland adenomas and keratoacanthomas.
Personal and demographic details of the patients including history of CRC in the family were collected and recorded. CRC patients with a strong family history of CRC among first- or second-degree relatives and who met the selection criteria were subjected to detailed pedigree analysis. Detailed pedigrees of these suspected cases of Lynch syndrome were prepared. A 5 mL blood sample was collected from identified Lynch syndrome patients after obtaining informed consent.
Paraffin-embedded tissue blocks from each resected bowel specimen containing carcinoma and preferably adjacent non-neoplastic colon were selected. Four µm-thick tissue sections were cut, dewaxed in xylene and rehydrated in graded alcohol concentrations to distilled water. Slides were placed in Tris-EDTA buffer at pH 9.0 in a pressure chamber for antigen retrieval for 2 min at 123 °C. Peroxidase blocking reagent (DakoCytomation) was used to block endogenous peroxidase activity. Slides were incubated with 150-200 μL of primary monoclonal mouse antibodies: MLH1 antibody (BD Biosciences Pharmingen, United States, Cat. No. 550838) at 1/50 dilution and MSH2 antibody (BD Biosciences Pharmingen, United States, Cat. No. 556349) at 1/200 dilution, Horseradish-peroxidase (HRP) labeled polymer, conjugated to secondary antibody and thereafter DAB substrate chromogen (DAKO Envision detection kit, peroxidase/DAB, Rabbit/Mouse) were applied. Slides were counterstained with hematoxylin.
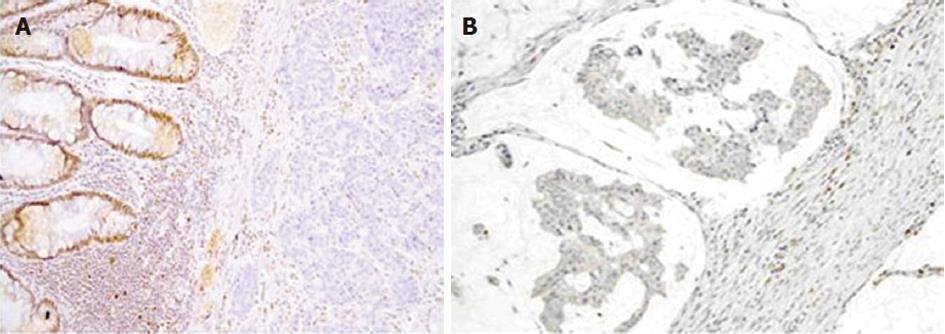
Normal expression of protein was defined as the presence of nuclear staining in colon cancer cells. Loss of staining in carcinoma with concurrent positive staining in nuclei of normal colon epithelial cells indicated absent expression of protein. Adjacent non-neoplastic colon and stromal inflammatory cells served as internal positive controls. The external positive control was normal colon.
Genomic DNA was extracted from blood using the commercially available DNA extraction kit, QIAamp DNA Blood Mini kit (Qiagen, Hilden, Germany). Specific primers were designed by using Primer3 software[9] to amplify all 19 exons of MLH1 gene and 16 exons of MSH2 gene. DNA was subjected to polymerase chain reaction (PCR) amplification of 19 exons of MLH1 gene and 16 exons of MSH2 gene using specific primers as listed in Tables 1 and 2, respectively, in a total reaction volume of 20 μL, consisting of 1X PCR reaction buffer (Applied Biosystem, Foster City, CA, United States), 1.875 μmol/L MgCl2 (Applied Biosystem, Foster City, CA, United States), 0.375 μmol/L dNTPs (Applied Biosystem, Foster City, CA, United States), 0.4 μmol/L each of forward and reverse primers, about 100 ng genomic DNA and 1 unit of AmpliTaq Gold DNA Polymerase (Applied Biosystem, Foster City, CA, United States). PCR was performed with an initial denaturation step at 96 °C for 5 min, followed by 40 cycles of denaturation at 95 °C for 1 min, annealing at temperatures as listed in Tables 1 and 2 for 1 min and extension at 72 °C for 1 min, followed by a final extension step at 72 °C for 7 min.
| MLH1 gene | Primer sequence (5’-3’) | Size (bp) | Ta (°C) |
| Exon 1 | 5’ cagagttgagaaatttgactgg 3’ | 339 | 50 |
| 5’ taagtcgtagcccttaagtgag 3’ | |||
| Exon 2 | 5’ tgtatgagcctgtaagacaaag 3’ | 315 | 50 |
| 5’ tagtttccagaacagagaaagg 3’ | |||
| Exon 3 | 5’ gggaattcaaagagatttgg 3’ | 373 | 50 |
| 5’ gaggtctctgcaggtaaaatag 3’ | |||
| Exon 4 | 5’ ctcatcgttgccacatatta 3’ | 319 | 50 |
| 5’ gtactcaagatctctgccaaaa 3’ | |||
| Exon 5 | 5’ tccccttgggattagtatctat 3’ | 306 | 50 |
| 5’ cccacaaaagccaatagtcat 3’ | |||
| Exon 6 | 5’ cactatcttaagacctcgcttt 3’ | 325 | 50 |
| 5’ gctaaaccttgaccagaaacta 3’ | |||
| Exon 7 | 5’ agaggagtctgtgttttgattc 3’ | 367 | 50 |
| 5’ atcataaccttatctccaccag 3’ | |||
| Exon 8 | 5’ atagtttgctggtggagataag 3’ | 306 | 50 |
| 5’ agcctgtgtatttgactaaagc 3’ | |||
| Exon 9 | 5’ agttagtttatgggaaggaacc 3’ | 438 | 50 |
| 5’ aagaaccagactccaacagtc 3’ | |||
| Exon 10 | 5’ atagtaagatagtgggctggaa 3’ | 432 | 62 |
| 5’ tacctgtaagaagggacagaac 3’ | |||
| Exon 11 | 5’ tatttgaacacactcagactcg 3’ | 476 | 62 |
| 5’ taggaacaacagcacaatacc 3’ | |||
| Exon 12 | 5’ atacagactttgctaccaggac 3’ | 475 | 50 |
| 5’ agagagaagatgcaagtgattc 3’ | |||
| Exon 13 | 5’ tgtcagataagcagtctaccag 3’ | 468 | 50 |
| 5’ ttagtaaaggaagaggagcttg 3’ | |||
| Exon 14 | 5’ cctcatttgtgttcgttttc 3’ | 411 | 50 |
| 5’ gtcgaacttggatttgaaac 3’ | |||
| Exon 15 | 5’ catgtttcagggattacttctc 3’ | 356 | 50 |
| 5’ ggctaccaaatgactattttgc 3’ | |||
| Exon 16 | 5’ gtatatgggggatgagtgttac 3’ | 319 | 50 |
| 5’ cagaagtataagaatggctgtc 3’ | |||
| Exon 17 | 5’ gtgctgtttgtcaggatgaa 3’ | 430 | 50 |
| 5’ tgaatattagtggggatgagag 3’ | |||
| Exon 18 | 5’ gtctgtgatctccgtttagaat 3’ | 358 | 50 |
| 5’ ctacatcagcagatggaagtaa 3’ | |||
| Exon 19 | 5’ caggacaccagtgtatgttg 3’ | 497 | 50 |
| 5’ ctgtgacatgttcaagacctc 3’ |
| MSH2 gene | Primer sequence (5’-3’) | Size (bp) | Ta (°C) |
| Exon 1 | 5’ gtctgcttatgattggttgc 3’ | 632 | 58 |
| 5’ actccgtgatcacaagttca 3’ | |||
| Exon 2 | 5’ gtggaactataagtccagtaagc 3’ | 457 | 50 |
| 5’ cagccaaactgcaacttttt 3’ | |||
| Exon 3 | 5’ agaatcgattgaacccttga 3’ | 554 | 62 |
| 5’ gtcaattaaagagcctttcc 3’ | |||
| Exon 4 | 5’ acatcatatcagtgtcttgcac 3’ | 302 | 50 |
| 5’ taatccatgtacctgattctcc 3’ | |||
| Exon 5 | 5’ gaaggaacaccaaggaaaat 3’ | 273 | 50 |
| 5’ ctgaaaaaggttaagggctctg 3’ | |||
| Exon 6 | 5’ ggtcttaggaagaggaactttt 3’ | 337 | 50 |
| 5’ gtataatcatgtgggtaactgc 3’ | |||
| Exon 7 | 5’ ttgagacttacgtgcttagttg 3’ | 391 | 50 |
| 5’ tctgaatgtgtcctaagagtga 3’ | |||
| Exon 8 | 5’ aactttggagacctactgtact 3’ | 353 | 50 |
| 5’ ccacaaaggtgctacaattag 3’ | |||
| Exon 9 | 5’ tacatcatcagcactgtaactg 3’ | 494 | 53 |
| 5’ ttgacagagatgtgaagtcatc 3’ | |||
| Exon 10 | 5’ cagctttcaagtcagaaact 3’ | 511 | 53 |
| 5’ aagaaagcttgactcttacctg 3’ | |||
| Exon 11 | 5’ tcacgtagtacacattgcttct 3’ | 365 | 61 |
| 5’ tatgtgcaagagtaacttccag 3’ | |||
| Exon 12 | 5’ caaatggggggattaaatgt 3’ | 377 | 50 |
| 5’ aaaacgttacccccacaaag 3’ | |||
| Exon 13 | 5’ gcagtaactctgtccacatctt 3’ | 432 | 62 |
| 5’ cttctcacaggacagagacata 3’ | |||
| Exon 14 | 5’ atgtttgtggcatatccttc 3’ | 499 | 50 |
| 5’ cagactgtgaattaaggggtaa 3’ | |||
| Exon 15 | 5’ ctaatgacaaggtgagaaggat 3’ | 338 | 50 |
| 5’ ccttcatcttagtgtcctgttt 3’ | |||
| Exon 16A | 5’ ggaactagacagtgcacacata 3’ | 428 | 62 |
| 5’ ctgggatttttcacgtagtaac 3’ | |||
| Exon 16B | 5’ attattcaggagttcctgtcc 3’ | 500 | 50 |
| 5’ ctctgagctatttgtttcttcc 3’ |
Amplicons were then visualized by gel electrophoresis in a 2% agarose gel. Three μL of each amplicon was mixed with 3 μL of wild-type sample, denatured at 95 °C for 3 min and then gradually reannealed by decreasing the sample temperature from 95 °C to 65 °C over a period of 30 min to allow the formation of heteroduplex. Mutation analysis was performed employing denaturing high performance liquid chromatography (dHPLC) using ProStar Helix System, Varian Inc, United States and samples were analyzed under the optimum melting temperature determined by using the melting program available online, http://insertion.stanford.edu/melt.html. The wild-type sample for all the exon regions was randomly selected and confirmed by DNA sequencing.
Samples which showed heteroduplex peaks were subjected to DNA sequencing to characterize the mutation. For this, amplicons were purified using the QIAGEN QIAamp PCR purification kit (Qiagen, Hilden, Germany). Purified amplicons were sequenced using BigDye Terminator v3.1 kit (Applied Biosystem, Foster City, CA, United States) on the automated ABI PRISM 3100 Genetic Analyzer (Applied Biosystem Inc, United States).
Protein expression of MLH1 and MSH2 antigens were evaluated in tissue samples from 34 Malaysian Lynch syndrome patients. Three out of 34 tissue samples (8.8%) showed absence of nuclear staining for MLH1 and four out of 34 tissue samples (11.8%) showed loss of nuclear staining for MSH2 in cancer epithelium cells. Figure 1A and B show the loss of nuclear staining in MLH1 and MSH2 protein, respectively, observed in tumor samples. Patients whose samples showed absence of MLH1 and MSH2 protein expression were subjected to mutation analysis of MLH1 and MSH2 genes to detect and characterize germline mutations, if present.
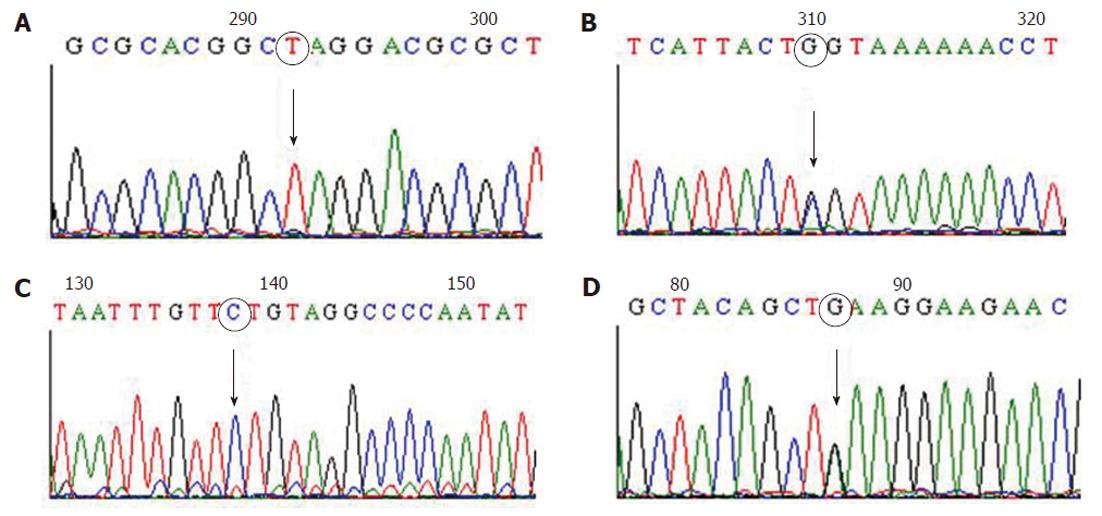
Nineteen exons of MLH1 gene and 16 exons of MSH2 gene were successfully amplified and screened by dHPLC. Samples which showed heteroduplex peaks by dHPLC were subjected to DNA sequencing to characterize the germline mutations. All four Lynch syndrome patients who showed absence of MSH2 protein expression by immunohistochemical analysis were found to harbor germline mutations. Three different types of mutations were identified in MSH2 gene among these four patients (Table 3). Of these, one was a substitution in exon 1 of MSH2 gene, c.142G > T, which resulted in a nonsense mutation (Glu48Stop) and is considered a deleterious mutation (Figure 2A). The second type was a transversion mutation, c.2005G > C (Figure 2B), which resulted in a missense mutation (Gly669Arg). This has not been reported before in any of Lynch syndrome mutation databases namely MMR Genes Variant Database (http://www.med.mun.ca/mmrvariants), International Society for Gastrointestinal Hereditary Tumours mutation database (http://www.insight-group.org) and Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/ac/index.php) and could be considered a novel mutation. This novel mutation was identified in two patients who showed absent expression of MSH2 protein by immunohistochemistry. The fourth patient showed a splice-site mutation, c.2006-6T > C (Figure 2C), which was adjacent to exon 13 of MSH2 gene in one of the patients.
| Gene | Patients ID | Age | Exon/intron region | Nucleotide change | Consequence |
| MSH2 | LS8 | 50 | Exon 12 | c.2005G > C | Missense mutation (G669R) |
| LS20 | 43 | Exon 12 | c.2005G > C | Missense mutation (G669R) | |
| LS37 | 41 | Intron 12 | c.2006-6T > C | Splice-site mutation | |
| LS41 | 49 | Exon 1 | c.142G > T | Nonsense mutation (E48X) |
However, no deleterious mutations were identified in any of the 19 exons or coding regions of MLH1 gene in the three Lynch syndrome patients who showed absence of MLH1 protein expression by immunohistochemical analysis, but MLH1 promoter polymorphism, -93G > A (rs1800734), was detected in 21 out of 34 patients (61.8%) (Figure 2D).
Germline mutations in DNA MMR genes particularly of MLH1 and MSH2 genes were reported to be responsible for the majority of Lynch syndrome cases[5,8]. The most crucial procedure in confirming Lynch syndrome is the sequencing analysis of DNA MMR genes to unravel the germline mutation harbored by the suspected patients. However, the procedure is problematic and challenging especially in large-scale studies, thus the application of a pre-screening method is essential. MSI analysis as well as immunohistochemical analysis of tumor tissue has been shown to be highly predictive for selecting candidates with defective MMR genes[10,11]. Immunohistochemical testing of tumor tissue in particular has been proven to be a good diagnostic tool, since it can pinpoint the MMR genes most eligible for germline mutation analysis of DNA MMR genes[12].
In our study consisting of 34 Malaysian Lynch syndrome patients, immunohistochemical analysis of tumor tissue was applied as a pre-screening method. Using immunohistochemistry, we identified 4 out of 34 patients (11.8%) who showed absent expression of MSH2 protein. One of them harbored a substitution in exon 1 of MSH2 gene, c.142G > T, which resulted in a nonsense mutation (Glu48Stop). This mutation can be considered as probably deleterious as it results in a premature stop codon and subsequently a nonfunctional and truncated protein. The mutation has been reported in Dutch hereditary nonpolyposis CRC family members who showed absent expression of MSH2 protein[13].
Interestingly, we identified a novel mutation, transversion mutation c.2005G > C, which resulted in a missense mutation (Gly669Arg) in two patients. Functional studies conducted by Acharya et al[14] showed that amino acid residues 658-670 is a highly conserved disordered loop region within Walker boxes in ATPase domain (domain V) which play a role in regulating mismatch binding and ATP-dependent clamp formation. Therefore, we strongly believe that changes in protein conformation and subsequent changes in biological properties of MSH2 protein are possible due to this mutation. Currently, we are unable to comment on the real impact of the novel mutation identified, and further functional studies are warranted to elucidate this issue.
In addition, we identified another splice-site mutation, c.2006-6T > C, which was adjacent to exon 13 of MSH2 gene in one of the patients. The location of this mutation which was in the region of the polypyrimidine tract prompted us to suggest that it might affect normal MSH2 mRNA splicing. Since the polypyrimidine tract plays a major role in promoting the assembly of spliceosome as well as polypyrimidine tract-binding protein[15], the mutation might result in aberrant mRNA splicing during the process of post-transcriptional modification. The skipping of exon 13 which encompasses the most conserved region of MSH2 gene might occur, leading to diminished MSH2 protein function. However, the impact of the splice-site mutation, c.2006-6T > C, in the pathogenesis of cancer is still controversial due to conflicting evidence. A few reports have shown that this mutation was associated with an increased risk of CRC in patients with ulcerative colitis[16], sporadic CRC[17] and non-Hodgkin lymphomas[18] as well as being described as a polymorphism[19]. It is also possible that the splice-site mutation is in linkage disequilibrium with the other unknown pathogenic mutation. Thus, further extensive studies are warranted to clarify the real impact of splice-site mutation towards the susceptibility to CRC.
Even though no deleterious mutations were identified in any of the 19 exons or coding regions of MLH1 gene in the three Lynch syndrome patients who showed absence of MLH1 protein expression by immunohistochemical analysis, the possibility of germline epimutation of MLH1 gene cannot be ruled out. On the other hand, the promoter polymorphism, -93G > A (rs1800734), which we identified in MLH1 gene in 61.8% of our patients has been reported to be located in the core promoter region, 93 nucleotides upstream from the transcription start site, in the potential region of transcription factor binding sites[20]. The high frequency of this promoter polymorphism, -93G > A, indicates the predominance of this variation among Malaysians. Functional studies have already demonstrated that the MLH1 promoter region from nucleotide position -184 to the transcription start site, in which the G > A alteration occurs, is essential for transcription of MLH1 gene[20]. The fact that the location of the promoter polymorphism is in the region of two potential transcription factor binding sites, GT-motif 2B and interleukin-6-regulated nuclear factor, could greatly enhance the chances of abnormal MLH1 transcription and expression and subsequently reduce the DNA repair capability[21,22]. On the other hand, there is a possibility that this promoter polymorphism may alternatively be in linkage disequilibrium with another polymorphism in the coding region or intron or with the MLH1 germline mutation itself which can affect MLH1 function.
The function of the MMR system is to maintain the fidelity of the genome during replication. The DNA MMR genes via its intranuclear protein, namely MMR protein, will correct nucleotide base mispairs and small insertions or deletions generated by slippage errors of DNA polymerase during replication. Germline mutation in one of these genes will cause inactivation of the MMR system where the MMR proteins would no longer be expressed resulting in failure to repair the replication errors that occur spontaneously during DNA replication. This defect leads to the accumulation of errors in repetitive DNA sequences (microsatellites) throughout the genome of tumors resulting in microsatellite instability.
In summary, immunohistochemical analysis for MMR protein expression in suspected cases of Lynch syndrome could be considered a pre-screening method and a good strategy prior to germline mutation screening of DNA MMR genes. Since no germline mutations were identified in MLH1 gene in Lynch syndrome patients who showed absence of MLH1 protein expression by immunohistochemical analysis, it is imperative to determine the methylation status of MLH1 promoter as it may play a role in the other molecular mechanism of Lynch syndrome. Germline mutation analysis of the other MMR genes especially PMS2 with the aim of characterizing the spectrum of DNA MMR gene mutation among Malaysian Lynch syndrome patients is also underway.
We would like to thank the patients and their family members who participated in this project. We would also like to thank the staff at Clinical Research Centre, Hospital Sultanah Bahiyah, Alor Setar, Kedah, Malaysia for their contribution to this study and Mariam Azmi, Advanced Medical and Dental Institute, Universiti Sains Malaysia, for her technical expertise in immunohistochemistry.
Lynch syndrome, the most common form of hereditary colorectal cancer is mainly due to germline mutations in a group of DNA mismatch repair (MMR) genes (MLH1, MSH2, MSH6, PMS1 and PMS2), and mutations in MLH1 and MSH2 genes alone account for more than 90% of the mutations identified in Lynch syndrome families. Data on germline mutations of DNA MMR genes are available from different parts of the world in varying frequencies but no reports are available from Malaysia.
Mutational analysis of DNA MMR genes is the most pivotal step in confirming Lynch syndrome cases in order to unravel the germline mutation harbored by suspected patients. Immunohistochemical analysis of MMR protein expression in suspected cases of Lynch syndrome can be considered a pre-screening method and a good strategy prior to germline mutation screening of DNA MMR genes.
There are a number of previous reports available on germline mutations in MMR genes in other populations with varying frequencies, but no reports are available from Malaysia. This is the first study to unravel the spectrum of germline mutations in DNA MMR genes in Malaysian Lynch syndrome patients. This study also focuses on the protein expression profile of MMR protein and germline mutations identified in MLH1 and MSH2 genes. The authors also identified one novel mutation, transversion mutation c.2005G > C, which resulted in a missense mutation (Gly669Arg). The possibility of germline epimutation of MLH1 gene cannot be ruled out since no deleterious mutations were identified in any of the 19 exons or coding regions of MLH1 gene in the three Lynch syndrome patients who showed absence of MLH1 protein expression by immunohistochemical analysis.
The data generated from this study will help to establish a MMR gene mutation database of Malaysian Lynch syndrome kindreds.
MLH1 and MSH2 genes are the genes which code for MMR protein and are major elements in the MMR system. The main function of the MMR system is to maintain the fidelity of the genome during replication, where the MMR protein will correct nucleotide base mispairs and small insertions or deletions generated by slippage errors of DNA polymerase during replication.
This is a very interesting article.
Peer reviewer: Andrada Seicean, MD, PhD, Third Medical Clinic Cluj Napoca, University of Medicine and Pharmacy Cluj Napoca, Romania, 15, Closca Street, Cluj-Napoca 400039, Romania
S- Editor Tian L L- Editor Webster JR E- Editor Li JY
| 1. | Aaltonen LA, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomäki P, Chadwick RB, Kääriäinen H, Eskelinen M, Järvinen H. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med. 1998;338:1481-1487. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 767] [Cited by in F6Publishing: 754] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 2. | Salovaara R, Loukola A, Kristo P, Kääriäinen H, Ahtola H, Eskelinen M, Härkönen N, Julkunen R, Kangas E, Ojala S. Population-based molecular detection of hereditary nonpolyposis colorectal cancer. J Clin Oncol. 2000;18:2193-2200. [PubMed] [Cited in This Article: ] |
| 3. | Goh KL, Quek KF, Yeo GT, Hilmi IN, Lee CK, Hasnida N, Aznan M, Kwan KL, Ong KT. Colorectal cancer in Asians: a demographic and anatomic survey in Malaysian patients undergoing colonoscopy. Aliment Pharmacol Ther. 2005;22:859-864. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 50] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 4. | Lawes DA, SenGupta SB, Boulos PB. Pathogenesis and clinical management of hereditary non-polyposis colorectal cancer. Br J Surg. 2002;89:1357-1369. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 24] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 5. | Dionigi G, Bianchi V, Rovera F, Boni L, Annoni M, Castano P, Villa F, Dionigi R. Genetic alteration in hereditary colorectal cancer. Surg Oncol. 2007;16 Suppl 1:S11-S15. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 9] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 6. | Lagerstedt Robinson K, Liu T, Vandrovcova J, Halvarsson B, Clendenning M, Frebourg T, Papadopoulos N, Kinzler KW, Vogelstein B, Peltomäki P. Lynch syndrome (hereditary nonpolyposis colorectal cancer) diagnostics. J Natl Cancer Inst. 2007;99:291-299. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 165] [Cited by in F6Publishing: 174] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 7. | Jacob S, Praz F. DNA mismatch repair defects: role in colorectal carcinogenesis. Biochimie. 2002;84:27-47. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 153] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 8. | Miyaki M, Konishi M, Tanaka K, Kikuchi-Yanoshita R, Muraoka M, Yasuno M, Igari T, Koike M, Chiba M, Mori T. Germline mutation of MSH6 gene as the cause of hereditary nonpolyposis colorectal cancer. Nat Genet. 1997;17:271-272. [DOI] [Cited in This Article: ] [Cited by in Crossref: 460] [Cited by in F6Publishing: 426] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 9. | Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. Bioinformatics Methods and Protocols: Methods in Molecular Biology. New Jersey: Humana Press, Totowa 2000; 365-386. [Cited in This Article: ] |
| 10. | Baudhuin LM, Burgart LJ, Leontovich O, Thibodeau SN. Use of microsatellite instability and immunohistochemistry testing for the identification of individuals at risk for Lynch syndrome. Fam Cancer. 2005;4:255-265. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 79] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 11. | Piñol V, Castells A, Andreu M, Castellví-Bel S, Alenda C, Llor X, Xicola RM, Rodríguez-Moranta F, Payá A, Jover R. Accuracy of revised Bethesda guidelines, microsatellite instability, and immunohistochemistry for the identification of patients with hereditary nonpolyposis colorectal cancer. JAMA. 2005;293:1986-1994. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 389] [Cited by in F6Publishing: 424] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 12. | Stormorken AT, Bowitz-Lothe IM, Norèn T, Kure E, Aase S, Wijnen J, Apold J, Heimdal K, Møller P. Immunohistochemistry identifies carriers of mismatch repair gene defects causing hereditary nonpolyposis colorectal cancer. J Clin Oncol. 2005;23:4705-4712. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 34] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 13. | Berends MJ, Hollema H, Wu Y, van Der Sluis T, Mensink RG, ten Hoor KA, Sijmons RH, de Vries EG, Pras E, Mourits MJ. MLH1 and MSH2 protein expression as a pre-screening marker in hereditary and non-hereditary endometrial hyperplasia and cancer. Int J Cancer. 2001;92:398-403. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 56] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 14. | Acharya S, Patterson K. Mutations in the conserved glycine and serine of the MutS ABC signature motif affect nucleotide exchange, kinetics of sliding clamp release of mismatch and mismatch repair. Mutat Res. 2010;684:56-65. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 15. | Roscigno RF, Weiner M, Garcia-Blanco MA. A mutational analysis of the polypyrimidine tract of introns. Effects of sequence differences in pyrimidine tracts on splicing. J Biol Chem. 1993;268:11222-11229. [PubMed] [Cited in This Article: ] |
| 16. | Brentnall TA, Rubin CE, Crispin DA, Stevens A, Batchelor RH, Haggitt RC, Bronner MP, Evans JP, McCahill LE, Bilir N. A germline substitution in the human MSH2 gene is associated with high-grade dysplasia and cancer in ulcerative colitis. Gastroenterology. 1995;109:151-155. [PubMed] [Cited in This Article: ] |
| 17. | Goessl C, Plaschke J, Pistorius S, Hahn M, Frank S, Hampl M, Görgens H, Koch R, Saeger HD, Schackert HK. An intronic germline transition in the HNPCC gene hMSH2 is associated with sporadic colorectal cancer. Eur J Cancer. 1997;33:1869-1874. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 32] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 18. | Paz-y-Miño C, Pérez JC, Fiallo BF, Leone PE. A polymorphism in the hMSH2 gene (gIVS12-6T& gt; C) associated with non-Hodgkin lymphomas. Cancer Genet Cytogenet. 2002;133:29-33. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 29] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 19. | Krüger S, Bier A, Plaschke J, Höhl R, Aust DE, Kreuz FR, Pistorius SR, Saeger HD, Rothhammer V, Al-Taie O. Ten novel MSH2 and MLH1 germline mutations in families with HNPCC. Hum Mutat. 2004;24:351-352. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 20. | Ito E, Yanagisawa Y, Iwahashi Y, Suzuki Y, Nagasaki H, Akiyama Y, Sugano S, Yuasa Y, Maruyama K. A core promoter and a frequent single-nucleotide polymorphism of the mismatch repair gene hMLH1. Biochem Biophys Res Commun. 1999;256:488-494. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 68] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 21. | Campbell PT, Curtin K, Ulrich CM, Samowitz WS, Bigler J, Velicer CM, Caan B, Potter JD, Slattery ML. Mismatch repair polymorphisms and risk of colon cancer, tumour microsatellite instability and interactions with lifestyle factors. Gut. 2009;58:661-667. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 90] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 22. | Chen H, Taylor NP, Sotamaa KM, Mutch DG, Powell MA, Schmidt AP, Feng S, Hampel HL, de la Chapelle A, Goodfellow PJ. Evidence for heritable predisposition to epigenetic silencing of MLH1. Int J Cancer. 2007;120:1684-1688. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 67] [Article Influence: 3.9] [Reference Citation Analysis (0)] |